Abstract
Picornavirus proteases cleave translation initiation factor eIF4G into a C-terminal two-thirds fragment (hereafter named p100) and an N-terminal one-third fragment, which interacts with the cap-binding factor eIF4E. As the timing of this cleavage correlates broadly with the shut-off of host cell protein synthesis in infected cells, a very widespread presumption has been that p100 cannot support capped mRNA translation. Through the use of an eIF4G-depleted reticulocyte lysate system, we show that this presumption is incorrect. Moreover, recombinant p100 can also reverse the inhibition of capped mRNA translation caused either by m7GpppG cap analogue, by 4E-BP1, which sequesters eIF4E and thus blocks its association with eIF4G, or by cleavage of endogenous eIF4G by picornavirus proteases. The concentration of p100 required for maximum translation of capped mRNAs is ∼4-fold higher than the endogenous eIF4G concentration in reticulocyte lysates. Our results imply that picornavirus-induced shut-off is not due to an intrinsic inability of p100 to support capped mRNA translation, but to the viral RNA outcompeting host cell mRNA for the limiting concentration of p100.
Keywords: cap analogue/4E-BP1/eIF4G cleavage/host cell shut-off/poliovirus
Introduction
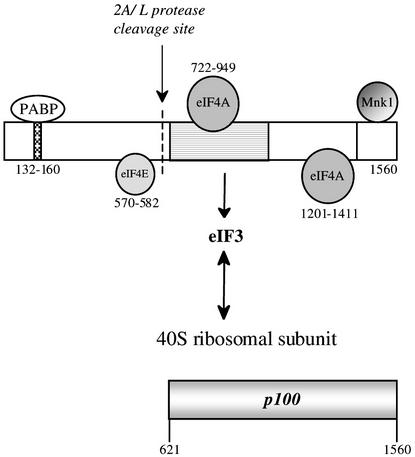
Of the numerous eukaryotic translation initiation factors, the one that plays the key role in promoting small ribosomal subunit binding to the mRNA is the eIF4F holoenzyme complex. The core complex consists of: (i) eIF4A, which has RNA helicase activity; (ii) eIF4E, which is the only factor with direct cap binding activity; and (iii) eIF4G, which appears to act as a scaffold, and in fact binds several other proteins (for a recent review see Gingras et al., 1999). Actually, there are two known species of eIF4G in mammalian cells, encoded by different genes (Gradi et al., 1998a). Here we focus on the more abundant form, eIF4GI (referred to simply as eIF4G), which is 1560 amino acid residues long according to the recently revised sequence (Gradi et al., 1998a). The first insights into the domain organization of eIF4G came from the observation that it is cleaved by picornavirus-encoded proteases into an N-terminal one-third fragment and a C-terminal two-thirds fragment (Lamphear et al., 1995), which is designated p100 throughout this report (Figure 1). Conceptually, eIF4G is also often considered as having three domains: the same N-terminal one-third fragment as defined by the site of cleavage by viral proteases, a central one-third domain and a C-terminal domain.
Fig. 1. Domain structure of eIF4G(I) and sites of interaction of other proteins with eIF4G. eIF4GI is depicted as a rectangle. The amino acid numbering of the sites on eIF4G for interaction with PABP and eIF4E are from Gingras et al. (1999), for eIF4A binding to the central domain from Lomakin et al. (2000), and for eIF4A and Mnk-1 interaction near the eIF4G C-terminus from Morino et al. (2000). Note, however, that the upstream eIF4A binding site is defined as amino acids 672–970 by Morino et al. (2000) and 672–876 by Korneeva et al. (2000, 2001). The eIF3 interaction domain is shown as a stippled rectangle, and has been defined as amino acids 697–1076 by Lomakin et al. (2000), 672–1065 by Morino et al. (2000) but 975–1065 by Korneeva et al. (2000). A single arrow is used to denote the sites at which eIF4G is cleaved by enterovirus 2A protease and foot-and-mouth disease virus (FMDV) L-protease; the two cleavage sites are actually 7 amino acid residues apart. Below is depicted the p100 fragment of eIF4GI used in this study.
