Abstract
PB1 domains are novel protein modules capable of binding to target proteins that contain PC motifs. We report here the NMR structure and ligand-binding site of the PB1 domain of the cell polarity establishment protein, Bem1p. In addition, we identify the topology of the PC motif-containing region of Cdc24p by NMR, another cell polarity establishment protein that interacts with Bem1p. The PC motif-containing region is a structural domain offering a scaffold to the PC motif. The chemical shift perturbation experiment and the mutagenesis study show that the PC motif is a major structural element that binds to the PB1 domain. A structural database search reveals close similarity between the Bem1p PB1 domain and the c-Raf1 Ras-binding domain. However, these domains are functionally distinct from each other.
Keywords: Bem1p/Cdc24p/NADPH oxidase/p67phox/p40phox
Introduction
Cellular proteins often function correctly and effectively by forming a multiprotein complex, in which components are tethered via modular specific interactions (Pawson, 1995; Pawson and Nash, 2000). In the budding yeast Saccharomyces cerevisiae, a complex containing the scaffold protein Bem1p, the small GTPase Cdc42p and its guanine nucleotide exchange factor (GEF) Cdc24p appears to be required for the establishment of cell polarity during budding in vegetative growth and mating directed by cell type-specific pheromones (Madden and Snyder, 1998; Chant, 1999). The molecular nature of interactions between the polarity proteins, however, has largely remained elusive. As is shown in the accompanying paper (Ito et al., 2001), it has become evident that Bem1p interacts directly with Cdc24p via novel modules, the PB1 domain of the former and the PC motif of the latter protein, and this interaction plays a crucial role in polarity establishment.
The interaction between Cdc24p and Bem1p requires the C-terminal 75 amino acid residues of Cdc24p (Ito et al., 2001), which involves the PC motif comprised of 20 amino acid residues with the consensus sequence of #XYXDEDGDX#X#XSDED/E#X (where # is hydrophobic and X is any amino acid) (Sumimoto et al., 1997; Nakamura et al., 1998). The name PC (phox and Cdc) is derived from the fact that the motif also occurs in mammalian p40phox (a cytosolic factor of the superoxide-generating NADPH oxidase in phagocytes) (Mizuki et al., 1998; Nakamura et al., 1998), which constitutively associates with the oxidase activator p67phox (Someya et al., 1993; Wientjes et al., 1993; Tsunawaki et al., 1994). The PC motif is also known as the OPR domain (Ponting, 1996).
It is now recognized that the PC motif is present in a variety of signaling proteins such as fission yeast scd1 (a homolog of Cdc24p) (Chang et al., 1994), MEK5 (a MAP kinase kinase implicated in epidermal growth factor-induced cell proliferation) (English et al., 1995; Zhou et al., 1995) and Zip (a protein linking the ζ isoform of protein kinase C, PKCζ, to RIP and/or potassium channels) (Puls et al., 1997; Gong et al., 1999; Sanz et al., 1999). Amino acid substitutions in the PC motif of Cdc24p and p40phox abrogate the interaction with their partners, Bem1p and p67phox, respectively, indicating direct involvement of the PC motif in molecular recognition (Nakamura et al., 1998; Ito et al., 2001).
Analysis of the amino acid sequences of the PC motif-binding regions of Bem1p and p67phox has led to identification of a novel protein-binding module, PB1 (phox and Bem) domain (Ito et al., 2001). The interaction between the PB1 domain of Bem1p and the PC motif of Cdc24p is crucial for polarity establishment of yeast during both budding and mating. The biological role of the association between p67phox and p40phox via the mutually interacting modules is still unclear, but is suggested to be physiologically relevant by the observation that an antibody capable of disrupting the interaction suppresses the NADPH oxidase activation in vitro (Tsunawaki et al., 1996). A database search has revealed that the PB1 domain also exists in various proteins from plants to mammals (Ito et al., 2001). Among them, the PB1 domain of PKCζ recognizes and interacts with the PC motif of the adaptor protein ZIP (Ito et al., 2001), an interaction that might be involved in recruitment of PKCζ to a signaling complex containing RIP and/or potassium channels.
Thus, the PB1 domain and the PC motif are likely to mediate a variety of functional protein–protein interactions. We report here the first three-dimensional (3D) structure of the PB1 domain derived from Bem1p, and identify the residues involved in the PC motif-binding site. In addition, we identified the secondary structure and the topology of the PC motif-containing region derived from Cdc24p. The PC motif-containing region is a structural domain that offers a scaffold to the PC motif. Based on the chemical shift perturbation experiment and the mutagenesis study, we revealed that the PC motif is a major structural element that binds to the PB1 domain.
Results and discussion
Structure of the Bem1p PB1 domain
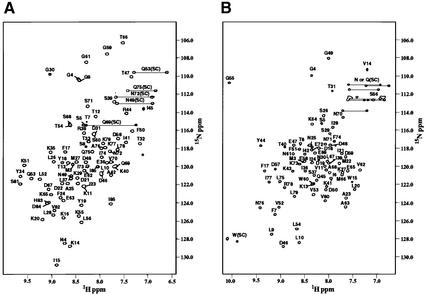
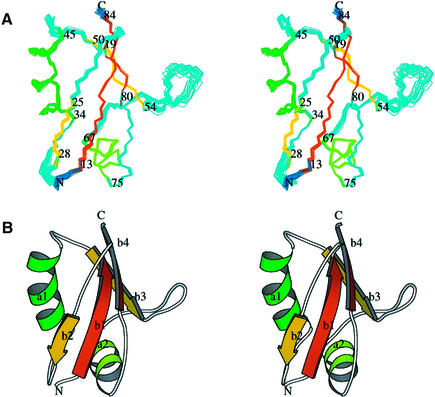
The boundaries of the Bem1p PB1 domain (designated as Bem PB1) were defined by the analyses using deletion mutants (Nakamura et al., 1998; Ito et al., 2001). Bem PB1 was expressed in Escherichia coli, and was purified as described in Materials and methods. As shown in Figure 1A, the 1H-15N HSQC spectrum of Bem PB1 is well dispersed, supporting that Bem PB1 is a structural domain. The structure of Bem PB1 was determined based on 1269 nuclear Overhauser effect (NOE)-derived interproton distances and 65 dihedral angle restraints obtained from a suite of triple-resonance multi-dimensional NMR spectra (Table I). A total of 200 conformers were calculated, and the 20 lowest energy conformers were selected. For the purposes of describing the structure, residues are numbered from 1 to 85, according to the sequence of the expressed portion of Bem1p. As the N-terminal extension 1–5 (Gly-Ala-Met-Gly-Ser) is derived from the TEV protease cleavage site (see Materials and methods), position 6 corresponds to residue 472 of intact Bem1p. Residues 1–11 and 56–61 are disordered due to lack of long-range NOEs. For residues 12–55 and 62–85, the root mean square deviation (r.m.s.d.) from the mean structure is 0.31 ± 0.04 Å for the Cα, C′ and N atoms, indicating that the backbone of Bem PB1 is well defined, as illustrated in Figure 2A.
