Abstract
Import of the ADP/ATP carrier (AAC) into mitochondria requires the soluble TIM10 complex to cross the intermembrane space. We report here that Tim9 and Tim10 purified from Escherichia coli can form a complex of the same size as the endogenous complex from yeast mitochondria. This shows that no other mitochondrial protein is required for the formation of the TIM10 complex. Co-expression of both proteins rendered Tim9 more soluble and allowed purification of the reconstituted complex in a single step. Urea/EDTA treatment of recombinant Tim10 allowed its import into tim10-ts mitochondria that lack endogenous Tim10 and cannot import AAC. In this way, we were able to (i) reconstitute the TIM10 complex in the intermembrane space and (ii) restore import of AAC to almost wild-type levels. The reconstituted TIM10 complex not only facilitated passage of AAC across the outer membrane but also ensured its accurate membrane insertion. We conclude that the TIM10 complex can be formed exclusively from Tim9 and Tim10 and that the reconstituted complex efficiently restores AAC import in a strain lacking the TIM10 complex.
Keywords: ADP/ATP carrier/mitochondria biogenesis/protein translocation/reconstitution/TIM10 complex
Introduction
Most mitochondrial proteins are synthesized in the cytosol as preproteins containing a mitochondrial targeting presequence at their N-terminus, which targets them to mitochondria (Schatz and Dobberstein, 1996; Voos et al., 1999; Herrmann and Neupert, 2000). These presequence-carrying preproteins are recognized mainly by the Tom20 receptor (Söllner et al., 1989; Moczko et al., 1992) and are then transferred to the TOM channel that contains the receptor protein Tom22 (Kiebler et al., 1993; Lithgow et al., 1994), the transfer protein Tom5 and the channel-forming protein Tom40 (Dietmeier et al., 1997). In a membrane potential-dependent reaction, preproteins cross the inner membrane via the TIM23 complex (composed of the homologous proteins Tim23 and Tim17) and are finally drawn into the matrix by the Tim44–hsp70 motor system that is powered by ATP hydrolysis in the matrix (Cyr, 1997; Moro et al., 1999). The presequence is removed upon import by the mitochondrial processing peptidase (Böhni et al., 1980; Conboy et al., 1982; McAda and Douglas, 1982; Miura et al., 1982). (Please note: throughout the text we adopt the convention whereby TIM and TOM refer to protein complexes and Tim and Tom to individual proteins.)
In contrast to matrix-targeted polypeptides, proteins of the inner membrane are mainly synthesized without a cleavable targeting presequence, and it is thought that targeting information is contained within internal sequences (Saraste and Walker, 1982; Adrian et al., 1986; Pfanner and Neupert, 1987; Endres et al., 1999; Wiedemann et al., 2001). These proteins are mainly recognized by the Tom70–Tom37 receptor complex and cross the outer membrane via the TOM channel. As the hydrophobic precursors exit the TOM channel, they interact with the 70 kDa ‘chaperone-like’ TIM10 complex comprising Tim9 and Tim10. This complex in the intermembrane space (IMS) is thought to keep the precursor in an insertion-competent conformation. Tim12, a protein homologous to Tim9 and Tim10, which is peripherally attached to the inner membrane, then takes over and catalyses membrane potential-dependent insertion into the membrane in the context of the 300 kDa TIM22 complex (Koehler et al., 1998b; Sirrenberg et al., 1998; Ryan et al., 1999). This complex contains the membrane proteins Tim22, Tim54 and Tim18, all of Tim12 and trace amounts of Tim9 and Tim10 (Sirrenberg et al., 1996; Kerscher et al., 1997, 2000; Koehler et al., 1998a, 2000). Another complex of similar size to the TIM10 complex was identified in the IMS and is composed of two other Tim proteins, Tim8 and Tim13, which are homologous to Tim9, Tim10 and Tim12. Tim8 and Tim13 are not essential for the import of carrier proteins and their function is still unclear, but deletion of this complex is synthetically lethal with a conditional mutation in Tim10. Import of only a subset of integral inner membrane proteins such as Tim23 is affected in Tim8 and Tim13 mutant strains (Leuenberger et al., 1999; Davis et al., 2000; Paschen et al., 2000). Although the Tim8/13 complex did not affect steady-state levels of Tim23 under normal growth conditions (Leuenberger et al., 1999; Davis et al., 2000) it was found that this complex binds during import to the N-terminal IMS domain of Tim23 (Davis et al., 2000; Paschen et al., 2000). Paschen et al. (2000) have shown that the interaction of the Tim8/13 complex with Tim23 becomes prominent at low levels of electrochemical potential, assigning thus a more specific role to the Tim8/13 complex. This might explain why this complex is apparently present in lower levels compared with TIM10 which seems to have a more general function (Leuenberger et al., 1999).
Tim9, Tim10 and Tim12 are all essential proteins and play a major role in the import of polytopic proteins. Despite the fact they are homologous, the TIM10 complex (containing the majority of Tim9 and Tim10) is in large excess over the membrane-embedded TIM22 complex (containing all of Tim12) (Koehler et al., 1998b; Adam et al., 1999; Tokatlidis and Schatz, 1999). We have proposed that this could be due to a ‘two-affinity system’. In this model, the TIM10 and TIM22 complexes would have different affinities for precursors: the TIM10 complex functions as a low-affinity chaperone-like system that interacts with a large number of precursors as they emerge from the TOM channel, whilst the TIM22 complex has a more stringent specificity and higher affinity for a subset of the precursors that are fed by the TIM10 complex (Tokatlidis and Schatz, 1999).
All carrier proteins have three similar repeats of ∼100 amino acids each, which would argue for three distinct internal targeting signals. Indeed, Endres et al. (1999) have shown that targeting signals are found in each of the three structurally related domains, but they suggested that only the third motif contained insertion information allowing proper membrane integration. However, a more recent study by Wiedemann et al. (2001) adopting a similar strategy using different deletion constructs of the ADP/ATP carrier (AAC) has shown that an interaction of all three separate modules of AAC is needed both for receptor recognition and also membrane translocation.