The N-terminal domain has the binding site for eIF4E, the cap-binding factor (Lamphear et al., 1995; Gingras et al., 1999), and also interacts with poly(A) binding protein (PABP) (Imataka et al., 1998). The C-terminal domain has a binding site for eIF4A (Lamphear et al., 1995; Morino et al., 2000; Korneeva et al., 2001) and also interacts with Mnk-1, a protein kinase specific for eIF4E (Figure 1). The all-important central domain has another eIF4A binding site (Imataka and Sonenberg, 1997), and also interacts with eIF3 (Lamphear et al., 1995). The binding of these two factors to the central domain exhibits some cooperativity (Korneeva et al., 2000). eIF3 also binds with high affinity to 40S ribosomal subunits (Hershey and Merrick, 2000), and since this binding appears not to be mutually exclusive with the eIF3–eIF4G interaction, in principle an eIF4G–eIF3–40S subunit tripartite interaction relay can occur and this is considered to be the key to 40S subunit delivery to the mRNA. With a capped mRNA translated by the scanning ribosome mechanism, the eIF4F complex is thought to bind to the cap via its eIF4E subunit, and to deliver the 40S subunit to a cap-proximal position on the mRNA via this eIF4G–eIF3–40S subunit interaction relay. Cleavage of the eIF4G by picornaviral proteases separates the cap-binding function, the eIF4E associated with the N-terminal one-third fragment of eIF4G, from the eIF4A helicase and the potential 40S subunit delivery system of the central domain, and this is presumed to be the cause of the inhibition of translation of capped cellular mRNAs in infected cells (Lamphear et al., 1995). Picornavirus RNA translation is not compromised by this cleavage event, for although it can be supported by the complete eIF4F complex (as occurs in the early stages of infection before eIF4G has been cleaved), the central domain of eIF4G appears to be sufficient and, in the one case examined in detail, encephalomyocarditis virus (EMCV) RNA, p100 binds directly to the viral RNA near the site of internal ribosome entry (Pestova et al., 1996a, b; Kolupaeva et al., 1998; Lomakin et al., 2000). The central domain of eIF4G, or the p100 fragment, can also support the translation of uncapped versions of normally capped mRNAs, and is actually much more efficient than the eIF4F holoenzyme complex (Ohlmann et al., 1995, 1996; De Gregorio et al., 1998). In contrast, the fact that the time at which cleavage of eIF4G is complete in infected cells correlates broadly with the timing of shut-off of host cell mRNA translation has led to the widespread presumption that p100 is not able to support translation of capped mRNAs by the scanning mechanism, and that the translation of such RNAs needs eIF4E, plus a sufficient length of eIF4G to include the eIF4E interaction site, as well as the central domain of eIF4G (Morino et al., 2000). We show here that, surprisingly, this is not the case, and that efficient translation of capped mRNA initiated at the correct 5′-proximal AUG can be effected by p100.
Results
Development of an eIF4G-depleted reticulocyte lysate system
Our initial aim was to prepare a rabbit reticulocyte lysate sufficiently depleted of eIF4G such that translation would be dependent on added eIF4G. Several affinity column depletion strategies were tried but found wanting, either because depletion was ineffective or because the recovery of EMCV IRES (‘internal ribosome entry site/segment’) activity on p100 add-back was poor. Success was achieved using a method adapted from one developed for depletion, but not tested in add-back assays, by Stassinopoulos and Belsham (2001); a poly(A)-tailed derivative of the high affinity binding site for eIF4G on the EMCV IRES, the J-K domain (Duke et al., 1992; Pestova et al., 1996b; Kolupaeva et al., 1998; Lomakin et al., 2000), bound to oligo-dT magnetic beads.