Fig. 1. 2D 1H-15N HSQC spectra of (A) Bem PB1 and (B) Cdc24p PCCR. The cross peaks are labeled with one-letter codes of amino acids and residue numbers (SC denotes side-chain resonances). Position 6 of Bem PB1 and Cdc24p PCCR corresponds to residue 472 of Bem1p and 780 of Cdc24p, respectively.
Table I. Structural statistics.
| Parameter | <SA> | (SA)r |
|---|---|---|
| R.m.s.ds from experimental distance constraints (Å) (1269) | 0.018 ± 0.001 | 0.018 |
| Number of distance constraint violations >0.3 Å | 0 (max. 0.22) | 0 (max. 0.20) |
| R.m.s.ds from experimental dihedral constraints (°) (65) | 0.36 ± 0.14 | 0.29 |
| Number of dihedral constraint violations >5.0° | 0 (max. 3.9) | 0 (max. 1.7) |
| FNOE (kcal/mol)a | 21.3 ± 1.2 | 19.8 |
| Fcdih (kcal/mol)a | 0.15 ± 0.09 | 0.08 |
| Frepel (kcal/mol)a | 14.1 ± 1.1 | 13.6 |
| FL–J (kcal/mol)a | –248.4 ± 20.2 | –217.2 |
| R.m.s.ds from idealized geometry | ||
| bonds (Å) (1369) | 0.005 ± 0.000 | 0.004 |
| angles (°) (2488) | 0.749 ± 0.006 | 0.723 |
| impropers (°) (674)b | 1.981 ± 0.004 | 1.967 |
| Ramachandran plotc | ||
| most favorable region | 74.2 | 72.7 |
| additionally allowed region | 21.6 | 19.5 |
| generously allowed region | 3.6 | 7.8 |
| disallowed region | 0.5 | 0.0 |
<SA> refers to the final set of simulated annealing conformers and (SA)r is the mean structure. The number of terms is given in parentheses.
aThe value of the square-well NOE potential, FNOE, is calculated with a force constant of 50 kcal/mol per Å2. The value of Fcdih is calculated with a force constant of 50 kcal/mol per rad2. The value of Frepel is calculated with a force constant of 4 kcal/mol per Å2 with the van der Waals radii scaled by a factor of 0.8 of the standard values used in the CHARMm empirical function.
bThe improper torsion term is used to maintain the planar geometry and chirality.
cThe program PROCHECK-NMR (Laskowski et al., 1996) was used to assess the quality of the conformers.
Fig. 2. The 3D structure of Bem PB1. The overall fold of amino acid residues 12–85 of Bem PB1 is depicted, with the N- and C-termini labeled. (A) Overlay of the 20 final conformers on the average coordinate position for the backbone (N, Cα and C′) atoms of residues 12–55 and 62–85. Residue numbers corresponding to the beginning and the end of each secondary structural element are shown. (B) Ribbon diagrams of the minimized mean structure of Bem PB1. Both figures were created using the program MOLSCRIPT (Kraulis, 1991). Two of the inner strands (β1 and β4) are parallel and can be seen in orange, with the other two strands (β2 and β3) running in an antiparallel manner shown in yellow. The first α-helix is shown in dark green and the second one is shown in light green. The notations of these secondary structural elements (a, α-helix; b, β-strand) are indicated.
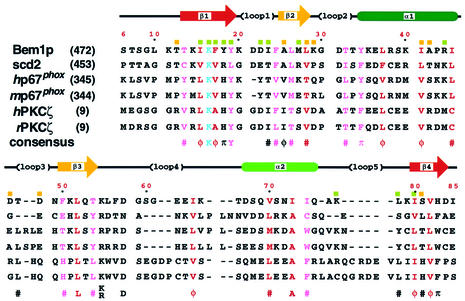
The Bem PB1 structure contains two α-helices (α1, residues 34–45; α2, 67–75) and a mixed β-sheet that consists of four strands (β1, residues 13–19; β2, 25–28; β3, 50–54; β4, 80–84) (Figure 2B). The inner two strands, β1 and β4, are parallel, while β2 and β3 run in an antiparallel manner to β1 and β4, respectively. The position of these secondary structural elements on the Bem PB1 sequence is indicated in Figure 3. Elements of the secondary structure were defined on the basis of NOE connectivities, hydrogen exchange data (see Materials and methods) and torsion angle analysis using the program PROCHECK-NMR (Laskowski et al., 1996). Then, the locations and the notations of the loops (loop 1, residues 20–24; loop 2, 29–33; loop 3, 46–49; loop 4, 55–66; loop 5, 76–79) were defined. The β-sheet has a convex surface, and α1 fits into the cavity formed by the sheet. A splayed corner of the architecture is sealed by α2.
Fig. 3. Sequence alignment of PB1 domains from selected proteins. The sequences were aligned allowing for the NMR-derived structure of Bem PB1. Prefix abbreviations are: h, human; m, mouse; r, rat. Residue numbers for the first amino acids are shown in parentheses on the left. Numbers along the top of the alignment refer to those used for Bem PB1. Secondary structural elements of Bem PB1 are shown above the sequences, as are the locations of the loops. Residues that seem to be buried in the hydrophobic core are shown in red and peripheral residues in pink. The absolutely conserved residues, corresponding to Lys16 in Bem PB1, are colored in light blue. The consensus sequence indicates the presence of identical amino acids (one-letter code) or conservative replacements (π: aromatics F, Y, W or H; φ: aliphatic hydrophobics A, I, L or V; #: C, M, T, π or φ) at least in five of the sequences. Residues showing large chemical shift changes upon ligand binding (ΔδNH ≥ 0.2 p.p.m.; ΔδN ≥ 1.0 p.p.m.) are indicated with green squares above the sequence for Bem1p; those showing moderate chemical shift changes (0.2 > ΔδNH > 0.1 p.p.m.; 0.1 > ΔδN > 0.5 p.p.m.) are labeled with yellow squares.