Earlier research has suggested that Tim10 and Tim12 recognize a conserved region after the first transmembrane domain, common to all mitochondrial metabolite carriers (Sirrenberg et al., 1998; Endres et al., 1999). More specifically, it was proposed that the interaction between TIM10 and AAC involves the charged residues of the zinc-finger of Tim10 and the opposite charged residues in the ‘carrier signature’ motif of AAC (Sirrenberg et al., 1998). However, TIM10 was found to mediate import of inner membrane proteins (like Tim11) that are unrelated to the metabolite carrier family (Leuenberger et al., 1999) and more recently Davis et al. (2000) have shown that charged residues in Tim23 are not required for an interaction with TIM10. These data would support a mechanism whereby the TIM10 complex might bind hydrophobic sequences with little, if any, sequence preference and could recognize more likely structural features of their substrates for import. Such a mode of action for the TIM10 complex is more in line with a chaperone-like function where the complex prevents aggregation of precursors during passage of the IMS. One other possible function is to create a ‘bridge’ between the translocases of the outer and the inner membrane (Koehler, 2000). This bridge would facilitate the precursor to cross the IMS due to an increasing affinity for the TIM22 complex.
Although several significant studies have been published on the import pathway of AAC and the functional importance of the TIM10 complex is undisputed, relatively few advances have been made on a thorough characterization of the TIM10 complex. In this paper, we provide evidence that Tim9 and Tim10 are the only two mitochondrial proteins that are essential for the formation of the TIM10 complex. We have reconstituted the TIM10 complex in vitro by co-expressing Tim9 and Tim10 in Escherichia coli cytosol. We have compared this reconstituted complex to the authentic mitochondrial complex and find that the two complexes have very similar properties in gel filtration and blue native gel electrophoresis (BN-PAGE) experiments. To examine the functionality of the reconstituted complex made by recombinant proteins, we developed an assay testing for restoration of AAC import in an AAC import-deficient strain that is lacking a functional, endogenous TIM10 complex. Import of recombinant Tim10 and Tim9 in this yeast strain resulted in (i) reconstitution of the TIM10 complex in organello (in the mitochondrial IMS) and (ii) restoration of import and insertion of AAC to almost wild-type levels, indicating that the recombinant proteins reconstituted a functional complex.
Results
Tim10 and Tim9 form a complex when co-expressed in the E.coli cytoplasm
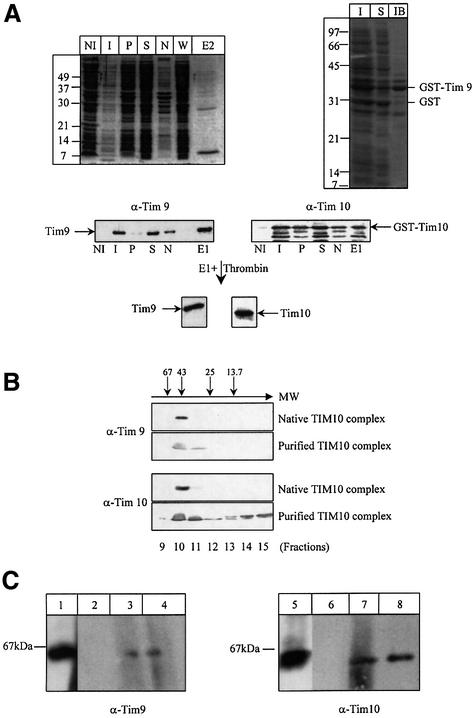
To test whether Tim9 and Tim10 could form a complex in the absence of any other mitochondrial protein, the two genes were cloned as an operon in the same expression vector and co-expressed E.coli. The gene encoding Tim9 was placed immediately downstream from a chimeric gene encoding GST–Tim10 (the glutathione S-transferase tag was used to facilitate purification). As a result, both proteins were produced from a single mRNA template, with the Shine–Dalgarno sequence between the two genes allowing reinitiation of translation. The expression of both proteins was confirmed by western blotting (Figure 1A). Although Tim9 is produced at lower levels than GST–Tim10, essentially all of Tim9 produced was in the soluble fraction after disruption of induced cells by a French Press. Interestingly, this was not the case when Tim9 was produced alone as a GST fusion using identical conditions of growth and induction: 75% of Tim9 was found in inclusion bodies and only 25% was soluble (Figure 1A, right panel, lanes S and IB). These data suggested that co-expression of Tim10 with Tim9 rendered Tim9 soluble and allowed an interaction between the two proteins to form a complex.
Fig. 1. Reconstitution and purification of the TIM10 complex in E.coli. (A) Left panel: the recombinant TIM10 complex was purified from an E.coli extract on glutathione–Sepharose beads. After washing, bound GST–Tim10–Tim9 was eluted either by adding 10 mM of reduced glutathione or by thrombin cleavage. The purification was followed by SDS–PAGE, Coomassie Blue staining and immunodecoration by Tim9 and Tim10 antibodies. NI: non-induced cells (25 µl); I: induced cells (5 µl); S: soluble fraction (5 µl, 1/4000 of total); P: pellet (5 µl, 1/4000 of total); N: non-bound (5 µl, 1/4000 of total); W: wash (25 µl, 1/6000 of total); E1: elution by reduced glutathione (10 µl, 1/300 of total); E2: elution by thrombin cleavage (10 µl). Right panel: the expression of GST–Tim9 was followed by SDS–PAGE and Coomassie Blue staining. I: total induced cells; S: soluble fraction and IB: inclusion bodies. (B) Gel filtration analysis: partially purified TIM10 complex eluted by cleavage with thrombin and IMS from purified wild-type mitochondria were loaded onto a Superdex-75 gel filtration column. Eluted fractions were analysed by SDS–PAGE and western blotting using anti-Tim9 and anti-Tim10 antibodies. Molecular weight markers are indicated by arrows (in kDa: ribonuclease 13.7, chymotrypsinogen A 25, ovalbumin 43 and albumin 67). (C) BN-PAGE: 400 µg of solubilized wild-type mitochondria (lanes 1 and 5) and 10 µl each of the fractions 9 (lanes 2 and 6), 10 (lanes 3 and 7), 11 (lanes 4 and 8), from the Superdex-75 gel filtration column were separated by BN-PAGE and blotted onto a PVDF membrane, followed by immunodecoration using antisera against Tim9 and Tim10.
To test for a direct interaction between Tim9 and Tim10, the soluble fraction, after disruption with French Press, was bound onto glutathione S–Sepharose beads. As shown in Figure 1A both proteins were concomitantly recovered in the bound fraction (lanes E1 and E2), as would be expected if they were in a complex.
