Abstract
Modular domains mediating specific protein–protein interactions play central roles in the formation of complex regulatory networks to execute various cellular activities. Here we identify a novel domain PB1 in the budding yeast protein Bem1p, which functions in polarity establishment, and mammalian p67phox, which activates the microbicidal phagocyte NADPH oxidase. Each of these specifically recognizes an evolutionarily conserved PC motif to interact directly with Cdc24p (an essential protein for cell polarization) and p40phox (a component of the signaling complex for the oxidase), respectively. Swapping the PB1 domain of Bem1p with that of p67phox, which abolishes its interaction with Cdc24p, confers on cells temperature- sensitive growth and a bilateral mating defect. These phenotypes are suppressed by a mutant Cdc24p harboring the PC motif-containing region of p40phox, which restores the interaction with the altered Bem1p. This domain-swapping experiment demonstrates that Bem1p function requires interaction with Cdc24p, in which the PB1 domain and the PC motif participate as responsible modules.
Keywords: Bem1p/Cdc24p/cell polarity/NADPH oxidase/p67phox
Introduction
It is now widely recognized that protein–protein interactions play key roles in various aspects of structural and functional organization of cells (Pawson, 1995; Pawson and Nash, 2000). Recent studies revealed that these interactions are often mediated by modules that are specialized for or dedicated to specific protein binding, e.g. SH2 and SH3 domains bind to phosphotyrosine-containing and proline-rich peptides, respectively. Com binatorial use of these modules has allowed cells to develop a variety of protein networks for fine regulation of various cellular activities. It also becomes evident that cells often physically integrate the components of the same pathway or cascade into a unit by forming a multiprotein complex, thereby ensuring specific and prompt signal transduction. For such a network, cells have constructed a novel class of proteins serving as scaffolds, on which relevant proteins are assembled to collaborate.
The establishment of cell polarity in the budding yeast is also governed by a complex protein machinery (Madden and Snyder, 1998; Chant, 1999). The yeast cell polarizes in two phases of its life; the budding in the vegetative growth phase and the formation of mating projection toward the partner at the mating. It is currently thought that an overlapping but not identical polarity establishment machinery functions in both cases. The core of the protein machinery for polarity establishment is comprised of the small GTPase Cdc42p, its guanine-nucleotide exchange factor Cdc24p and an SH3 domain-containing protein, Bem1p, although the complex also contains other components. The complex is assumed to receive a positional cue at each stage, one from the intrinsic signal from the bud site and the other from the mating pheromone receptor, and transmits the signal downstream to induce reorganization of cytoskeleton. Accordingly, the complex has been shown to interact not only with the upstream regulators like Bud1p/Rsr1p in budding and Ste4p in mating but also with downstream effectors for cytoskeletal reorganization (O’Shea and Herskowitz, 2000).
Among the proteins involved in the complex, Bem1p appears to function as a scaffold for others, since it lacks any catalytic domain but interacts, both physically and genetically, with various proteins involved in the polarity establishment and mating signal transduction as well as those of the cytoskeleton (Chenevert et al., 1992; Peterson et al., 1994; Leeuw et al., 1995; Zheng et al., 1995; Bender et al., 1996; Lyons et al., 1996; Matsui et al., 1996; Park et al., 1997; Butty et al., 1998). The molecular architecture of the Bem1p-centered complex has to be elucidated for understanding the mechanism underlying polarity establishment, but it is poorly understood at present: non-SH3 regions of Bem1p have remained unpursued for their potential involvement in protein binding.
Here we present data to show that the C-terminal portion of Bem1p comprises a novel protein-binding module termed the PB1 domain, which recognizes the PC motif to interact directly with Cdc24p. A domain-swapping strategy with a mammalian PB1 domain and PC motif pair demonstrates that they function as mutually interacting modules to mediate interaction between Bem1p and Cdc24p, and that this interaction is crucial for the polarity establishment in both budding and mating. We also show that the novel PB1 domain and PC motif modules occur in other signaling systems and appear to regulate a variety of cellular processes.
Results
Bem1p recognizes the PC motif to associate with Cdc24p
Two-hybrid analyses of Bem1p and Cdc24p revealed that they form a complex via a C-terminal tail-to-tail interaction. The minimum essential region for this binding was mapped to amino acid residues 472–551 of Bem1p, the C-terminal extremity; further deletion from either end completely abolished the two-hybrid interaction (Figure 1A). The pinpointed region was also confirmed by an in vitro pull-down binding assay (Figure 1A). For Cdc24p, it was previously shown that its C-terminal 75 amino acid region is capable of binding to Bem1p (Peterson et al., 1994). We thus prepared deletion mutants of the C-terminal 75 amino acid portion of Cdc24p. As shown in Figure 2A, further deletion from either the N- or the C-terminal end resulted in an abolished or severely compromised interaction with Bem1p, respectively. These results indicate that the C-terminal 75 amino acids of Cdc24p are required for its stable association with Bem1p.
Fig. 1. Pinpointing the Cdc24p-binding region on Bem1p. Various portions of Bem1p were tested for binding to Cdc24p by two-hybrid assays (A) and in vitro pull-down assays (B). PB2 represents a novel domain conserved among p47phox, p40phox, Bem1p and various other signaling proteins (Sumimoto et al., 1997; Ago et al., 1999). It is also called the PX domain (Ponting, 1996).