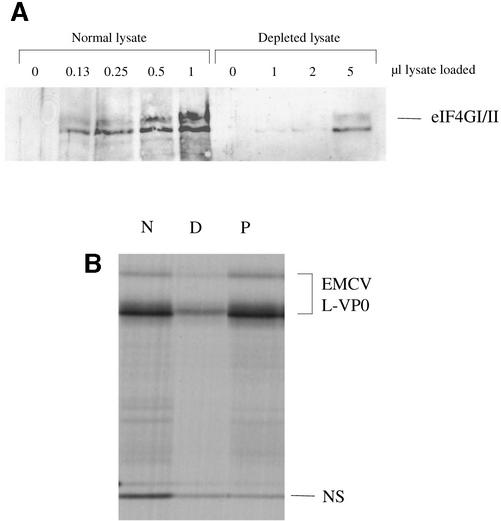
A western blot, using an antibody raised against a peptide epitope in the C-terminal one-third domain of eIF4GI (which reacts equally with human and rabbit eIF4GI, and has been observed to cross-react with eIF4GII), shows that almost complete depletion (∼95%) of eIF4G was achieved (Figure 2A); the amount of residual eIF4G in 5 µl of depleted lysate corresponds to the signal given by 0.25 µl of the parent lysate. Depletion very severely compromised translation of both cistrons of a dicistronic mRNA with the EMCV IRES, but complete rescue of translation of the IRES-dependent cistron occurred on add-back of p100 (Figure 2B). (As is shown later, the lack of rescue of upstream cistron translation is due to competition by the EMCV IRES for functional interaction with p100.)
Fig. 2. Affinity depletion of eIF4G from reticulocyte lysates. (A) The indicated volumes of parent (normal) lysate and eIF4G-depleted lysate were subjected to SDS–PAGE and western blotting with anti-eIF4G antiserum and peroxidase-conjugated secondary antibody. Detection was by ECL. (B) Capped dicistronic mRNA with an upstream cistron coding for influenza virus NS1, an EMCV IRES, and EMCV coding sequences for L-VP0 (∼55 kDa) as downstream cistron, was translated at a final concentration of 25 µg/ml in the parent (non-depleted) lysate (N), in the depleted lysate (D) or in depleted lysate supplemented with 20 µg/ml recombinant p100 (P). Radiolabelled translation products were separated by gel electrophoresis, and the resulting autoradiograph is shown. Of the two L-VP0 products, the major (smaller) one results from translation of transcripts of the cDNA template linearized at the StuI site, and the minor (larger) protein is the translation product resulting from incomplete linearization: it is initiated at the same site as the smaller product, and is terminated at an in-frame termination codon that lies just within the vector sequences and beyond the StuI site.
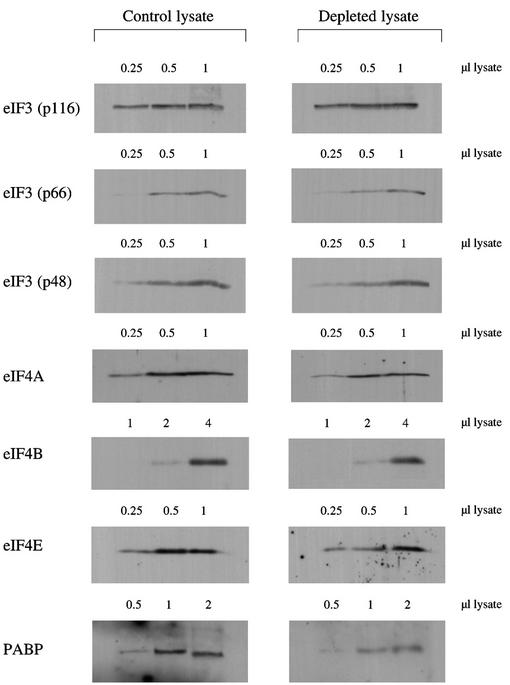
Western blotting was also used to assess the degree of depletion of some other factors. A representative set of blots is shown in Figure 3, although not surprisingly there was some batch to batch variation. A consistent observation was that remarkably little eIF3 was removed, typically <10% and never >20%. There was greater depletion of eIF4A, in the range 30–40%, similar to the results of Stassinopoulos and Belsham (2001), but even so the residual eIF4A will still be in large excess over the added p100 (Rau et al., 1996). Although some depletion of eIF4B was expected since this binds to the J-K domain of the aphthovirus IRES, which is very similar to the EMCV IRES (Meyer et al., 1995), in fact eIF4B appeared to be depleted to a lesser extent than eIF4A (Figure 3). Depletion of eIF4E seldom exceeded ∼30% and was even less in most batches, consistent with previous results showing that only ∼25% of the eIF4E in a reticulocyte lysate is actually associated with eIF4G (Rau et al., 1996). Of all the factors tested, PABP was the most severely depleted (typically 50–60%).