To verify that Tim9 did not simply aggregate and/or bound non-specifically on the beads, we specifically eluted GST–Tim10 with 10 mM reduced glutathione. We found that Tim9 co-eluted with GST–Tim10 in this way (Figure 1A, lane E1). To further test the association of Tim9 with Tim10, we cleaved the GST–Tim10 bound to the beads with thrombin. As there is a specific thrombin cleavage site in the sequence between GST and Tim10, this treatment should specifically release Tim10 from the beads. We found that Tim9 was concomitantly released by thrombin cleavage, providing strong evidence for a specific interaction between the two proteins. Gel filtration analysis of this fraction after thrombin cleavage suggested that the complex of co-expressed Tim9 and Tim10 had the same size as the endogenous TIM10 complex from the IMS (Figure 1B). Essentially all of Tim9 is found co-migrating with Tim10 mainly in fraction 10 (and traces in fraction 11). The gel filtration allowed us to remove the excess of Tim10. The ratio in between Tim10 and Tim9 was 50:1 in the cytosol and decreased to 2:1 in fraction 10 after gel filtration (as estimated by semiquantitative western blotting). Fraction 10 therefore contains almost the same amount of Tim10 and Tim9 as a complex (given that the error in our estimation of Tim9 and Tim10 was of the order of 20%). This is comparable to the authentic mitochondrial complex that was also mainly present in fraction 10. Some recombinant Tim10 was also found in fractions 14 and 15 (which correspond to a molecular weight of ∼10 kDa). This most likely reflects the fact that Tim10 was produced in excess over Tim9 in this expression system, and the excess Tim10 molecules were present in an unassembled form as monomers in fractions 14 and 15. In comparison, in the endogenous Tim10 complex, most of the Tim9 and Tim10 proteins (that are in stoichiometric amounts) are assembled in a complex that migrates in fraction 10. To further substantiate the formation of a complex of the recombinant proteins, BN-PAGE was performed on the recombinant TIM10 complex (Figure 1C) and we found that the recombinant TIM10 complex migrated as the endogenous, authentic complex from mitochondria. Taken together, these data show that Tim9 and Tim10 can form a complex in the E.coli cytoplasm without any other mitochondrial protein.
Import of purified Tim10 into tim10-ts mitochondria
To test whether recombinant Tim10 was functional we first wanted to check whether it could be imported into purified mitochondria. To this end, we used tim10-ts mitochondria since they are devoid of Tim10 when grown at the non-permissive temperature. The import of Tim10 in these mitochondria can thus be monitored using Tim10-specific antisera without any cross-reaction from endogenous protein material. tim10-ts mitochondria still contain some Tim9 but, in contrast to wild-type mitochondria, Tim9 remains associated with the pellet when they are osmotically shocked to release the IMS (data not shown). This could be explained by one of the following hypotheses: in the absence of its partner Tim10, Tim9 is either redistributed to bind to other Tim component(s) in the TIM22 membrane complex that also contains Tim12, or it simply aggregates and co-sediments in a non-specific manner with the membrane pellet. In either case, a similar behaviour for Tim9 is seen in the co-expression of the two proteins in a recombinant form, where co-expression and association with Tim10 renders Tim9 more soluble.
Import of purified Tim10 alone in a native form was unsuccessful. As Tim10 has been proposed to be a zinc-finger protein (Sirrenberg et al., 1996), we thought that zinc bound to the protein may induce very tight folding of the protein thereby inhibiting its import. To unfold Tim10 and facilitate its import, we denatured the protein with 8 M urea, but still no imported Tim10 protein could be detected by western blotting, suggesting that urea denaturation alone was not sufficient to render Tim10 translocation-competent. To overcome this and chelate the zinc ion, we then denatured Tim10 with 8 M urea in the presence of 10 mM EDTA and 10 mM β-mercaptoethanol (to prevent disulfide bonds). By this harsh treatment combining a strong chelator and a reducing agent, we expected Tim10 to become unfolded and import-competent. Such strong denaturing conditions are often used for unfolding zinc-binding proteins, as they are very stable. Using this treatment we imported Tim10 again in tim10-ts mitochondria. After import, we were able to detect by western blotting a specific signal for imported Tim10 (Figure 2). Interestingly, the amount of imported Tim10 seems to reach a maximum level after 15 min of import at 30°C, close to the in vivo reported levels for Tim10 in wild-type mitochondria (∼1 µg Tim10 per mg of mitochondria, see Sirrenberg et al., 1997; Tokatlidis and Schatz, 1999).
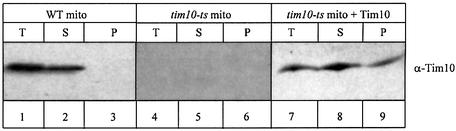
Fig. 2. Import of recombinant, purified Tim10 into mitochondria. Purified Tim10 was denatured and imported into mitochondria. Surface-bound precursor was removed by trypsin treatment. Mitoplasts were generated by osmotic shock to release the intermembrane space fraction in the supernatant. Samples were analysed by SDS–PAGE and western blotting. T: total, intact mitochondria, S: IMS and P: pellet after mitoplasting.
Imported Tim10 can be immunoprecipitated with Tim9
In its native state, Tim10 is in a complex with Tim9 in the IMS. As the protein had to be denatured before import, we wanted to verify that the imported species was properly refolded and assembled in the IMS in its native complex with Tim9. To address this, we scaled-up import using 75 µg of Tim10 and 5 mg of tim10-ts mitochondria (under identical import conditions as above, ratio Tim10/mitochondria equal to 1.5 µg of Tim10/200 µg of mitochondria). After 15 min of import at 30°C, mitochondria were harvested, resuspended in 5 ml of breaking buffer and treated with 0.2 mg/ml of trypsin. Mitochondria were then osmotically shocked in a hypotonic medium to create mitoplasts and release the IMS in the supernatant fraction after centrifugation. As shown in Figure 2 (lanes 8 and 9) most of Tim10 was recovered in the soluble fraction suggesting that the protein is imported in the IMS.