Fig. 2. Role for the PC motif in the association of Cdc24p with Bem1p. (A) Pinpointing the Bem1p-binding region on Cdc24p. The C-terminal 75 aa region, which was previously shown to bind to Bem1p (Peterson et al., 1994), was further deleted from either the N- or the C-terminus and examined for two-hybrid interaction with Bem1p. (B) The PC motifs from various proteins are aligned with a tentative consensus sequence shown on the top, in which hydrophobic residues are represented by #. ZIP is also known as p62, p60 or ORCA. (C) Effects of PC motif mutations examined by the yeast two-hybrid assay. A DNA-binding domain fusion plasmid pGBK-Bem1p-(472–551) was co-transformed with an activation domain fusion plasmid pGADg-Cdc24p-(780–854) (WT) or pGADg-Cdc24p-(780–854, D824K/D831R) (mt) into PJ69-2A (Clontech) and SFY526 (Clontech). Following selection on SC-Trp-Leu plates, PJ69-2A transformants were tested for Ade-, His-independent growth (top) and SFY526 cells were examined for β-galactosidase activity by the filter assay using X-gal as the substrate (bottom). (D) Effects of PC motif mutations examined by in vitro pull-down assays. The purified GST–Bem1p-(464–551) fusion protein was incubated with purified MBP–Cdc24p-(780–854) (WT) or MBP–Cdc24p-(780–854, D824K/D831R) (mt). Subsequently, proteins were precipitated with glutathione–Sepharose (left) or amylose resin (right), and were resolved on SDS–PAGE followed by Coomassie Brilliant Blue staining.
It is interesting that the pinpointed Bem1p-binding region of Cdc24p contains a PC (Phox and Cdc) motif, a characteristic sequence found in a variety of signaling proteins from yeast to human (Sumimoto et al., 1997; Nakamura et al., 1998) (Figure 2B). In addition to budding yeast Cdc24p (Miyamoto et al., 1987), the motif is found in fission yeast scd1 (a homolog of Cdc24p) (Chang et al., 1994), mammalian p40phox (an SH3 domain-containing protein in the NADPH oxidase complex) (Wientjes et al., 1993; Mizuki et al., 1998), MEK5 (a MAP kinase kinase implicated in epidermal growth factor-induced cell proliferation) (English et al., 1995; Zhou et al., 1995) and ZIP (a protein linking the ζ isoform of protein kinase C to RIP and/or potassium channels (Puls et al., 1997; Gong et al., 1999; Sanz et al., 1999) (Figure 2B). The ZIP is also known as p62, p60 or ORCA, a protein associated with the tyrosine kinase Lck, the cytokine receptor EBI3 or the orphan nuclear receptor COUP-TFII, respectively (Devergne et al., 1996; Joung et al., 1996; Marcus et al., 1996). While others also recognized this evolutionarily conserved sequence and termed it OPR (Ponting, 1996), no clues had been available for their roles until we experimentally demonstrated that the one in p40phox is required for interaction with p67phox, another SH3 domain-containing protein in the oxidase complex (Nakamura et al., 1998).
These results prompted us to examine whether the PC motif of Cdc24p is involved in interaction with Bem1p. We prepared a construct bearing mutations in the conserved residues of the motif. Both two-hybrid analysis (Figure 2C) and an in vitro binding assay using purified recombinant proteins (Figure 2D) showed that the mutant Cdc24p fails to bind to Bem1p, indicating that the PC motif is required for Cdc24p to interact directly with Bem1p.
Binding partners for PC motif constitute PB1 domain
The PC motif plays a key role in the two interactions, one between Cdc24p and Bem1p and the other between p40phox and p67phox. Thus, the proteins recognizing the PC motif may share some common structural features. To clarify the molecular nature of binding partners for PC motif-containing regions (PCCRs) through sequence comparison, we pinpointed the region of p67phox that can recognize the PC motif to bind to p40phox. For this, we constructed a series of deletion mutants for the region between the two SH3 domains of p67phox, which had been shown to interact with p40phox (Nakamura et al., 1998), and tested them for binding to p40phox by the two-hybrid assay as well as an in vitro pull-down binding assay (Figure 3). Consequently, the minimum essential region for binding to p40phox was mapped to amino acid residues 345–427 of p67phox (Figure 3).
Fig. 3. Pinpointing the p67phox domain that interacts with the PCCR of p40phox. The inter-SH3 region of p67phox, which had been shown to bind to the PCCR of p40phox (Nakamura et al., 1998), was variously deleted from either the N- or the C-terminus and tested for binding to p40phox by two-hybrid assays (A) or in vitro pull-down assays (B).