Fig. 3. Western blotting assessment of degree of depletion of other factors. The indicated volumes (µl) of parent lysate and eIF4G-depleted lysate were resolved by SDS–PAGE, and after blotting the gels, the blots were probed with antisera against eIF3, eIF4A, eIF4B, eIF4E and PABP, as indicated. Horseradish peroxidase-conjugated secondary antibodies were used (against goat IgGs in the case of the eIF3 and eIF4B blots, and rabbit IgG in all other cases), and detection was by ECL.
However, functional assays provide a much more meaningful criterion for the status of the eIF4G-depleted system than western blotting. In order to provide a stringent test for recovery of translation on addition of p100, we have generally used mRNA at near-saturating concentrations, yet, as shown below, p100 effected very good recovery of translation of several different mRNAs when each was tested separately in the absence of any competitor mRNA. The only exceptions were RNAs with the poliovirus or rhinovirus IRES, which could not be rescued by addition of p100 together with eIF4A, 4B, 4E and PABP, tested either singly or in combinations. Evidently the procedure depletes the lysate of an additional, and as yet unidentified factor, which is uniquely required for the function of these type I picornavirus IRESs.
Since good recovery was observed with all other mRNAs despite the high RNA concentration, it follows that other components of the translation machinery cannot have been depleted to the extent that any of them has become severely limiting. We find that in the absence of added p100, no stimulation of translation of any mRNA species occurred on addition of eIF4A, 4B, 4E or PABP, either singly or in combination, to the depleted system. With some batches of depleted lysate, the stimulatory effect of p100 on translation of the capped mRNAs described below (especially globin mRNA) was modestly enhanced by co-addition of eIF4A plus eIF4B, but eIF4E or PABP addition had no consistent influence, either alone or together with eIF4A, 4B and p100 (data not shown). We conclude that this depleted lysate is a valid functional assay system for testing the activity of eIF4G fragments.
Translation of capped mRNA in the eIF4G-depleted system
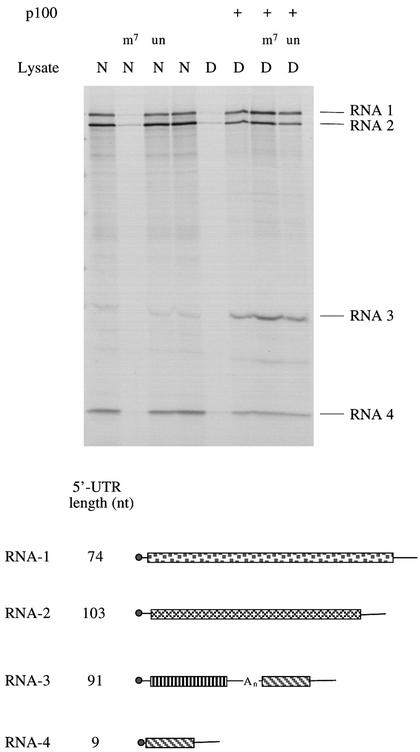
We then used this eIF4G-depleted system to study the translation of capped dicistronic transcripts with a hepatitis A virus (HAV) IRES, and were surprised that translation of the upstream (scanning-dependent) cistron was strongly stimulated by addition of p100 to the depleted lysate (Ali et al., 2001). Since p100 can markedly stimulate translation of uncapped mRNAs in normal (non-depleted) reticulocyte lysate (Ohlmann et al., 1995, 1996; De Gregorio et al., 1998), and because capping of in vitro transcripts is far from 100% efficient, in fact only ∼70% in our hands (Dasso and Jackson, 1989), our initial assumption was that this was actually a stimulation of translation of just the uncapped transcripts in the preparation. To test whether this presumption was correct it was obviously