Bem PB1 has a well defined hydrophobic core composed of side chains from 21 amino acids with an r.m.s.d. of 0.71 ± 0.07 Å from the average coordinates of all heavy atoms. In Figure 3, the residues that appear buried in the hydrophobic core are colored in red and peripheral residues in pink. Most of these residues are located on the secondary structural elements. Pro43 is located on α1 and forms a kink that enables Ile45 to contribute to the hydrophobic core. Ile45 δ methyl protons are high-field shifted to –0.45 p.p.m. due to ring current effects from the nearby aromatic rings of Tyr19 and Phe50. A hydroxyl proton of Thr32 is buried within the core and gives rise to a resolved 1H resonance in slow exchange with solvent water on the NMR chemical shift time scale. NOEs are observed between the hydroxyl proton and backbone amide protons of Lys29 and Thr32. Residues 6–11 are not well defined in the structure. Therefore, Thr12 and Ile85 define the boundaries of the structural domain (see Figure 2).
Comparison with other PB1 domains
Figure 3 provides a comparison of the amino acid sequence of Bem PB1 with those from fission yeast scd2 (a homolog of Bem1p; Chang et al., 1994), human p67phox, mouse p67phox, human PKCζ and rat PKCζ. These amino acid sequences were aligned allowing for the positions of conserved and/or type-conserved residues in all the secondary structural elements. The lysine residue, colored in light blue in Figure 3, is strictly conserved in all the PB1 domains. Substitutions of Ala for Lys16 of Bem PB1 (corresponding to Lys482 of full-length Bem1p) and Lys355 of human p67phox completely abolish interaction with their cognate target molecules (Ito et al., 2001). It should be noted that insertions or deletions are not located in the regions corresponding to the secondary structural elements, but are located on the exposed loops, especially loops 3, 4 and 5 (Figure 3).
Bem PB1 (residues 12–85) shares sequence identity ranging from 23% (human PKCζ) to 13% (scd2), at which levels structural homology can not be predicted (Chothia and Lesk, 1986). However, considering the conserved hydrophobic core and the function of the PB1 domains as the PC motif-binding regions, the structure reported here can define the global fold for this novel domain, and our structure will be useful for modeling of other PB1 domains.
Electrostatic surface potential of Bem PB1
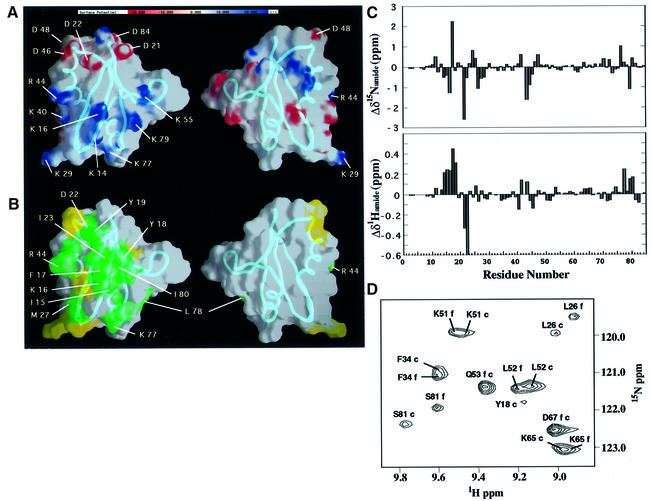
The electrostatic surface potential of Bem PB1 is shown in Figure 4A. A cluster of basic residues (Lys14, Lys16, Lys29, Lys40, Arg44, Lys55, Lys77 and Lys79) is present around Lys16, while an acidic residue-rich region (Asp21, Asp22, Asp46, Asp48 and Asp84) is located close to variable loops 1 and 3. These two regions flank a surface that includes several neutral or hydrophobic residues (shown on the left in Figure 4A), while the opposite surface of the domain (shown on the right in Figure 4A) has no characteristic features on the electrostatic surface potential. All eight side chains in the basic cluster (Figure 4A, left) are solvent exposed and structurally unconstrained, suggesting that, in complex with the PC motif, the extremities of those basic residues would interact with several acidic residues on the PC motif.
Fig. 4. Electrostatic surface potential and putative ligand-binding site of Bem PB1. (A) Electrostatic surface potential of Bem PB1. Charged residues in proximity to the putative ligand-binding region are indicated with one-letter amino acid codes and residue numbers. (B) Proposed Cdc24p PCCR-binding site of Bem PB1. Residues showing large (green) and moderate (yellow) chemical shift changes upon ligand binding are mapped on the Bem PB1 surface. Bem PB1 is presented in the same orientation as in Figure 2 (left) or rotated by 180° (right). The main chains are shown as tubes. These figures were prepared using the program GRASP (Nicholls et al., 1991). (C) Backbone amide 1H and 15N chemical shift changes for the amino acid residues of the 15N-labeled Bem PB1 upon binding to Cdc24p PCCR as observed in 2D 1H-15N HSQC spectra. Note Δδ1H of Ile23 is –1.16 p.p.m. (D) Representative region of the 2D 1H-15N HSQC spectra of Bem PB1 in the free state (labeled with f) and the complex with unlabeled Cdc24p PCCR (labeled with c). Molar ratio of 15N-labeled Bem PB1 to unlabeled Cdc24p PCCR was 2:1.
Identification of the PC motif-binding surface
The 75 C-terminal amino acid residues of Cdc24p are the minimum region required for binding to Bem1p (Peterson et al., 1994; Ito et al., 2001). We referred to this region as Cdc24p PCCR (PC motif-containing region). Cdc24p PCCR was expressed in E.coli, and purified as described in Materials and methods. The 1H-15N HSQC spectrum of Cdc24p PCCR shows well dispersed resonances, supporting that Cdc24p PCCR is also a structural domain (Figure 1B).