Then, we aligned the two pinpointed regions as well as the corresponding regions of scd2 (fission yeast homolog of Bem1p) (Chang et al., 1994) and mouse p67phox (Mizuki et al., 1998) to reveal the modest homology, as shown in Figure 4A. Furthermore, we used the PSI-BLAST program, which is highly sensitive to detect weak but biologically relevant sequence similarities (Altschul et al., 1997), to search for additional proteins bearing homology with the pinpointed regions. Consequently, the proteins we identified were hypothetical ones of Arabidopsis as well as the N-terminus of the isoforms of atypical protein kinase C (PKCζ and PKCι/λ), a region of unknown function but of high conservation in this PKC subfamily (Figure 4). To test the functional relevance of the alignment, we examined the effect of substitution for the invariant lysine residue, which is solely conserved among the aligned sequences (Figure 4A). Both a two-hybrid system and an in vitro pull-down assay showed that the substitutions of alanine for K482 of Bem1p and K355 of p67phox completely abolish the interaction with their respective targets, indicating the functional importance of these residues as well as the significance of the alignment (Figure 4B and C). Therefore, we designate the homologous region the PB1 (Phox and Bem1) domain after its first discovery in p67phox and Bem1p.
Fig. 4. PB1 domain as the binding partners for PCCRs. (A) Alignment of PB1 domains. Pinpointed regions in Figures 1 and 3 are aligned with the corresponding regions of fission yeast scd2 and mouse p67phox. The lower portion shows additional PB1 domains revealed by PSI-BLAST search. They include members of the atypical protein kinase C family from various species and anonymous hypothetical proteins in the Arabidopsis genome (indicated by their DDBJ/EMBL/Genbank accession Nos). A tentative consensus sequence is shown on the top, where # stands for hydrophobic residues. (B) Effect of PB1 domain mutation in Bem1p. The conserved K482 residue of Bem1p was replaced with alanine, and tested for binding to Cdc24p using two-hybrid (upper) and in vitro pull-down assays (lower). (WT, wild type; mt, mutant). (C) Effect of PB1 domain mutation in p67phox. The conserved K355 residue was replaced with alanine, and tested for binding to p40phox using two-hybrid (upper) and in vitro pull-down assays (lower). (WT, wild type; mt, mutant).
We next examined whether the PB1 domain of Bem1p can interact with the PCCR of p40phox, but failed to detect the interaction in vitro (Figure 5) as well as by the two-hybrid assay (data not shown). Similarly, no binding was observed between the PB1 domain of p67phox and the PCCR of Cdc24p (Figure 5). Thus, each PB1 domain specifically recognizes its intrinsic target among PC motif-containing proteins, as is the case for protein–protein interactions via other modular domains. Such specificity may be defined by the structural elements in the flanking regions, because various chimeric constructs in which the PC motif of Cdc24p is replaced with that of p40phox failed to interact with p67phox (our unpublished observation). While the interaction between the PB1 domain and the PC motif is highly specific, cross-interactions were observed among orthologs; both PB1 domains of Bem1p and its fission yeast homolog scd2 bind not only the PCCR of Cdc24p but also that of its fission yeast homolog scd1 (Figure 5).
Fig. 5. Specificity of interactions between PB1 domains and PCCRs. PB1 domains from p67phox, PKCζ, Bem1p and scd2 were expressed and purified as GST fusions. On the other hand, PCCRs from p40phox, ZIP, Cdc24p and scd1 were expressed and purified as MBP fusions. All the possible combinations between these PB1 domains and PCCRs were examined for interactions by in vitro pull-down assays using glutathione–Sepharose.
The PB1 domain is essential for Bem1p function
To address the biological role for the PB1 domain of Bem1p, we replaced wild-type BEM1 with bem1K482A (see Materials and methods), which encodes a mutant Bem1p with the K482A substitution in the PB1 domain, a mutation leading to defective interaction with Cdc24p (Figure 4). The yeast cells bearing bem1K482A displayed temperature-sensitive growth and a bilateral mating defect (Figure 6). While one may argue that the mutation induces instability or insolubility of the protein, we assume it unlikely, because the mutant protein displays a two-hybrid interaction with Ste20p comparable to that of wild-type protein (see below; Table I). The deletion of BEM1 causes a temperature-sensitive growth defect (Chenevert et al., 1992; Lyons et al., 1996) or lethality (Leberer et al., 1996). The C-terminal portion of Bem1p is reported to be responsible for the bilateral mating defect (Leeuw et al., 1995) and, at least in the genetic background of the strain that we used, the deletion of the Bem1p C-terminal portion also confers the temperature-sensitive growth phenotype (Matsui et al., 1996). The similar phenotypes shared by bem1K482A and bem1Δ cells indicate that the PB1 domain plays a major role in Bem1p function presumably mediated by the interaction with Cdc24p.
Fig. 6. Effect of a PB1 domain mutation abolishing the binding of Bem1p to Cdc24p. Left panel: schematic representation of mutants and interactions. Middle panel: the wild-type cells (strain BYM2, sector 1) and bem1K482A cells (strain BYM1, sector 2) were streaked on an SC-Ura plate and incubated at 37°C for 2 days. Right panel: the bem1K482A MATa cells (strain BYM1, patch 1) and MATa wild-type cells (strain BYM2, patch 2) were replica-plated to lawns of bem1K482A MATα cells (strain BYM3) and incubated at 25°C overnight. The plate was then replica-plated to SC-Leu-Ura, on which only diploid cells were able to grow, and incubated for 3 days at 25°C. DH, Dbl-homology domain; PH, pleckstrin-homology domain.