necessary to use an mRNA preparation that is 100% capped, and for this purpose we chose RNA prepared from Brome Mosaic Virus (BMV). As expected, the efficiency of BMV RNA translation in the depleted lysate was much lower than in the parent lysate, but, much to our surprise, addition of p100 to the depleted lysate stimulated the translation of all four BMV RNAs (Figure 4). (As shown later in Figure 6, this concentration of p100 has very little impact on the translation of BMV RNA in the parent, non-depleted lysate.) Remarkably, while BMV RNA translation in the parent lysate is highly sensitive to inhibition by m7GpppG cap analogue (but not the GpppG control), translation in a depleted lysate supplemented with p100 is completely resistant to such inhibition, and is actually slightly stimulated by cap analogue (Figure 4), most probably because the binding of residual eIF4E to the caps weakly antagonizes the ability of p100 to drive capped mRNA translation. Thus, this stimulation by p100 is quite independent of any eIF4E–cap interactions, and it appears that the RNA is being translated by a ‘cap-independent’ mechanism, despite the fact that all four BMV RNAs are capped.
Fig. 4. p100 can support the translation of capped BMV RNAs by a mechanism that is operationally cap independent. BMV RNA was translated at a final concentration of 20 µg/ml in either normal (parent) lysate (N) or eIF4G-depleted lysate (D), with the following additions where indicated: (m7), 0.4 mM m7GpppG cap analogue with 0.32 mM additional MgCl2; (un), 0.4 mM unmethylated GpppG with 0.32 mM additional MgCl2; (p100), 20 µg/ml (200 nM) recombinant p100. Radiolabelled translation products were separated by gel electrophoresis, and the resulting autoradiograph is shown. The products encoded by BMV RNA-1, RNA-2, RNA-3 and RNA-4 are indicated. The schematic diagram depicts the four BMV RNAs, approximately to scale, and the lengths of the 5′-UTRs (Ahlquist et al., 1981, 1984; Dasgupta and Kaesberg, 1982). Open reading frames are shown as rectangles, UTRs as lines. BMV RNA-3 is functionally monocistronic, but structurally dicistronic; the silent downstream open reading frame is identical to the ORF of RNA-4, and the intercistronic spacer has a 16–21 residue oligo(A) tract.
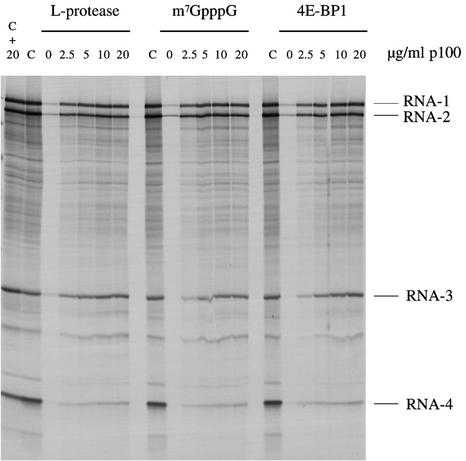
Fig. 6. p100 can reverse the inhibition of capped mRNA translation caused by either FMDV L-protease, m7GpppG cap analogue or 4E-BP1. BMV RNA was translated at a final concentration of 20 µg/ml in either control (untreated) lysate (C), control lysate supplemented with 20 µg/ml recombinant p100 (C + 20), or lysate subjected to one of the following regimes, and then supplemented with 0, 2.5, 5, 10 or 20 µg/ml recombinant p100 as indicated: pre-incubation for 10 min at 30°C with recombinant FMDV L-protease; supplementation with 0.4 mM m7GpppG (with 0.32 mM additional MgCl2); or pre-incubated for 10 min at 30°C with 10 µg/ml recombinant 4E-BP1. Radiolabelled translation products were separated by gel electrophoresis, and the resulting autoradiograph is shown.