Molecular interaction between Cdc24p PCCR and Bem PB1 was analyzed by NMR using unlabeled Cdc24p PCCR and 15N-labeled Bem PB1. Upon addition of aliquots of the Cdc24p PCCR solution, a pair of resonances in the 1H-15N HSQC spectrum was observed corresponding to the free and the bound states of Bem PB1, indicating that the complex formation is in slow exchange on the NMR chemical shift time scale (Figure 4D). We assigned the resonances of Bem PB1 in complex with Cdc24p PCCR independently of the free form by the analysis of a suite of triple-resonance multi-dimensional NMR spectra. The chemical shift difference of each resonance between the Cdc24p PCCR bound and unbound forms of Bem PB1 was plotted in Figure 4C. Each residue was classified according to the degree of perturbations on the HSQC spectrum, which monitors amide proton and nitrogen chemical shifts. Strong (ΔδNH ≥ 0.2 p.p.m.; ΔδN ≥ 1.0 p.p.m.) and moderate (0.2 > ΔδNH > 0.1 p.p.m.; 1.0 > ΔδN > 0.5 p.p.m.) chemical shift changes upon ligand binding were observed for 12 residues (Ile15, Lys16, Phe17, Tyr18, Tyr19, Asp21, Ile23, Met27, Arg44, Lys77, Leu78 and Ile80) and nine residues (Thr12, Ala25, Leu28, Lys29, Ile41, Ala42, Asp46, Asp48 and Ser81), respectively (Figures 3 and 4C). In addition, minor chemical shift perturbation was observed over the whole molecule, indicating the global conformation change. When the residues with appreciable chemical shift changes by complex formation (Figure 4C) were mapped on the structure of Bem PB1, they covered the basic, acidic and neutral residue clusters on the surface of the molecule as shown in Figure 4B (left), while such residues are not present on the opposite surface (Figure 4B, right). The clearly identified PCCR-binding surface corresponds to the putative ligand-binding region around β1, which includes the conserved Lys16 residue.
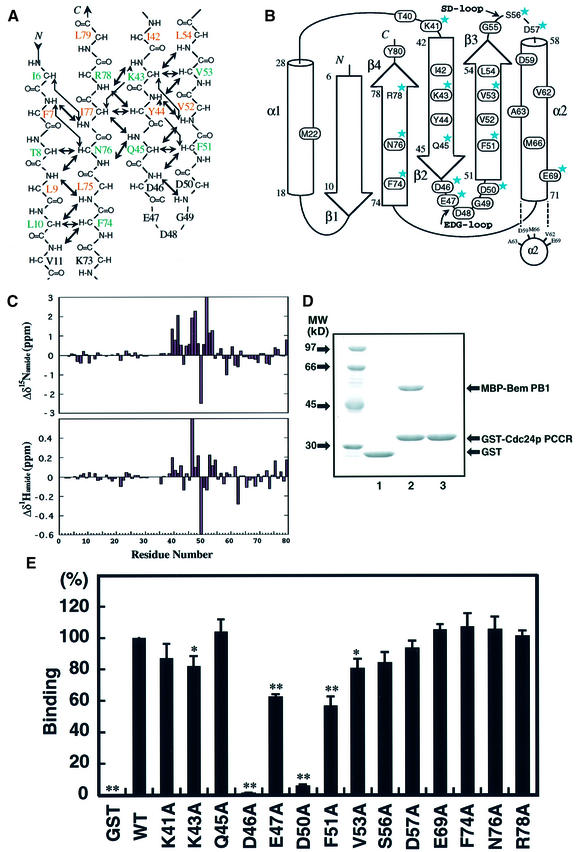
Topology of Cdc24p PCCR
In order to elucidate the details of the interaction between Bem PB1 and Cdc24p PCCR, we first investigated the solution structure of Cdc24p PCCR in an unbound state. Residues are numbered from 1 to 80, according to the sequence of the expressed portion of Cdc24p, where position 6 corresponds to residue 780 of full-length protein. By 3D NMR analysis, we assigned the main chain resonances of Cdc24p PCCR. On the basis of both NOE connectivities observed among the main chain CαH and NH resonances, and CSI analysis (Wishart and Sykes, 1994), we determined the secondary structure and the topology of Cdc24p PCCR (Figure 5A and B). Cdc24p PCCR was identified as a structural domain comprising two α-helices (α1, 18–28; α2, 58–71) and an antiparallel β-sheet formed by four β-strands (β1, 6–10; β2, 42–45; β3, 51–54; β4, 74–78). The PC motif (residues 39–66) is assigned to the region spanning from β2 to the midpoint of α2. It should be noted that the PC motif forms a core in the structural domain of Cdc24p PCCR. The consensus acidic residues (Asp46, Glu47, Asp48, Gly49 and Asp50) in the PC motif are located on the β-hairpin loop between β2 and β3, designated as the EDG-loop. Another loop also rich in acidic residues (Gly55, Ser56, Asp57 and Glu58), named the SD-loop, is located between β3 and α2 (Figure 5B and F). Next, we identified the residues on Cdc24p PCCR that are responsible for binding to Bem PB1. In a manner similar to the previous section, we added aliquots of unlabeled Bem PB1 to 15N-labeled Cdc24p PCCR in order to observe the chemical shift difference of individual residues between the free and the bound states of Cdc24p PCCR. Since the exchange process between the free and the bound states was slow compared with the NMR chemical shift time scale, the resonance assignments of the bound form were made independently of the free form. Interestingly, the residues with large chemical shift difference are mainly located on the PC motif (Figure 5C). We mapped the residues with chemical shift difference of ΔδNH ≥ 0.1 p.p.m. and ΔδN ≥ 0.5 p.p.m. on the topology of Cdc24 PCCR (the residues enclosed by an oval in Figure 5B), which are localized on the region containing the PC motif and its succeeding region. Thus, these regions can be identified as the binding surface to Bem PB1.