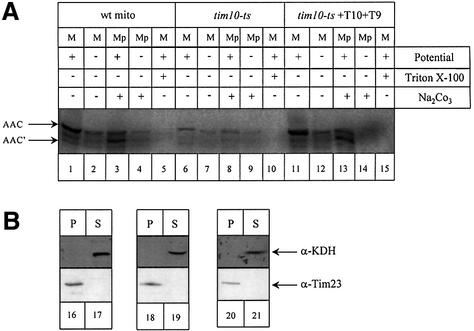
To verify that the imported Tim10 was indeed in a complex with Tim9, the components of the IMS were subjected to analysis by immunoprecipitation. The imported Tim10 and the endogenous Tim9 formed a complex and were co-immunoprecipitated (Figure 3, lanes IP10 and IP9). Although the efficiency of the immunoprecipitation was low (∼5–10% of the material present) it was specific. A control antiserum against cytochrome b2, an abundant IMS protein, did not immunoprecipitate either Tim9 or Tim10 supporting the specificity of the assay (lane ‘Cont’). Pre-immune antisera also gave a clean pattern ensuring specificity (lanes C9 and C10 for pre-immune Tim9 and pre-immune Tim10 antisera respectively). In Figure 3B western blotting controls for the mitoplasting treatment are presented using a matrix (ketoglutarate dehydrogenase, KDH), an inner membrane (Tim11), an outer membrane (porin) and an IMS protein (cyt b2).
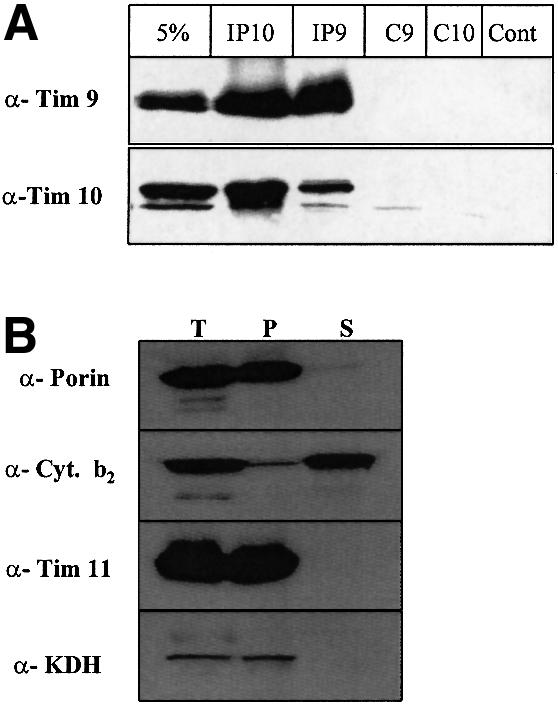
Fig. 3. The TIM10 complex can be reconstituted in the IMS by imported purified Tim10. (A) Seventy-five micrograms of purified Tim10 were denatured. Tim10 was imported first for 15 min into 5 mg of tim10-ts mitochondria. Mitoplasts were generated by osmotic shock. The supernatant fraction (which contains the IMS soluble proteins) was subjected to immunoprecipitation with anti-Tim10 (IP10), anti-Tim9 (IP9), pre-immune Tim9 (C9), pre-immune Tim10 (C10) and cytochrome b2 (Cont) antisera. ‘5%’ corresponds to 5% of the starting immunoprecipitated material. (B) Western blot control of mitoplast treatment (200 µg total mitochondria) using marker proteins. T: total intact mitochondria, P: pellet and S: supernatant. As markers we used porin (outer membrane), cytochrome b2 (IMS), Tim11 (inner membrane) and KDH (matrix).
This work shows that the denaturated imported Tim10 was associated with the native, endogenous Tim9 present in tim10-ts mitochondria to form a reconstituted TIM10 complex.
The reconstituted TIM10 complex is active
As tim10-ts mitochondria are deficient in the import of AAC, we decided to assay for the restoration of AAC import after the reconstitution of the TIM10 complex. These results are shown in Figure 4.
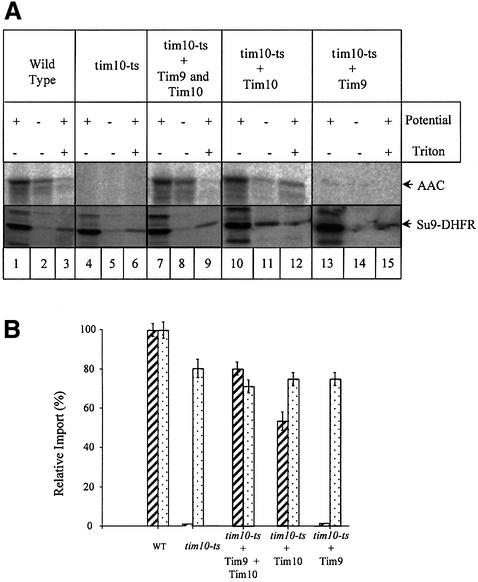
Fig. 4. Restoration of import of AAC in tim10-ts mitochondria. (A) Twenty microlitres of radioactive translated AAC or Su9–DHFR were imported into 200 µg each of different types of mitochondria: wild-type (lanes 1–3), tim10-ts (lanes 4–6), tim10-ts supplemented with purified Tim10 and Tim9 (lanes 7–9), tim10-ts supplemented only with purified Tim10 (lanes 10–12) and tim10-ts supplemented only with Tim9 (lanes 13–15). Import conditions were as described in the text. Import products were separated by SDS–PAGE and detected using a Phosphor Imager. (B) Quantification of AAC (hashed bars) and Su9–DHFR (dotted bars) was performed with the AIDA200 software from three separate experiments carried out under identical conditions. One hundred percent was taken as the amount of AAC or Su9-DHFR imported into wild-type mitochondria.
In tim10-ts mitochondria (Figure 4, lanes 4–6), there is no import for AAC even in the presence of electrochemical potential as expected (Koehler et al., 1998b; Sirrenberg et al., 1998). A small amount of imported AAC was detected in uncoupled mitochondria, which is most likely to be due to non-specific insertion of AAC as both wild-type and tim10-ts mitochondria present this same pattern.
When import of Tim10 preceded import of AAC (Figure 4, lanes 10–12), ∼50% of the level of AAC import in wild-type mitochondria was observed. Trypsin treatment after import showed that AAC was protected suggesting that AAC had completely crossed the outer membrane and was not blocked in the TOM channel where it would be accessible to external proteases (Pfanner and Neupert, 1987).
We then checked for import of AAC after both Tim10 and Tim9 were imported into mitochondria (Tim10 was imported first followed by Tim9 under the same import conditions). In this case, the restoration of AAC import was even more efficient and reached ∼80% of the level of wild-type mitochondria (Figure 4, lanes 7–9).
Finally, we also checked whether import of Tim9 alone could have the same effect as import of Tim10 alone. Under the same import conditions, purified Tim9 was imported in tim10-ts mitochondria in the same way as purified Tim10. As expected, no restoration of AAC import was observed, suggesting that Tim9 alone cannot substitute, not even partially, for the function of the TIM10 complex.