Table I. Interaction between Bem1p and Ste20p.
| DBD fusion | AD fusion | Two-hybrid interaction |
|---|---|---|
| Bem1p (140–551) | Ste20p (1–497) | + |
| Bem1p (283–551) | Ste20p (1–497) | – |
| Bem1p (472–551) | Ste20p (1–497) | – |
| Bem1p (140–256) | Ste20p (1–497) | + |
| Bem1p (140–256) | Ste20p (69–497) | + |
| Bem1p (140–256) | Ste20p (196–497) | + |
| Bem1p (140–256) | Ste20p (338–497) | + |
| Bem1p (140–256) | Ste20p (1–29; 435–497) | + |
| Bem1p (140–256) | Ste20p (69–434) | – |
| Bem1p (140–256) | Ste20p (338–434) | – |
| Bem1p (140–327) | Ste20p (1–497) | – |
| Bem1p (140–463)-p67phox (335–427) | Ste20p (1–497) | + |
| Bem1p (140–551, K482A) | Ste20p (1–497) | + |
However, since Bem1p interacts with various other proteins involved in cell polarization and signal transduction for the mating process (Peterson et al., 1994; Leeuw et al., 1995; Zheng et al., 1995; Bender et al., 1996; Lyons et al., 1996; Matsui et al., 1996; Park et al., 1997; Butty et al., 1998), it is formally possible that the bem1K482A mutation causes these phenotypes by affecting interactions with proteins other than Cdc24p. In this context, it should be noted that both the second SH3 domain and C-terminal portion of Bem1p were reported to be necessary for its binding to the N-terminal half of Ste20p (Leeuw et al., 1995). Unexpectedly, we found that the second SH3 domain of Bem1p [amino acids (aa) 140–256] is necessary and sufficient to show a two-hybrid interaction with Ste20p and that the interaction is dependent on a proline-rich sequence on the latter protein (Table I). Furthermore, we showed that Bem1p (K482A) does interact with Ste20p (Table I). These results indicate that the phenotype induced by K482A mutation is not due to the defective interaction with Ste20p.
Domain swapping revealed the role for PB1–PC-mediated interaction between Bem1p and Cdc24p
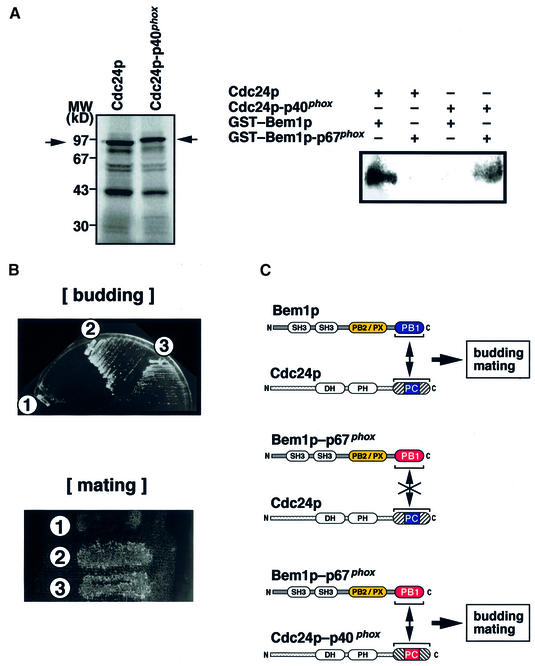
To demonstrate the role of the PB1 domain-mediated interaction with Cdc24p in Bem1p function unequivocally, we designed the following experiment using a ‘domain-swapping’ strategy. First, we constructed a chimeric gene, bem1-p67phox, encoding a mutant Bem1p whose PB1 domain is replaced with that of p67phox. As expected, this chimeric protein, in contrast to Bem1p, can not bind to Cdc24p (Figure 7A). The yeast cells bearing the bem1-p67phox gene, instead of the BEM1 gene, displayed temperature-sensitive growth and a mating defect (Figure 7B), as did the bem1K482A mutant (Figure 6).
Fig. 7. Domain swapping to prove the roles for the interaction between Bem1p and Cdc24p. (A) Interaction between Bem1p-p67 and Cdc24-p40. Radiolabeled Cdc24p and Cdc24p-p40phox (left) were incubated with GST–Bem1p or GST–Bem1p-p67phox. The proteins precipitated with glutathione–Sepharose were subjected to SDS–PAGE and visualized by autoradiography (right). (B) The defects caused by PB1 domain-swapped bem1 are suppressed by the expression of Cdc24p bearing compensatory alterations in the PC motif-containing region. Upper panel: the bem1-p67 cells (strain BYM4) carrying YEpCDC24 (sector 1) or YEpCDC24-p40 (sector 2) and wild-type cells (strain BYM2) carrying a control plasmid (pYO324, a high-copy-number-plasmid with the TRP1 marker; Ohya et al., 1991) (sector 3) were streaked on an SC-Trp-Ura plate and incubated at 37°C for 2 days. Lower panel: patches of the three yeast cells as above were replica-plated to lawns of bem1K482A MATa cells (strain BYM3) and incubated at 25°C overnight. The plate was then replica-plated to SC-Trp-Leu-Ura, on which only diploid cells were able to grow, and incubated for 3 days at 25°C. (C) Summary of the domain-swapping experiment.