Fig. 5. NMR and mutational analysis of Cdc24p PCCR and sequence alignment of PC motifs. (A) Interstrand backbone NOE connectivities of Cdc24p PCCR. Interstrand NOEs observed in 3D 15N-edited and 13C-edited NOESY spectra are marked by arrows. The residues with their side chains above the sheet are colored in green, while the residues with their side chains below the sheet are colored in brown. The residues in the loop are in black. (B) Topology of Cdc24p PCCR. Strands and helices are represented by arrows and cylinders. Residues showing large chemical shift changes upon Bem PB1 binding (ΔδNH > 0.1 p.p.m.; ΔδN > 0.5 p.p.m.) are enclosed by an oval. Residues into which mutations were introduced are labeled with stars. (C) Plots showing backbone amide 1H and 15N chemical shift changes of 15N-labeled Cdc24p PCCR upon binding to the unlabeled Bem PB1. Δδ1H of Glu47 is 1.47 p.p.m., and Δδ15N of Val52 is 3.28 p.p.m. 2D 1H-15N HSQC signals of Ala2, Glu12, Asn16, His32, Asn33, Asn34 and Asn35 are not assigned either due to broadening or overlapping in the free and/or the Bem PB1-bound form. Therefore, the chemical shift changes of these residues are not plotted. (D) Complex formation between Cdc24p PCCR and Bem PB1 as examined by in vitro pull-down assays. MBP–Bem PB1 (140 pmol) was incubated with 450 pmol of GST alone (lane 1), MBP–Bem PB1 with GST–Cdc24p PCCR (lane 2) and MBP alone with GST–Cdc24p PCCR (lane 3). Proteins pulled down with glutathione–Sepharose-4B beads were subjected to SDS–PAGE and stained with Coomassie Blue. (E) Effects of amino acid substitutions of Cdc24p PCCR on the binding to Bem PB1 examined by in vitro pull-down assays. MBP–Bem PB1 was incubated with GST–Cdc24p PCCR mutant proteins carrying the indicated amino acid substitution. Proteins were pulled down with glutathione–Sepharose-4B beads and analyzed in a similar manner to that in (D). The amounts of bound Bem PB1 were estimated and plotted as described in Materials and methods. The values represent the mean ± SD of three independent experiments. The amount of wild-type Cdc24p PCCR is set as 100%. Statistical significance is reported as ** or *, corresponding to p < 0.001 or 0.01, respectively, between mutant and wild type. (F) Amino acid sequence of Cdc24p PCCR and multiple sequence alignment of the PC motifs from selected proteins. Prefix abbreviations are: h, human; r, rat. Residue numbers for the first amino acid residues are shown in parentheses on the left. Numbers along the top of the alignment refer to those for Cdc24p PCCR. The consensus sequence indicates the presence of identical amino acids (one-letter code) or conservative replacements (#: hydrophobics). Secondary structural elememts of Cdc24p PCCR are shown above the sequences together with the locations and the notations of the loops. Effects of mutations on the individual residues are summarized with the following symbols above the sequences: circle, single-point mutation (from E); triangle, single-point mutation (Nakamura et al., 1998); red, strong; pink, moderate; yellow, weak; black, no effect.
Specificity determinants for complex formation between the PB1 domain and the PC motif
We have made an extensive alanine-scanning mutagenesis for those residues that showed appreciable chemical shift changes upon PB1–PCCR complex formation and appear to project their side chains to the protein surface (residues for which alanine-scanning mutagenesis were applied are labeled with stars in Figure 5B). The binding affinity of the Cdc24p PCCR and its mutants toward Bem PB1 was measured by in vitro binding assay as described in Materials and methods (Figure 5D). The results of the mutagenesis study are summarized in Figure 5E. The D46A and D50A mutants showed a significant decrease in binding affinity, and a moderate decrease was observed for E47A and F51A, and a slight decrease for K43A and V53A mutants, while other mutants showed essentially similar affinity toward the PB1 binding compared with the wild type. Asp46 and Asp50 are located on the EDG-loop and are primarily important for binding to Bem PB1, while the residues on the SD-loop did not show any decrease in binding affinity. Lys43, Phe51 and Val53 are located on β2 and β3. Taking NMR and mutagenesis studies together, Asp46 and Asp50, highly conserved on the EDG-loop, are generally responsible for binding between the PB1 domain and the PC motif, while the residues on β2 and β3 are not conserved in the PC motif and appear to play a role in determining the specificity of the PC motif-containing regions toward the cognate pair of the PB1 domains.
The sequences of the PC motif identified in several proteins are aligned in Figure 5F with the sequence of Cdc24p PCCR. The acidic residues on the EDG- and the SD-loops are highly conserved in almost all the PC motifs. The intensive site-directed mutagenesis applied to the PC motif in p40phox revealed that the SD-loop as well as the EDG-loop plays a critical role in binding to the PB1 domain of p67phox (Nakamura et al., 1998; Figure 5F), which is in contrast to the case of Cdc24p PCCR. The EDG-loop is generally important for PCCR–PB1 complex formation through binding to the basic cluster that involves the conserved lysine residue.
Comparison of the PB1 domain with other structures
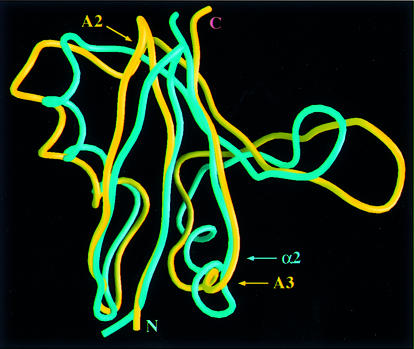
An automated search of the DALI database (Holm and Sander, 1996) revealed an unexpected structural similarity between Bem PB1 and the Ras-binding domain (RBD) of the Ser/Thr-specific protein kinase c-Raf1 (referred to as Raf; Nassar et al., 1996). An overlay of these structures is shown in Figure 5. These domains have identical topologies and a very similar organization of secondary structures with an r.m.s.d. of 2.2 Å for backbone atoms of 69 structurally equivalent residues, even though the overall sequence similarity is below the level of significance (Chothia and Lesk, 1986). The 3D folds of these domains are similar to that of ubiquitin (Vijay-Kumar et al., 1987), and the ubiquitin fold has been classified as a protein superfold, the ubiquitin α/β roll (Orengo et al., 1994). The major structural differences between Bem PB1 and Raf RBD are found in the following regions shown in Figure 6; the small 310-helix A2 (residues 93–95) in Raf RBD is absent in Bem PB1, and another 310-helix A3 (residues 118–121) in Raf RBD is replaced with the long α2 helix in Bem PB1 (Nassar et al., 1995). However, these regions are isolated from the ligand-binding surface.
Fig. 6. Comparison of the structures of Bem PB1 and Raf RBD. Superposition of the backbone structures of Bem PB1 (blue) and Raf RBD (yellow). Residues 12–85 of Bem PB1 are overlaid with residues 56–131 of the Raf RBD (Nassar et al., 1996) on the basis of the structurally equivalent residues determined by the DALI database search (Holm and Sander, 1996) using Bem PB1 mean coordinates. The N- and C-termini, the 310 helices (A2 and A3) of Raf RBD and the α-helix (α2) of Bem PB1 are indicated. This figure was generated using GRASP (Nicholls et al., 1991). The coordinates of Raf RBD were obtained from the Brookhaven Protein Data Bank (entry 1GUA).