As a control, the same experiments were performed with radioactive translated Su9–DHFR (dehydrofolate reductase), a matrix-targeted fusion protein that uses the TIM23 channel for import. As expected, no significant difference in the import characteristics was observed in any type of mitochondria used (wild-type, tim10-ts or supplemented tim10-ts mitochondria).
These results conclusively show that the reconstituted TIM10 complex restored the import of AAC into tim10-ts mitochondria close to the levels observed for wild-type mitochondria. This data provides for the first time an in vitro reconstitution of the TIM10 complex from purified components.
Imported radioactive AAC is in the membrane fraction
By using this in vitro reconstitution system, we were able to import AAC completely across the outer membrane, as it remained protease-protected. However, the function of the TIM10 complex is to ensure translocation of AAC not only across the outer membrane via the TOM channel but onto the TIM22 complex for proper membrane integration as well. We therefore investigated the proper membrane insertion of AAC by a combination of mitoplasting treatment in the presence of Proteinase K and sodium carbonate extraction after import in the tim10-ts supplemented tim10-ts mitochondria (Figure 5).
Fig. 5. Imported AAC in the reconstituted system is properly embedded at the inner membrane. (A) Import of 35S-labelled AAC was performed under the same conditions as in Figure 4 (for details see Materials and methods) in wild-type (lanes 1–5), tim10-ts (lanes 6–10) and tim10-ts supplemented (lanes 11–15) mitochondria. After import, mitochondria (M) were swollen and the mitoplast pellet (Mp) corresponding to the membrane fraction was treated with Proteinase K and PMSF followed by a sodium carbonate extraction. Samples were analysed by SDS–PAGE and autoradiography. AAC: full-length AAC, AAC′: degraded AAC. (B) As a control, samples were checked by western blotting with antibodies against KDH, a matrix protein and Tim23 an inner membrane protein.
Both in wild-type and tim10-ts mitochondria, the imported AAC was largely protease-resistant, and was cleaved only to a slightly smaller fragment (Figure 5, AAC′) by Proteinase K when the outer membrane was selectively disrupted to generate mitoplasts (Figure 5A, lanes 3 and 13). In contrast, in intact mitochondria the AAC is not degraded (Figure 5A, lanes 1 and 11). This degradation pattern of AAC by Proteinase K upon mitoplasting is characteristic of the proper insertion of the AAC into the inner membrane (Wachter et al., 1992). These AAC′ species were sodium carbonate inextractable, which verified the fact that they are embedded in the membrane. The combination of these data showed that the supplemented tim10-ts mitochondria were able to properly insert the AAC into the inner membrane. As a control we checked for the localization of the soluble matrix protein KDH and the inner membrane protein Tim23 in all fractions (Figure 5B).
This result shows that the reconstituted TIM10 complex from purified proteins fulfils the function of the native TIM10 complex, namely proper translocation of AAC across the outer membrane and its accurate membrane integration in the context of the TIM22 complex.
Discussion
Tim9 and Tim10 proteins are essential components of the carrier import pathway in mitochondria and they exert their function by associating to form a soluble complex in the IMS of mitochondria (Koehler et al., 1998b). Until now, it was unknown whether any other mitochondrial proteins were involved in the formation of this complex. We report here that the TIM10 complex can be formed without the help of any other mitochondrial protein. We were able to reconstitute the native TIM10 complex by co-expressing both proteins in the E.coli cytoplasm. We have obtained similar results (S.Vial, P.Luciano and K.Tokatlidis, data not shown) by separately expressing and purifying the two proteins and then allowing complex formation. The reconstituted complex obtained by co-expression of the two proteins was compared with the native, authentic mitochondrial complex in terms of both their structural and functional properties, which were found to be very similar. Two independent methods, gel filtration and BN-PAGE indicate that the reconstituted complex has the same size as the authentic mitochondrial complex. Although the size of the complex (both the authentic and the reconstituted one) measured by gel filtration (40–50 kDa) is slightly smaller than that estimated by BN-PAGE (∼70 kDa), this could be due to the presence of detergent in BN-PAGE as opposed to gel filtration which was performed under completely detergent-free conditions. However, an accurate estimation of the size of the complex cannot be made from these data alone and we are investigating this point further by using additional techniques.
In the cell, proteins imported into mitochondria must be maintained in an unfolded, translocation-competent state. To mimic these in vivo conditions, proteins imported in an in vitro assay are usually denatured by urea. Import of urea-denatured Tim10 failed, suggesting that this treatment is not sufficient to render Tim10 competent for translocation. The zinc ion common to the ‘tiny Tim’ proteins presumably keeps these proteins in a compact, folded structure and inhibits their import. When we added β-mercaptoethanol and EDTA to remove the zinc ion we succeeded in importing Tim10. Since all of our assays indicate that recombinant Tim10 is equivalent to the native protein, this suggests that the ‘tiny Tims’ are imported in a monomeric form, without a bound zinc molecule in vivo and that metal binding occurs after import in the IMS. The mechanism for this is unknown and will be of great interest to determine, as zinc binding is apparently essential at least for the interaction of Tim10 and Tim12 with AAC during import (Sirrenberg et al., 1996).
Previous attempts to establish a functionality assay whereby the TIM10 complex (either purified from mitochondria or the reconstituted one) binds directly to AAC in vitro (tested by co-immunoprecipitation and BN-PAGE) have been so far unsuccessful (our own unpublished data). This was probably due to the fact that AAC must be in a very specific conformation to interact with the TIM10 complex. Presumably this conformation is specifically induced by passage through the TOM channel and/or contact sites between the outer and the inner mitochondrial membranes. In agreement with this notion, Wiedemann et al. (2001) recently reported that AAC is translocated through the TOM channel in a loop form, enabling a spatial interaction of all three AAC domains that is essential for proper translocation. These conditions are very difficult to accurately ‘mimic’ in vitro in a mitochondria-free assay. To overcome this, we applied a different strategy: we tried to reconstitute the TIM10 complex in tim10-ts mitochondria that are severely impaired in AAC import, and then test whether import of AAC can be restored to wild-type levels. In this way we were able to show that (i) the TIM10 complex can be reconstituted exclusively from Tim10 and Tim9 in the IMS, just like in the E.coli cytoplasm, and (ii) this is the minimal functional unit that can restore import of AAC. This type of assay allowed us to obtain for the first time, positive data on restoration of import of AAC (until now, all data published were based on showing impairment of AAC import in specific tim mutants). Also, it allowed us to assess whether the bacterially produced, reconstituted complex is functional in AAC import in physiological conditions ‘in organello’. Although binding data in completely mitochondria-free, in vitro conditions are still lacking, the assay we are reporting here, allows us to further examine the functionality of the TIM10 complex; by examining deleted versions of Tim9 and Tim10, we can assess which domains of Tim9 and Tim10 are important for assembly of the complex on the one hand and interaction with AAC on the other. Such studies are under way in our laboratory to extend our current findings both in the E.coli cytosol and in the mitochondrial IMS.