Next, we examined whether the defects caused by the chimeric bem1-p67phox gene can be suppressed by expression of a mutant Cdc24p that harbors the PCCR of p40phox and thereby can interact with Bem1p-p67phox (Figure 7A). The defects were indeed suppressed in transformants expressing the modified Cdc24p bearing the entire C-terminal portion of p40phox (aa 234–339) in place of its authentic PCCR; the yeast cells can grow at 37°C and mate as efficiently as their parental control strain (Figure 7B). In contrast, wild-type Cdc24p, incapable of binding to the chimeric Bem1p-p67phox in vitro (Figure 7A), failed to suppress these defects in vivo (Figure 7B).
Thus, the defects caused by the alteration of the PB1 domain of Bem1p were suppressed only if the interaction with Cdc24p is restored by the compensatory alterations in the PCCR of Cdc24p (Figure 7C). These results indicate that the PB1 domain and the PC motif function as mutually interacting modules to mediate the physical association of Bem1p and Cdc24p in vivo, and, moreover, that the interaction per se is crucial for Bem1p function.
Discussion
Bem1p is a protein harboring two SH3 domains and assumed to function as a scaffold for the proteins involved in cell polarity establishment, pheromone signaling cascade and cytoskeletal organization. Accordingly, it has been shown to interact, directly or indirectly, with a variety of signaling and cytoskeletal proteins such as actin, Boi1p, Boi2p, Bud1p/Rsr1p, Cdc24p, Cdc42p, Far1p, Ste5p and Ste20p (Peterson et al., 1994; Leeuw et al., 1995; Zheng et al., 1995; Bender et al., 1996; Lyons et al., 1996; Matsui et al., 1996; Park et al., 1997; Butty et al., 1998). However, the structural basis for these interactions has been poorly analyzed except for the second SH3 domain, which is responsible for the binding to Boi1p and Boi2p (Bender et al., 1996; Matsui et al., 1996). In this study, we show that the C-terminal end region of Bem1p interacts directly with the PCCR of Cdc24p (Figures 1 and 2) and shares a modest homology with a region of p67phox, which is minimally required for binding to the PCCR of p40phox (Figure 3). Based on these findings, here we identify a novel module, termed PB1 domain, which recognizes the PC motif to mediate specific protein–protein interactions (Figures 4 and 5). The identity as a novel domain is confirmed by determination of the three-dimensional structure of the PB1 domain of Bem1p; it has a compact structure containing two α-helices and a mixed β-sheet composed of four strands (Terasawa et al., 2001).
Identification of the structural units mediating the interaction between Bem1p and Cdc24p has led us to reveal its role in polarity establishment of the budding yeast. We first introduced a mutation leading to substitution of alanine with K482 of Bem1p, the solely conserved residue among the PB1 domains (Figures 4 and 6). The substitution results not only in abolished interaction with the PCCR of Cdc24p (Figure 4) but also in impaired budding and mating (Figure 6). Direct involvement of K482 in contact with PCCR of Cdc24p is indicated by the observation that PCCR binding induces a strong chemical shift change at this residue (Terasawa et al., 2001). In addition, the K482A substitution would not be expected to disturb the correct integrity of the PB1 domain, since the side chain of K482 is exposed to solvent and does not interact with other amino acid residues of this domain (Terasawa et al., 2001). Taken together, the experiments using this site-directed mutant indicate that the interaction between Bem1p and Cdc24p plays a crucial role in polarity establishment. Furthermore, identification of two mutually interacting, but not cross-reacting, PB1–PC pairs (Figure 5) enabled a unique domain-swapping experiment to demonstrate unequivocally the functional importance of the interaction; defects in both budding and mating caused by the swapping of the PB1 domain of Bem1p with that of p67phox, which abolishes its interaction with Cdc24p, are suppressed by a mutant Cdc24p harboring the PC motif-containing region of p40phox, which restores the interaction with the altered Bem1p (Figure 7). Thus, the data obtained here imply that a complex containing the two proteins Cdc24p and Bem1p is a prerequisite for both budding and mating; Cdc24p can be targeted towards the budding site or mating projection through the interaction with Bud1p/Rsr1p (Zheng et al., 1995; Park et al., 1997) or Ste4p (Nern and Arkowitz, 1998, 1999; Shimada et al., 2000), respectively, but, without association with Bem1p, fails to reorganize the cytoskeleton and hence to establish polarity.
One may argue a potential role for the PB1 domain in association of Bem1p with Ste20p, based on a previous report showing that Bem1p-s1 (aa 1–298) and Bem1p-s2 (aa 1–518), each lacking the PB1 domain, fail to associate with Ste20p in vivo (Leeuw et al., 1995). In contrast to this, we show here that the PB1 domain per se (aa 464–551) does not bind to Ste20p, and the second SH3 domain of Bem1p (aa 140–256) is necessary and sufficient to interact with Ste20p (Table I). The discrepancy may be due to the possibility that the C-terminal truncations that occurred in Bem1p-s1 and Bem1p-s2 somehow affect the overall conformation of the proteins, which presumably would prevent the second SH3 domain from interacting efficiently with Ste20p. The possibility is likely to be supported by the fact that both we and others failed to detect the interaction between Ste20p and another construct for the SH3 domain with a different C-terminal truncation point (aa 140–327) (Leeuw et al., 1995; Table I). On the other hand, Bem1p (K482A) and Bem1p-p67phox are both capable of interacting with Ste20p (Table I). It should be noted that these proteins retain the identical domain architecture to that of native Bem1p (i.e. SH3-SH3-PB2/PX-PB1), which is likely to render the second SH3 domain in a state capable of binding to Ste20p. Taken together, Bem1p appears to interact with Ste20p via the second SH3 domain but not via the PB1 domain.