The residues Gln66, Lys84 and Arg89 in Raf, which primarily determine the binding affinity to Ras (Block et al., 1996), are structurally equivalent to Asp22, Lys40 and Arg44 in Bem PB1, respectively. Moreover, Park et al. (1997) have reported direct binding of Bem1p to the GDP-bound form of Bud1p, a Ras-like GTPase. Furthermore, Bokoch et al. (1991) suggest that a Ras-related protein Rap1A may participate in regulation of the NADPH oxidase system and that this function of Rap1A may be altered by phosphorylation. Accordingly, using the yeast two-hybrid system and an in vitro binding assay using purified proteins (Ago et al., 1999), we have examined the interactions between the PB1 domains of Bem1p, p67phox and PKCζ, and Ras-related proteins, Bud1p, Rap1A and Ras in both GDP- and GTP-bound forms. However, we could detect no positive signal in any of the possible combinations (data not shown). Similarly, the PC motif-containing regions of Cdc24p, p40phox and ZIP did not bind to the RBD of Raf and RalGDS (a guanine nucleotide dissociation stimulator specific for the Ral GTPase) (data not shown), which also have similar folds to that of Bem PB1 (Huang et al., 1998). These data suggest that the PB1 domain is not a subfamily of the RBD set, but a functionally distinct domain.
Conclusions
The present report describes the first structure determination of a novel protein-binding motif, the PB1 domain from Bem1p and the identification of the topology of the PC motif-containing region from Cdc24p. The Bem PB1 structure will be helpful for modeling of other PB1 domains and the PC motif-containing region. Identification of the ligand-binding sites located on Bem PB1 and Cdc24p PCCR from the chemical shift perturbation studies enables us to elucidate the binding interface between the PB1–PC motif complex. The PC motif forms a core of PCCR and is mainly responsible for the binding to the PB1 domain. This study provides the insights into the structural basis for the molecular recognition between the PB1 domain and the PC motif-containing region. The 3D fold of Bem PB1 is remarkably similar to that of Raf RBD. However, the PB1 domains do not bind to the Ras-related proteins, nor do the PC motif-containing regions interact with the Ras-binding domains. Therefore, the PB1 domains are structurally and functionally distinct protein modules.
Materials and methods
Preparation of Bem PB1 and Cdc24p PCCR
Bem PB1 gene (residues 472–551) and Cdc24p PCCR gene (residues 780–854) were amplified by PCR from yeast genomic DNA and then cloned into the BamHI–EcoRI sites of pProEX-HTa vector (Gibco-BRL). Bem PB1 and Cdc24p PCCR were expressed in E.coli BL21 DE3 with His6 fused at the N-terminus. For unlabeled protein, the transformed cells were grown in Luria–Bertani media. We also prepared uniformly 13C/15N- and 15N-labeled proteins by growing cells in M9 media containing 15NH4Cl (1 g/l) and CELTONE-CN or -N powder (1 g/l; Martek) with [U-13C]glucose (2 g/l) or unlabeled glucose. After cell lysis, the His6 fusion protein was first purified by affinity chromatography on a Ni-NTA Superflow resin (Qiagen), followed by the removal of the His6 tag by TEV protease cleavage (Gibco-BRL). Resource S cation affinity and Resource RPC reverse-phase columns (Biotech Pharmacia) were used for further purification of Bem PB1, while a Mono Q anion exchange column (Biotech Pharmacia) was used for further purification of Cdc24p PCCR. The identity and integrity of the protein were confirmed by N-terminal sequencing, MALDI/TOF MS spectra and SDS–PAGE.
The purified protein was dialyzed and concentrated to 2 mM in H2O/D2O (9/1) or D2O buffers containing 50 mM potassium phosphate pH 6.3, 150 mM NaCl and 1 mM NaN3.
NMR spectroscopy
All NMR measurements were carried out at 25°C on Varian Unity-plus 600 or Unity-Inova 500 spectrometers equipped with three rf channels and a triple-resonance pulsed-field gradient probe with an actively shielded z gradient coil. The sequential assignments of 1H, 13C and 15N chemical shifts were achieved mainly by through-bond heteronuclear correlations along the backbones and the side chains with the following 3D pulse sequences: HN(CO)CA, HNCA, CBCA(CO)NH, CBCANH, HBHA(CO)NH, C(CO)NH and HC(C)H-TOCSY (Clore and Gronenborn, 1994). Inter-proton NOEs were derived from 3D 15N- edited and 13C-edited NOESY spectra (Clore and Gronenborn, 1994) recorded with a mixing time of 75 ms. The pulse sequences of HN(CO)CA, HNCA, CBCANH, HBHA(CO)NH, C(CO)NH and 15N- edited NOESY were modified incorporating pulsed-field gradient and sensitivity enhancement methods (Kay et al., 1992; Muhandiram and Kay, 1994). We determined φ-angle restraints based on the 3JHN,Hα coupling constants measured in a 3D HNHA spectrum (Kuboniwa et al., 1994). Stereospecific assignments of Hβ methylene protons and χ1-angle restraints were determined based on the 3JN,Hβ and 3JCO,Hβ coupling constants measured in 3D HNHB and HN(CO)HB spectra (Archer et al., 1991; Grzesiek et al., 1992). Slowly exchanging amide protons were identified from a series of 2D 1H-15N HSQC spectra recorded after buffer exchange from H2O to D2O.
Structure calculations
The program NMRPipe (Delaglio et al., 1995) was used to process the NMR data, and the transformed spectra were converted to FELIX 95.0 (Molecular Simulations Inc., San Diego, CA) format using in-house software. The resonances were peak-picked using the in-house program described in Hatanaka et al. (1994). Semiautomatic assignment of the NOESY spectra was performed with the program XPLOR-UP (Hatanaka et al., 1994) to generate an unbiased set of distance restraints. Ambiguous NOE crosspeaks were assigned in an iterative manner, with reference to the calculated conformers (Terasawa et al., 1994; Hatanaka et al., 1996; Kohda et al., 1996). Upper limits of distance restraints were calculated as kI–1/6, where I is the peak intensity and k is a constant adjusted in each NOESY spectrum, and relaxed by 0.5 Å allowing for mobility. The lower limits of distance restraints were all set to 1.8 Å. After 10 cycles of iterative NOESY peak assignment and structure calculation, a total of 500 intraresidue (|i – j| = 0), 253 sequential (|i – j| = 1), 193 medium-range (2 ≤ |i – j| ≤ 5) and 323 long-range (|i – j| ≥ 6) distance restraints were used in the final structure calculations. This included 42 restraints from hydrogen bonds implied by hydrogen–deuterium exchange experiments and local secondary structure.