In the experiments reported here we found that import of Tim10 alone restored the level of AAC import to ∼50% of that in wild-type mitochondria. A possible explanation might be that imported Tim10 could form a complex with the endogenous Tim9 in tim10-ts mitochondria. Most of Tim9 in tim10-ts mitochondria remains associated with the membrane probably because it lacks its partner Tim10. Import of Tim10 into these mitochondria presumably might render part of Tim9 population more soluble through the formation of their complex. This would be corroborated by the in vitro experiments where co-production of Tim10 allows Tim9 to be more soluble. Interestingly, a further increase in the efficiency of AAC import to reach 80% of wild-type levels was obtained when we imported both Tim10 and Tim9. The simplest explanation for this is probably that the imported Tim9 associated rapidly with Tim10 leading to an increase in the number of functional TIM10 complexes available in the IMS. Semi-quantitative western blotting showed that the amount of Tim10 that could be imported was roughly similar to that found under steady-state conditions in wild-type mitochondria, which also explains the very high levels of AAC import restoration obtained.
Trypsin digestion, sodium carbonate extraction and mitoplast generation in the presence of Proteinase K in our reconstitution assay show that (i) the imported AAC is protected from added proteases and therefore crosses the outer membrane completely and (ii) it is properly inserted at the inner membrane. In terms of the import mechanism, this means that the reconstituted complex of Tim9 and Tim10 is assembled to a functional form and therefore correctly interacts with the Tim12 in the context of the TIM22 complex that catalyses insertion of AAC in the membrane.
The functional reconstitution of the TIM10 complex reported here identified this complex as the minimal functional unit in the import of AAC and presumably other polytopic proteins following the carrier pathway. It should allow us to address mechanistic details of this translocation pathway in a more defined in vitro system.
Materials and methods
One step cloning of the GST–Tim10–Tim 9 operon
A 312 bp DNA fragment containing at its 5′ end one EcoRI site, the entire tim10 gene and two Shine–Dalgarno sequences at its 3′ end was amplified by PCR (Boehringer-Mannhein). We used pGEX4.1 GST–tim10 plasmid as template and the following 5′ and 3′ primers: 5′-Cg gAA TTC ATg TCT TTC TTA ggT TTC and 3′-CCA Tgg TCT AgA TCC TgT TTC CTg CTA AAA CTT ACC ggC TgC GTT (the underlined, the italic and the bold-type sequence correspond to the two Shine–Dalgarno sequences, the new restriction site and the 3′ coding region of the tim10 gene respectively).
A 294 bp DNA fragment containing at its 5′ end two Shine–Dalgarno, the entire tim9 gene and one HindIII site at its 3′ end was amplified by PCR (Boehringer-Mannhein). We used pGEX4.1 GST–tim10 plasmid as template and the following 5′ and 3′ primers: 5′- Cag gAA ACA ggA TCT AgA CCA Tgg ATg gAC gCA TTg AAC TCC AAA and 3′-CCC AAg CTT TTA TCg GCC CAA gCC TTg (the underlined, the italic and the bold-type sequence correspond to the two Shine–Dalgarno, the new restriction site and the 3′ coding region of the tim10 gene, respectively).
The two PCR fragments were mixed together and amplified by using the 5′-Tim10 primer and the 5′-Tim9 primer. The PCR product (582 bp) was digested by EcoRI and HindIII and cloned into the pGEX4.1 plasmid to produce the pGEX4.1 GST–Tim10–Tim9 plasmid. The plasmid obtained was sequenced and used for the expression of both proteins.
Purification of the TIM10 complex
Five hundred millilitres of BL21 (pGEX4.1 GST–Tim10–Tim9) were grown at 30°C in LB medium to an A600nm = 0.3 and induced for 4 h with 1 mM IPTG. Cells were harvested, washed and resuspended in 25 ml of lysis buffer [50 mM Tris–HCl pH 8, 150 mM NaCl, 0.5 mM phenylmethyl sulfonylfluoride (PMSF), 1 mM dithiothreitol (DTT), 10 mM β-mercaptoethanol). The cells were disrupted by French-Press at 1000 p.s.i., centrifuged for 20 min at 20 000 g, 4°C and the soluble part containing the complex was incubated overnight at 4°C with 3 ml of glutathione–Sepharose B beads pre-incubated with the lysis buffer. The beads were washed with 10 volumes of wash buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 0.5 mM PMSF, 1 mM DTT) and the complex was eluted either with 3 ml of wash buffer containing 10 mM reduced-glutathione or 30 U of thrombin for 10 h at 4°C. The eluted complex was concentrated 10× with microcon 3 (Amicon) and loaded at 0.2 ml/min onto a gel filtration column (Superdex-75 prep grade, Amersham Pharmacia Biotech) pre-equilibrated with 50 ml of wash buffer.
Gel filtration
Superdex-75 prep grade gel filtration column (Amersham Pharmacia Biotech) was equilibrated with 50 ml of buffer A (50 mM Tris–HCl pH 8, 150 mM NaCl, 0.5 mM PMSF, 1 mM DTT) on an AKTA-Purifier 100 system at a flow rate of 0.2 ml/min. The complex eluted from GS-Trap beads was concentrated down to 500 µl on a microcon 3 (Amicon) and loaded onto the Superdex-75 gel filtration column. Fractions (1 ml) were collected, loaded on a 15% SDS–PAGE gel and immunodecorated with anti-Tim9 and anti-Tim10 antisera.
Blue native gel electrophoresis
The assembly and integrity of the TIM10 complex was confirmed by BN-PAGE. One µl of sample buffer [100 mM bis–Tris pH 7.0, 500 mM 6-aminocaproic acid, 5% (w/v) Coomassie Brilliant Blue G250] was added to 10 µl of the fractions from the gel filtration Superdex-75 and to 400 µg of solubilized mitochondria. Samples were loaded onto a 6–16% polyacrylamide gradient gel (Schagger and Von Jagow, 1991). After electrophoresis, the gel was soaked in transfer buffer (100 mM Tris, 200 mM glycine, 0.025% SDS, 20% methanol) and then transferred via semidry blotting (Bio-Rad) onto polyvinylidene difluoride (PVDF) membranes. Immunodecoration was performed by standard techniques using the Amersham enhanced chemiluminescence system.