What is rather surprising is that although translation initiation is operationally cap independent, it still shows a strong preference for the 5′-proximal AUG. There is remarkably little ‘noise’ of incomplete products initiated at internal sites, indeed hardly any increase in incomplete products over what is seen with the parent (non-depleted) lysate (Figure 4). Thus, the functional interaction of p100 with the mRNA must be such that it delivers the 40S ribosomal subunit to a cap-proximal position. [The lack of incomplete translation products appears to be variable between different mRNA species, since with other mRNAs there was a distinct increase in the yield of such products (W.Wood, I.K.Ali, L.McKendrick, S.J.Morley and R.J.Jackson, unpublished observations), although the overwhelming majority of initiation events were still at the 5′-proximal AUG.]
Also of interest is the fact that the relative yield of the four BMV RNA translation products is subtly different in the depleted lysate supplemented with p100 as compared with the parent lysate. Rescue of translation of RNA-3 is particularly efficient, with the product yield often higher than in the parent lysate, while rescue of RNA-4, which has a 5′-UTR of only 9 nt, is rather inefficient (Figure 4). Of the two large RNAs, rescue of translation is invariably rather better for RNA-1 than RNA-2, even though the first 47 nt of the two 5′-UTR sequences differ in only three positions (Ahlquist et al., 1984). These reproducible observations imply that the relative functional affinity of p100 for the 5′-ends of the four RNAs is not the same as the relative affinity of binding of the eIF4F holoenzyme complex to the RNAs via eIF4E–cap interaction.
Effect of p100 concentration
We next studied the dependency of this rescue on p100 concentration. The results (Figure 5A) show that a discernible rescue of BMV RNA translation can be elicited with 2.5 µg/ml p100 (25 nM), and that 10 µg/ml (100 nM) achieves near-saturation. In order to give some physiological perspective to these data, we needed to estimate the amount of endogenous eIF4G in the parent lysate. This aim dictated the use of western blotting, but one complication in such an approach is that intact eIF4G and p100 may not be equally reactive to the antibody or may not transfer with the same efficiency; it is often observed that cleavage of eIF4G by picornavirus proteases (into an N-terminal one-third fragment and a C-terminal two-thirds fragment equivalent to p100) results in an increase in the total signal on the blot. Therefore, we took normal (non-depleted) lysate, and treated it with foot-and-mouth disease virus (FMDV) L-protease to cleave the endogenous eIF4G. The p100 western blot signal given by this protease-treated parent lysate was compared with the signals obtained with depleted lysate supplemented with various concentrations of recombinant p100, under conditions where the signal was shown to be proportional to p100 concentration. The result (Figure 5B) suggested that the concentration of endogenous eIF4G in the parent non-depleted lysate is equivalent to not less than 2.5 µg/ml p100 (25 nM). Thus, detectable rescue of capped BMV RNA translation in the depleted lysate can be achieved by addition of p100 at a concentration equivalent to that of endogenous eIF4G in the parent lysate, and complete rescue requires at most 4-fold greater amounts. Given that our recombinant p100 expressed in bacteria is more likely to be less active than the endogenous factor (rather than more active), it is clear that the rescue we are seeing cannot be dismissed as having no physiological relevance.