A total of 200 conformers were calculated on the basis of 1269 distance, 39 φ and 26 χ1 dihedral angle constraints using program X-PLOR version 3.1 (Brünger, 1993). The mean structure was obtained by averaging the coordinates of the 20 selected conformers that had been superimposed in advance to the best converged conformers, and then minimized under the restraints. The coordinates of the mean structure have been deposited in the Brookhaven Protein Data Bank (1IPG code).
Ligand titration
The initial sample for the Cdc24p PCCR titration experiment consisted of 2 mM 15N-labeled Bem PB1, 50 mM potassium phosphate pH 6.3, 150 mM NaCl and 1 mM NaN3 in H2O/D2O (9/1) buffer. A stock solution of unlabeled Cdc24p PCCR was concentrated to 1.5 mM in the same aqueous buffer. Aliquots of the Cdc24p PCCR stock solution were added in a stepwise manner to the 15N-labeled Bem PB1 sample. Similarly, aliquots of Bem PB1 stock solution were added to the 15N-labeled Cdc24p PCCR sample to observe the titration behavior of the Cdc24p PCCR resonances.
An in vitro pull-down binding assay using purified proteins
The cDNAs for Bem PB1 were ligated to pMAL (New England Biolabs). The cDNAs encoding point mutants of the C-terminal region of Cdc24p were obtained by PCR-mediated site-directed mutagenesis. The PCR products were ligated to pGEX-6P (Amersham Pharmacia Biotech). All the constructs were sequenced to confirm their identities. Glutathione S-transferase (GST)- and maltose-binding protein (MBP)-tagged proteins were expressed in E.coli strain BL21 and purified by glutathione– Sepharose-4B (Amersham Pharmacia Biotech) or Amylose resin (New England Biolabs), respectively, as previously described (Noda et al., 2001). Pull-down binding assays were performed as previously described (Noda et al., 2001). Briefly, a pair of a GST fusion and an MBP-tagged protein was incubated in 500 µl of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4 and 1.5 mM KH2PO4 pH 7.4) containing 0.5% Triton X-100, and precipitated by glutathione–Sepharose-4B. After washing six times with the same buffer containing 0.5% Triton X-100, bound proteins were eluted with 10 mM glutathione. The eluates were subjected to 10% SDS–PAGE and visualized with Coomassie Blue. To estimate the amounts of protein on the gel, densitometric analysis was carried out using the software NIH Image. Student’s t-test was used for estimation of statistical significance. A p value <0.01 was considered statistically significant.
Acknowledgments
Acknowledgements
We are grateful to Dr Daisuke Kohda of Biomolecular Engineering Research Institute for his stimulating advice. We also thank Dr Masataka Horiuti, Mrs Etsuko Ebisui, Dr Satoru Yuzawa and Mr Masashi Yokochi in the Tokyo Metropolitan Institute of Medical Science for their useful comments. This work was supported by a Grant-in-Aid from the Ministry of Education, Science and Culture of Japan and CREST of Japan Science and Technology.
References
- Ago T., Nunoi,H., Ito,T. and Sumimoto,H. (1999) Mechanism for phosphorylation-induced activation of the phagocyte NADPH oxidase protein p47phox. J. Biol. Chem., 274, 33644–33653. [DOI] [PubMed] [Google Scholar]
- Archer S.J., Ikura,M., Torchia,D.A. and Bax,A. (1991) An alternative 3D NMR technique for correlating backbone 15N with side chain Hβ resonances in larger proteins. J. Magn. Reson., 95, 636–641. [Google Scholar]
- Block C., Janknecht,R., Herrmann,C., Nassar,N. and Wittinghofer,A. (1996) Quantitative structure–activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nature Struct. Biol., 3, 244–251. [DOI] [PubMed] [Google Scholar]
- Bokoch G.M., Quilliam,L.A., Bohl,B.P., Jesaitis,A.J. and Quinn,M.T. (1991) Inhibition of Rap1A binding to cytochrome b558 of NADPH oxidase by phosphorylation of Rap1A. Science, 254, 1794–1796. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1993) X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Chang E.C., Barr,M., Wang,Y., Jung,V., Xu,H.-P. and Wigler,M.H. (1994) Cooperative interaction of S.pombe proteins required for mating and morphogenesis. Cell, 79, 131–141. [DOI] [PubMed] [Google Scholar]
- Chant J. (1999) Cell polarity in yeast. Annu. Rev. Cell Dev. Biol., 15, 365–391. [DOI] [PubMed] [Google Scholar]
- Chothia C. and Lesk,A.M. (1986) The relation between the divergence of sequence and structure in proteins. EMBO J., 5, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1994) Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol., 239, 349–363. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- English J.M., Vanderbilt,C.A., Xu,S., Marcus,S. and Cobb,M.H. (1995) Isolation of MEK5 and differential expression of alternatively spliced forms. J. Biol. Chem., 270, 28897–28902. [DOI] [PubMed] [Google Scholar]
- Gong J., Xu,J., Bezanilla,M., van Huizen,R., Derin,R. and Li,M. (1999) Differential stimulation of PKC phosphorylation of potassium channels by ZIP1 and ZIP2. Science, 285, 1565–1569. [DOI] [PubMed] [Google Scholar]
- Grzesiek S., Ikura,M., Clore,G.M., Gronenborn,A.M. and Bax,A. (1992) A 3D triple-resonance NMR technique for qualitative measurement of carbonyl-HβJ couplings in isotopically enriched proteins. J. Magn. Reson., 96, 215–221. [Google Scholar]
- Hatanaka H., Oka,M., Kohda,D., Tate,S., Suda,A., Tamiya,N. and Inagaki,F. (1994) Tertiary structure of erabutoxin b in aqueous solution as elucidated by two-dimensional nuclear magnetic resonance. J. Mol. Biol., 240, 155–166. [DOI] [PubMed] [Google Scholar]
- Hatanaka H., Ogura,K., Moriyama,K., Ichikawa,S., Yahara,I. and Inagaki,F. (1996) Tertiary structure of destrin and structural similarity between two actin-regulating protein families. Cell, 85, 1047–1055. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–602. [DOI] [PubMed] [Google Scholar]
- Huang L., Hofer,F., Martin,G.S. and Kim,S.-H. (1998) Structural basis for the interaction of Ras with RalGDS. Nature Struct. Biol., 5, 422–426. [DOI] [PubMed] [Google Scholar]