Purification of wild-type and tim10-ts mitochondria
A 10 or 20 l fermentor with lactate medium (Glick and Pon, 1995) was inoculated with 200 ml or 400 ml of a wild-type (D273–10B) or tim 10-ts (Koehler et al., 1998b) saturated preculture. Cells were grown for 16 or 32 h at 30 or 25°C and shifted at 37°C for 8 h for the tim10-ts strain. Cells were then harvested and mitochondria were prepared as described by Daum et al. (1982) and Glick and Pon (1995).
Import experiments
Two hundred micrograms of purified mitochondria were incubated in import buffer (100 mM HEPES pH 7.1, 1.2 M sorbitol, 4 mM KH2PO4, 100 mM KCl, 20 mM MgCl2, 10 mM l-methionine, 2 mg/ml fatty acid-free bovine serum albumin) in the presence of 5 mM NADH or 5 µg/ml valinomycin for 5 min (Tokatlidis, 1999). Twenty microlitres of 35S-labelled AAC were then added and import was performed for 10 min at 30°C. Mitochondria were harvested by centrifugation and incubated with 200 µl breaking buffer containing 0.1 mg/ml trypsin for 20 min on ice. Trypsin was inhibited by SBTI (soybean trypsin inhibitor) at 0.2 mg/ml (10 min on ice). Mitochondria were then re-isolated by centrifugation and resuspended in 50 µl sample buffer and loaded on a 15% SDS–PAGE gel. Import products were visualized by autoradiography and quantified by densitometry (FUJI Bas station version 1.3). Total imported precursor into wild-type mitochondria was taken as the 100% basis for quantification of import.
In vitro reconstitution of import
Mitochondria from a tim10-ts strain were purified as described above and were incubated first with 1.5 µg of Tim10 for 15 min at 30°C and then with 1.5 µg of Tim9. Both Tim9 and Tim10 were precipitated with ammonium sulfate, resuspended in 8 M urea in 50 mM HEPES pH 7.4, 10 mM EDTA, 10 mM β-mercaptoethanol and left for 1 h at room temperature before import. This mixture was diluted 20× (final urea concentration 0.4 M and 0.5 mM β-mercaptoethanol) upon import. Finally, 20 µl of 35S-labelled AAC were added and import was performed for 10 min at 30°C. Mitochondria were then re-isolated and analysed by autoradiography. Relative amounts of radioactive AAC were determined by densitometry and compared with the level of import into wild-type mitochondria.
Immunoprecipitation
Seventy-five micrograms of urea-denaturated Tim10 were imported into 5 mg of tim10-ts mitochondria for 15 min at 30°C. Mitochondria were then pelleted, resuspended in 10 ml of breaking buffer containing 0.2 mg/ml trypsin and left on ice for 20 min. Trypsin was inhibited by adding 100 µl of 100 mg/ml of SBTI. Mitochondria were harvested and resuspended in 150 µl of breaking buffer. They were then swollen by adding 1.350 ml of 20 mM HEPES pH 7.4, 1 mM DTT and left on ice for 45 min. After centrifugation, the supernatant corresponding to the IMS was incubated overnight with Tim9, Tim10, pre-immune and Cyt b2 antisera bound onto Protein A–Sepharose beads. Beads were then washed 6 times with 1× phosphate-buffered saline, resuspended in 100 µl of sample buffer and loaded on a 15% polyacrylamide SDS–PAGE gel. Immunoprecipitation products were revealed by antisera against Tim9 and Tim10.
Sodium carbonate extraction
Mitochondria from a tim10-ts strain were incubated with 1.5 µg of Tim10 for 15 min at 30°C and then with 1.5 µg of Tim9. Then, 20 µl of 35S-labelled AAC were added and import was performed for 10 min at 30°C. Mitochondria were then treated by trypsin as described above. After centrifugation, the mitochondrial pellet was resuspended in 100 µl of 0.1 M sodium carbonate, left on ice for 30 min and spun for 15 min at 55 000 r.p.m. (TL100) at 4°C. The pellet fraction was treated with Proteinase K (0.1 mg/ml) for 15 min on ice. Proteinase K was inhibited by 1 mM PMSF. The pellet and the supernatant fractions were loaded onto a 15% SDS–PAGE gel. Import products were visualized by autoradiography and quantified by densitometry (FUJI Bas station version 1.3). Total imported precursor into wild-type mitochondria was taken as the 100% basis for quantification of import.
Acknowledgments
Acknowledgements
We would like to thank Dr V.Geli, Professor S.High, Professor C.Stirling, Dr P.Woodman and Dr P.Savory for helpful discussions and comments on the manuscript, and Dr C.Koehler for the tim10-ts strain. Research in our laboratory is supported by grants from the Wellcome Trust, the BBSRC, the Leverhulme Trust and the Royal Society. K.T. is a Lister Institute Research Fellow. S.V. was supported by a Swiss National Science Foundation and an EU Marie-Curie postdoctoral fellowship. D.R.R. was supported by a Wellcome Trust Career Development Fellowship.
References
- Adam A., Endres,M., Sirrenberg,C., Lottspeich,F., Neupert,W. and Brunner,M. (1999) Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J., 15, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian G.S., McCammon,M.T., Montgomery,D.L. and Douglas,M.G. (1986) Sequences required for delivery and localization of the ADP/ATP translocator to the mitochondrial inner membrane. Mol. Cell. Biol., 6, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhni P., Gasser,S., Leaver,C. and Schatz,G. (1980) A matrix-localized mitochondrial processing peptidase cytoplasmically-made precursors to mitochondrial proteins. In Kroon,A.M. and Saccone,C. (eds), The Organization and Expression of the Mitochondrial Genome. Elsevier/North Holland, Amsterdam, pp. 423–433.