In addition to the clearly demonstrated case of Bem1p and Cdc24p, other pairs of PB1 domain and PC motif also seem to mediate functional protein–protein interactions in a variety of biological processes. The association of p67phox and p40phox (Nakamura et al., 1998; Figure 3), also mediated by these modules, is implicated in activation of the superoxide-producing NADPH oxidase (Tsunawaki et al., 1996). The PB1 domain of PKCζ is necessary and sufficient to interact with the PCCR of the adaptor protein ZIP (Figure 5); it is conceivable that this interaction may be required for recruitment of the kinase to a signaling complex containing RIP or potassium channels (Gong et al., 1999; Sanz et al., 1999). It is also interesting to note that two isoforms of MEK5, one bearing and the other lacking the PC motif, show different intracellular localization (English et al., 1995). This PC motif might play a role in differential cellular localization of MEK5 isoforms by mediating interaction with an as yet unidentified PB1 domain-containing protein. Thus, the PB1 domain and the PC motif may participate in various aspects of cellular regulation as a mutually interacting unit to mediate functional protein–protein interactions.
Currently increasing evidence has revealed that a variety of cellular proteins function effectively in multiprotein complexes formed via specific interactions. In this study, we focused our attention on the interaction between the scaffold protein Bem1p and Cdc24p in the protein complex for cell polarity establishment of the budding yeast. To reveal the biological role of this interaction, we took two strategies: one is the introduction of an amino acid substitution that leads to a defective interaction without affecting the protein integrity, and the other is domain swapping. The latter strategy is quite useful, because a suppressor mutant can be readily designed to test the role of the interaction as demonstrated in this study. It can also be used in the case with a difficulty in generating a functionally defective protein with the correct structural integrity, which is often lost by amino acid substitutions or deletions. The domain-swapping strategy would thus provide a general and robust method to dissect a focused interaction from the other ones in a multiprotein complex and decipher its biological significance, when the interaction is mediated by modular domains, as is often the case in most signaling proteins.
Materials and methods
Plasmid construction
DNA fragments encoding various portions of Cdc24p and Bem1p were amplified from yeast genomic DNA, and cloned into pGEX2T (Pharmacia), pMALc2g (a modified version of pMALc2; NEB), pGBK [a derivative of pGBT9 (Clontech) bearing the Kanr marker] and pGADg (a modified version of pGAD424; Clontech) plasmids as described previously (Ito et al., 1996; Nakamura et al., 1998; Ago et al., 1999). Site-directed mutagenesis was performed with an inverse-PCR-mediated procedure.
A K482A mutation was introduced into YIpUBEM1, a plasmid bearing BEM1 flanked by URA3 (Matsui et al., 1996), by means of site-directed mutagenesis to obtain YIpUBEM1K482A. The PB1 domain-encoding region of YIpUBEM1 was replaced with a PCR fragment encoding the PB1 domain of p67phox to obtain YIpUBEM1-p67.
The fragment encoding the C-terminal 75 aa of Cdc24p was replaced on YEpCDC24 (Miyamoto et al., 1987) with DNA fragments encoding the C-terminal portion of p40phox (234–339) preceded by a (Gly)5-linker to obtain YEpCDC24-p40. Similar constructs were also prepared on pCITE4a (Novagen) for in vitro transcription and translation.
All the plasmids were subjected to DNA sequencing prior to use for confirmation of their precise construction.
Two-hybrid assays
For the yeast two-hybrid screening, we used a yeast genomic library and PJ69-4A as described (James et al., 1996). For individual two-hybrid assays, we co-transformed PJ69-2A and SFY526 strains (Clontech) with various combinations of pGBK/pGBTg and pGADg plasmids. The activation of ADE2 and HIS3 reporter genes was examined by Ade-, His-independent growth of PJ69-2A transformants, whereas the activation of lacZ was visualized by the filter assay in SFY526 transformants.
In vitro binding assays
Purification of the recombinant proteins and in vitro pull-down assays using glutathione–Sepharose (Pharmacia) for glutathione S-transferase (GST) fusion proteins and amylose resin (NEB) for maltose-binding protein (MBP) fusion proteins were performed as described previously (Nakamura et al., 1998; Ago et al., 1999).
Radiolabeled Cdc24p and Cdc24p-p40phox were expressed using an in vitro transcription and translation system (Ota et al., 1998), and tested for the binding to GST–Bem1p or GST–Bem1p-p67phox proteins in vitro.