- Ito T., Matsui,Y., Ago,T., Ota,K. and Sumimoto,H. (2001) Novel modular domain PB1 recognizes PC motif to mediate functional protein–protein interactions. EMBO J., 20, 3938–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay L.E., Keifer,P. and Saarinen,T.P. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc., 114, 10663–10665. [Google Scholar]
- Kohda D., Morton,C.J., Parkar,A.A., Hatanaka,H., Inagaki,F.M., Campbell,I.D. and Day,A.J. (1996) Solution structure of the link module: a hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell, 86, 767–775. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Kuboniwa H., Grzesiek,S., Delaglio,F. and Bax,A. (1994) Measurement of HN-HαJ couplings in calcium-free calmodulin using new 2D and 3D water-flip-back methods. J. Biomol. NMR, 4, 871–878. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Madden K. and Snyder,M. (1998) Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol., 52, 687–744. [DOI] [PubMed] [Google Scholar]
- Mizuki K., et al. (1998) Functional modules and expression of mouse p40phox and p67phox, SH3-domain-containing proteins involved in the phagocyte NADPH oxidase complex. Eur. J. Biochem., 251, 573–582. [DOI] [PubMed] [Google Scholar]
- Muhandiram D.R. and Kay,L.E. (1994) Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B, 103, 203–216. [Google Scholar]
- Nakamura R., Sumimoto,H., Mizuki,K., Hata,K., Ago,T., Kitajima,S., Takeshige,K., Sakaki,Y. and Ito,T. (1998) The PC motif: a novel and evolutionarily conserved sequence involved in interaction between p40phox and p67phox, SH3 domain-containing cytosolic factors of the phagocyte NADPH oxidase. Eur. J. Biochem., 251, 583–589. [DOI] [PubMed] [Google Scholar]
- Nassar N., Horn,G., Herrmann,C., Scherer,A., McCormick,F. and Wittinghofer,A. (1995) The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature, 375, 554–560. [DOI] [PubMed] [Google Scholar]
- Nassar N., Horn,G., Herrmann,C., Block,C., Janknecht,R. and Wittinghofer,A. (1996) Ras/Rap effector specificity determined by charge reversal. Nature Struct. Biol., 3, 723–729. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Noda Y., Takeya,R., Ohno,S., Naito,S., Ito,T. and Sumimoto,H. (2001) Human homologues of the Caenorhabditis elegans cell polarity protein PAR6 as an adaptor that links the small GTPases Rac and Cdc42 to atypical protein kinase C. Genes Cells, 6, 107–119. [DOI] [PubMed] [Google Scholar]
- Orengo C.A., Jones,D.T. and Thornton,J.M. (1994) Protein superfamilies and domain superfolds. Nature, 372, 631–634. [DOI] [PubMed] [Google Scholar]
- Park H.-O., Bi,E., Pringle,J.R. and Herskowits,I. (1997) Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl Acad. Sci. USA, 94, 4463–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. (1995) Protein modules and signaling networks. Nature, 373, 573–580. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Nash,P. (2000) Protein–protein interactions define specificity in signal transduction. Genes Dev., 14, 1027–1047. [PubMed] [Google Scholar]
- Peterson J., Zheng,Y., Bender,L., Myers,A., Cerione,R. and Bender,A. (1994) Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J. Cell Biol., 127, 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting P.C. (1996) Novel domains in NADPH oxidase subunits, sorting nexins and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci., 5, 2353–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls A., Schmidt,S., Grawe,F. and Stabel,S. (1997) Interaction of protein kinase C ζ with ZIP, a novel protein kinase C-binding protein. Proc. Natl Acad. Sci. USA, 94, 6191–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Sanchez,P., Lallena,M.J., Diaz-Meco,M.T. and Moscat,J. (1999) The interaction of p62 with RIP links the atypical PKCs to NF-κB activation. EMBO J., 18, 3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya A., Nagaoka,I. and Yamashita,T. (1993) Purification of the 260 kDa cytosolic complex involved in the superoxide production of guinea pig neutrophils. FEBS Lett., 330, 215–218. [DOI] [PubMed] [Google Scholar]
- Sumimoto H., Ito,T., Hata,K., Mizuki,K., Nakamura,R., Kage,Y., Nakamura,M., Sakaki,Y. and Takeshige,K. (1997) Membrane translocation of cytosolic factors in activation of the phagocyte NADPH oxidase: role of protein–protein interactions. In Mihara,K. and Hamasaki,N. (eds), Membrane Proteins: Structure, Function and Expression Control. S. Karger AG, Basel, Switzerland, pp. 235–245.
- Terasawa H., Kohda,D., Hatanaka,H., Nagata,K., Higashihashi,N., Fujiwara,H., Sakano,K. and Inagaki,F. (1994) Solution structure of human insulin-like growth factor II; recognition sites for receptors and binding proteins. EMBO J., 13, 5590–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunawaki S., Mizunari,H., Nagata,M., Tatsuzawa,O. and Kuratsuji,T. (1994) A novel cytosolic component, p40phox, of respiratory burst oxidase associates with p67phox and is absent in patients with chronic granulomatous disease who lack p67phox. Biochem. Biophys. Res. Commun., 199, 1378–1387. [DOI] [PubMed] [Google Scholar]
- Tsunawaki S., Kagara,S., Yoshikawa,K., Yoshida,L.S., Kuratsuji,T. and Namiki,H. (1996) Involvement of p40phox in activation of phagocyte NADPH oxidase through association of its carboxyl-terminal, but not its amino-terminal, with p67phox. J. Exp. Med., 184, 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg,C.E. and Cook,W.J. (1987) Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol., 194, 531–544. [DOI] [PubMed] [Google Scholar]
- Wientjes F.B., Hsuan,J.J., Totty,N.F. and Segal,A.W. (1993) p40phox, a third cytosolic component of the activation complex of the NADPH oxidase to contain src homology 3 domains. Biochem. J., 296, 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S. and Sykes,B.D. (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR, 4, 171–180. [DOI] [PubMed] [Google Scholar]
- Zhou G., Bao,Z.Q. and Dixon,J.E. (1995) Components of a new human protein kinase signal transduction pathway. J. Biol. Chem., 270, 12665–12669. [DOI] [PubMed] [Google Scholar]