- Conboy J.G., Fenton,W.A. and Rosenberg,L.E. (1982) Processing of preornithine transcarbamylase requires a zinc-dependent protease localized to the mitochondrial matrix. Biochem. Biophys. Res. Commun., 105, 1–7. [DOI] [PubMed] [Google Scholar]
- Cyr D.M. (1997) Coupling chemical energy by the hsp70/tim44 complex to drive protein translocation into mitochondria. J. Bioenerg. Biomembr., 29, 29–34. [DOI] [PubMed] [Google Scholar]
- Daum G., Bohni,P.C. and Schatz,G. (1982) Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem., 257, 13028–13033. [PubMed] [Google Scholar]
- Davis A.J., Sepuri,N.B., Holder,J., Johnson,A.E. and Jensen,R.E. (2000) Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol., 150, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmeier K., Honlinger,A., Bomer,U., Dekker,P.J., Eckerskon,C., Lottspeich,F., Kubrick,M. and Pfanner,N. (1997) Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature, 388, 195–200. [DOI] [PubMed] [Google Scholar]
- Endres M., Neupert,W. and Brunner,M. (1999) Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J., 18, 3214–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.S. and Pon,L.A. (1995) Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol., 260, 213–223. [DOI] [PubMed] [Google Scholar]
- Herrmann J.M. and Neupert,W. (2000) Protein transport into mitochondria. Curr. Opin. Microbiol., 3, 210–214. [DOI] [PubMed]
- Kerscher O., Holder,J., Srinivasan,M., Leung,R.S. and Jensen,R.E. (1997) The Tim54p–Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol., 139, 1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O., Sepuri,N. and Jensen,R.E. (2000) Tim18p is a new component of the Tim54p–Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell, 11, 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M., Keil,P., Schneider,H., Van der Klei,I.J., Pfanner,N. and Neupert,W. (1993) The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from the receptors to the general insertion pore. Cell, 74, 483–492. [DOI] [PubMed] [Google Scholar]
- Koehler C.M. (2000) Protein translocation pathways of the mitochondrion. FEBS Lett., 476, 27–31. [DOI] [PubMed] [Google Scholar]
- Koehler C.M., Merchant,S., Oppliger,W., Schmid,K., Jarosch,E., Dolfini,L., Junne,T., Schatz,G. and Tokatlidis,K. (1998a) Tim9, an essential partner subunit of Tim10 for the import of mitochondrial carrier proteins. EMBO J., 17, 6477–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C.M., Jarosch,E., Tokatlidis,K., Schmid,K., Schweyen,R.J. and Schatz,G. (1998b) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science, 279, 369–373. [DOI] [PubMed] [Google Scholar]
- Koehler C.M., Murphy,M.P., Bally,N.A., Leuenberger,D., Oppliger,W., Dolfini,L., Junne,T., Schatz,G. and Or,E. (2000) Tim18p, a new subunit of the TIM22 complex that mediates insertion of imported proteins into the yeast mitochondrial inner membrane. Mol. Cell. Biol., 20, 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger D., Bally,N., Schatz,G. and Koehler,C. (1999) Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J., 17, 4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow T., Junne,T., Suda,K., Gratzer,S. and Schatz,G. (1994) The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc. Natl Acad. Sci. USA, 91, 11973–11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAda P. and Douglas,M.G. (1982) A neutral metallo-endo-protease involved in the processing of an F1-ATPase subunit precursor in mitochondria. J. Biol. Chem., 257, 3177–3182. [PubMed] [Google Scholar]
- Miura S., Mori,M. and Tatibana,M. (1982) A mitochondrial protease that cleaves the precursor of ornithine carbamyl-transferase precursor into mitochondria: purification and properties. Eur. J. Biochem., 122, 641–647. [PubMed] [Google Scholar]
- Moczko M., Dietmeier,K., Sollner,T., Segui,B., Steger,H.F., Neupert,W. and Pfanner,N. (1992) Identification of the mitochondrial receptor complex in Saccharomyces cerevisiae. FEBS Lett., 310, 265–268. [DOI] [PubMed] [Google Scholar]
- Moro F., Sirrenberg,C., Schneider,H.C., Neupert,W. and Brunner,M. (1999) The TIM17.23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J., 18, 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen S.A., Rothbauer,U., Kaldi,K., Bauer,M.F., Neupert,W. and Brunner,M. (2000) The role of the TIM8–13 complex in the import of Tim23 into mitochondria. EMBO J., 19, 6392–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N. and Neupert,W. (1987) Distinct steps in the import of ADP/ATP carrier into mitochondria. J. Biol. Chem., 262, 7528–7536. [PubMed] [Google Scholar]
- Ryan M.T., Muller,H. and Pfanner,N. (1999) Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem., 274, 20619–20627. [DOI] [PubMed] [Google Scholar]
- Saraste M. and Walker,J.E. (1982). Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett., 144, 250–254. [DOI] [PubMed] [Google Scholar]
- Schagger H. and Von Jagow,G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem., 199, 223–231. [DOI] [PubMed] [Google Scholar]
- Schatz G. and Dobberstein,B. (1996) Common principles of protein translocation across membranes. Science, 271, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Bauer,M.F., Guiard,B., Neupert,W. and Brunner,M. (1996) Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature, 384, 582–585. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Endres,M., Becker,K., Bauer,M.F., Walther,E., Neupert,W. and Brunner,M. (1997) Functional cooperation and stoichiometry of protein translocases of the outer and inner membranes of mitochondria. J. Biol. Chem., 272, 29963–29966. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Endres,M., Folsch,H., Stuart,R.A., Neupert,W. and Brunner,M. (1998) Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature, 391, 912–915. [DOI] [PubMed] [Google Scholar]
- Söllner T., Griffiths,G., Pfaller,R., Pfanner,N. and Neupert,W. (1989) MOM19, an import receptor for mitochondrial precursor proteins. Cell, 59, 1061–1070. [DOI] [PubMed] [Google Scholar]
- Tokatlidis K. (1999) Directing proteins to mitochondria by fusion to mitochondrial targeting signals. Methods Enzymol., 327, 305–317. [DOI] [PubMed] [Google Scholar]
- Tokatlidis K. and Schatz,G. (1999) Biogenesis of mitochondrial inner membrane proteins. J. Biol. Chem., 274, 35285–35288. [DOI] [PubMed] [Google Scholar]
- Voos W., Martin,H., Krimmer,T. and Pfanner,N. (1999) Mechanisms of protein translocation into mitochondria. Biochim. Biophys. Acta, 1422, 235–254. [DOI] [PubMed] [Google Scholar]
- Wachter C., Schatz,G. and Glick,B.S. (1992) Role of ATP in the intramitochondrial sorting of cytochrome c1 and the adenine nucleotide translocator. EMBO J., 13, 4787–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N., Pfanner,N. and Ryan,M.T. (2001) The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J., 20, 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]