Construction of yeast bem1 mutants
The yeast strains used were BYM1 (MATa bem1K482A:URA3 ura3 trp1 leu2 his3 ade2), BYM2 (MATa BEM1:URA3 ura3 trp1 leu2 his3 ade2), BYM3 (MATα bem1K482A:LEU2 ura3 trp1 leu2 his3 ade2) and BYM4 (MATa bem1-p67:URA3 ura3 trp1 leu2 his3 ade2). BYM1 was constructed by transforming wild-type MATa cells, a segregant of W303 cells (Sutton et al., 1991), with YIpUBEM1K482A to replace the BEM1 locus with bem1K482A. BYM2 was constructed as above but using YIpUBEM1, a plasmid for replacing the BEM1 locus with BEM1 flanked by URA3 marker, for transformation. BYM3 was constructed by transformation of MATα cells (a MATα segregant of W303 cells) with YIpUBEM1K482A to replace the BEM1 locus with bem1K482A followed by the replacement of URA3 with LEU2 using a ura3-disruption plasmid. BYM4 was constructed by transforming wild-type MATa cells with YIpUBEM1-p67phox to replace the BEM1 locus with bem1-p67phox.
Acknowledgments
Acknowledgements
We are grateful to Profs Y.Sakaki and A.Toh-e (University of Tokyo) for their support and encouragement, and Prof. K.Mihara (Kyushu University) for rabbit reticulocyte lysate for in vitro translation and technical advice. We also thank Dr T.Taylor (RIKEN) for his helpful comments on the manuscript, and N.Aoyama, R.Ozawa (University of Tokyo) and Y.Kage (Kyushu University) for their excellent technical assistance. This work was partly supported by grants from the Ministry of Education, Science, Sports and Culture, Japan, and Science and Technology Agency, Japan.
References
- Ago T., Nunoi,H., Ito,T. and Sumimoto,H. (1999) Mechanism for phosphorylation-induced activation of the phagocyte NADPH oxidase protein p47phox: triple replacement of serines 303, 304, 328 with aspartates disrupts the SH3 domain-mediated intramolecular interaction in p47phox, thereby activating the oxidase. J. Biol. Chem., 274, 33644–33653. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender L., Lo,H.S., Lee,H., Kokojan,V., Peterson,J. and Bender,A. (1996) Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in yeast. J. Cell Biol., 133, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butty A.-C., Pryciak,P.M., Huang,L.S., Herskowitz,I. and Peter,M. (1998) The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science, 282, 1511–1516. [DOI] [PubMed] [Google Scholar]
- Chang E.C., Barr,M., Wang,Y., Jung,V., Xu,H.-P. and Wigler,M.H. (1994) Cooperative interaction of S.pombe proteins required for mating and morphogenesis. Cell, 79, 131–141. [DOI] [PubMed] [Google Scholar]
- Chant J. (1999) Cell polarity in yeast. Annu. Rev. Cell Dev. Biol., 15, 365–391. [DOI] [PubMed] [Google Scholar]
- Chenevert J., Corrado,K., Bender,A., Pringle,J.R. and Herskowitz,I. (1992) A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature, 356, 77–79. [DOI] [PubMed] [Google Scholar]
- Devergne O., Hummel,M., Koeppen,H., Le Beau,M.M., Nathanson,E.C., Kieff,E. and Birkenbach,M. (1996) A novel interleukin-12 p40-related protein induced by latent Epstein–Barr virus infection in B lymphocytes. J. Virol., 70, 1143–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J.M., Vanderbilt,C.A., Xu,S., Marcus,S. and Cobb,M.H. (1995) Isolation of MEK5 and differential expression of alternatively spliced forms. J. Biol. Chem., 270, 28897–28902. [DOI] [PubMed] [Google Scholar]
- Gong J., Xu,J., Bezanilla,M., van Huizen,R., Derin,R. and Li,M. (1999) Differential stimulation of PKC phosphorylation of potassium channels by ZIP1 and ZIP2. Science, 285, 1565–1569. [DOI] [PubMed] [Google Scholar]
- Ito T., Nakamura,R., Sumimoto,H., Takeshige,K. and Sakaki,Y. (1996) An SH3 domain-mediated interaction between the phagocyte NADPH oxidase factors p40phox and p47phox. FEBS Lett., 385, 229–232. [DOI] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung I., Strominger,J.L. and Shin,J. (1996) Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc. Natl Acad. Sci. USA, 93, 5991–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Chenevert,J., Leeuw,T., Harcus,D., Herskowitz,I. and Thomas,D.Y. (1996) Genetic interactions indicate a role for Mdg1p and the SH3 domain protein Bem1p in linking the G-protein mediated yeast pheromone signalling pathway to regulators of cell polarity. Mol. Gen. Genet., 252, 608–621. [DOI] [PubMed] [Google Scholar]
- Leeuw T., Fourest-Lieuvin,A., Wu,C., Chenevert,J., Clark,K., Whiteway,M., Thomas,D.Y. and Leberer,E. (1995) Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science, 270, 1210–1213. [DOI] [PubMed] [Google Scholar]
- Lyons D.M., Mahanty,S.K., Choi,K.-Y., Manandhar,M. and Elion,E.A. (1996) The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 4095–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K. and Snyder,M. (1998) Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol., 52, 687–744. [DOI] [PubMed] [Google Scholar]
- Marcus S.L., Winrow,C.J., Capone,J.P. and Rachubinski,R.A. (1996) A p56lck ligand serves as a coactivator of an orphan nuclear hormone receptor. J. Biol. Chem., 271, 27197–27200. [DOI] [PubMed] [Google Scholar]
- Matsui Y., Matsui,R., Akada,R. and Toh-e,A. (1996) Yeast src homology region 3 domain-binding proteins involved in bud formation. J. Cell Biol., 133, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Ohya,Y., Ohsumi,Y. and Anraku,Y. (1987) Nucleotide sequence of the CLS4 (CDC24) gene of Saccharomyces cerevisiae. Gene, 54, 125–132. [DOI] [PubMed] [Google Scholar]
- Mizuki K. et al. (1998) Functional modules and expression of mouse p40phox and p67phox SH3-domain-containing proteins involved in the phagocyte NADPH oxidase complex. Eur. J. Biochem., 251, 573–582. [DOI] [PubMed] [Google Scholar]
- Nakamura R., Sumimoto,H., Mizuki,K., Hata,K., Ago,T., Kitajima,S., Takeshige,K., Sakaki,Y. and Ito,T. (1998) The PC motif: a novel and evolutionarily conserved sequence involved in interaction between p40phox and p67phox, SH3 domain-containing cytosolic factors of the phagocyte NADPH oxidase. Eur. J. Biochem., 251, 583–589. [DOI] [PubMed] [Google Scholar]
- Nern A. and Arkowitz,R.A. (1998) A GTP-exchange factor required for cell orientation. Nature, 391, 195–198. [DOI] [PubMed] [Google Scholar]
- Nern A. and Arkowitz,R.A. (1999) A Cdc24p–Far1p–Gβγ protein complex required for yeast orientation during mating. J. Cell Biol., 144, 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y., Goebl,M., Goodman,L.E., Petersen-Bjorn,S., Friesen,J.D., Tamanoi,F. and Anraku,Y. (1991) Yeast CAL1 is a structural and functional homologue to the DPR1 (RAM) gene involved in ras processing. J. Biol. Chem., 266, 12356–12360. [PubMed] [Google Scholar]
- O’Shea E.K. and Herskowitz,I. (2000) The ins and outs of cell-polarity decisions. Nature Cell Biol., 2, E39–E41. [DOI] [PubMed] [Google Scholar]
- Ota K., Sakaguchi,M., von Heijne,G., Hamasaki,N. and Mihara,K. (1998) Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol. Cell, 2, 495–503. [DOI] [PubMed] [Google Scholar]
- Park H.O., Bi,E., Pringle,J.R. and Herskowitz,I. (1997) Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl Acad. Sci. USA, 94, 4463–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. (1995) Protein modules and signalling networks. Nature, 373, 573–580. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Nash,P. (2000) Protein–protein interactions define specificity in signal transduction. Genes Dev., 14, 1027–1047. [PubMed] [Google Scholar]
- Peterson J., Zheng,Y., Bender,L., Myers,A., Cerione,R. and Bender,A. (1994) Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J. Cell Biol., 127, 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P. (1996) Novel domains in NADPH oxidase subunits, sorting nexins and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci., 5, 2353–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls A., Schmidt,S., Grawe,F. and Stabel,S. (1997) Interaction of protein kinase C zeta with ZIP, a novel protein kinase C-binding protein. Proc. Natl Acad. Sci. USA, 94, 6191–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Sanchez,P., Lallena,M.J., Diaz-Meco,M.T. and Moscat,J. (1999) The interaction of p62 with RIP links the atypical PKCs to NF-κB activation. EMBO J., 18, 3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y., Gulli,M.-P. and Peter,M. (2000) Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nature Cell Biol., 2, 117–124. [DOI] [PubMed] [Google Scholar]
- Sumimoto H., Ito,T., Hata,K., Mizuki,K., Nakamura,R., Kage,Y., Nakamura,M., Sakaki,Y. and Takeshige,K. (1997) Membrane translocation of cytosolic factors in activation of the phagocyte NADPH oxidase: role of protein–protein interactions. In Hamasaki,N. and Mihara,K. (eds), Membrane Proteins: Structure, Function and Expression Control. S.Karger AG, Basel, Switzerland, pp. 235–245.
- Sutton A., Immanuel,D. and Arndt,K.T. (1991) The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol., 11, 2133–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa H., Noda,Y., Ito,T., Hatanaka,H., Ichikawa,S., Ogura,K., Sumimoto,H. and Inagaki,F. (2001) Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif. EMBO J., 20, 3947–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunawaki S., Kagara,S., Yoshikawa,K., Yoshida,L.S., Kuratsuji,T. and Namiki,H. (1996) Involvement of p40phox in activation of phagocyte NADPH oxidase through association of its carboxyl-terminal, but not its amino-terminal, with p67phox. J. Exp. Med., 184, 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes F.B., Hsuan,J.J., Totty,N.F. and Segal,A.W. (1993) p40phox, a third cytosolic component of the activation complex of the NADPH oxidase to contain src homology 3 domains. Biochem. J., 296, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Bender,A. and Cerione,R.A. (1995) Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J. Biol. Chem., 270, 626–630. [DOI] [PubMed] [Google Scholar]
- Zhou G., Bao,Z.Q. and Dixon,J.E. (1995) Components of a new human protein kinase signal transduction pathway. J. Biol. Chem., 270, 12665–12669. [DOI] [PubMed] [Google Scholar]