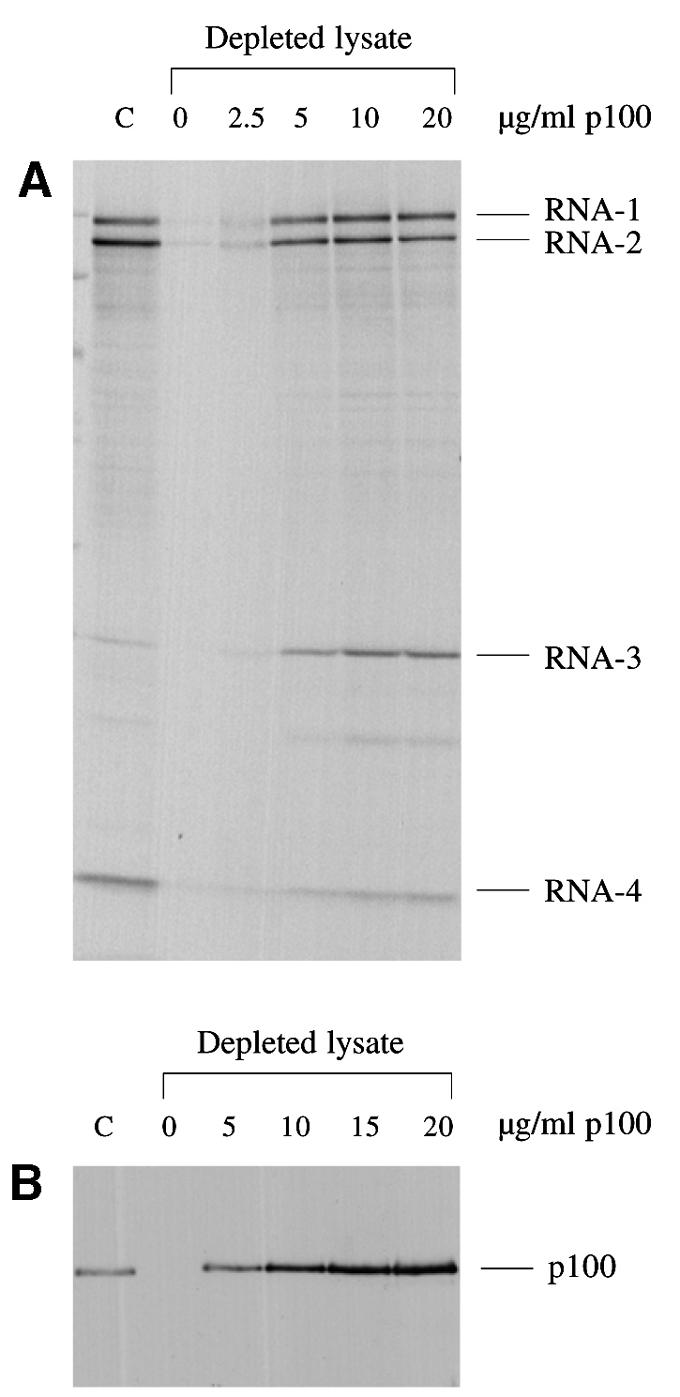
Fig. 5. The dependency of capped mRNA translation on p100 concentration, in relation to the concentration of endogenous eIF4G in the parent (non-depleted) lysate. (A) BMV RNA was translated at a final concentration of 20 µg/ml in either the control (non-depleted) lysate (C) or the eIF4G-depleted lysate supplemented with 0, 2.5, 5, 10 or 20 µg/ml recombinant p100, as indicated. Radiolabelled translation products were separated by gel electrophoresis, and the resulting autoradiograph is shown. (B) Western blot analysis. eIF4G-depleted lysate was supplemented with 0, 5, 10, 15 or 20 µg/ml recombinant p100, as indicated. Parent (non-depleted) lysate (C) was pre-incubated for 10 min at 30°C with in vitro expressed FMDV L-protease (see Materials and methods). Aliquots (equivalent to 1 µl lysate) were separated by gel electrophoresis, which was blotted and the blot probed with anti-eIF4G antiserum.
p100 can reverse the inhibition of capped mRNA translation caused by cap analogue, 4E-BP1 or cleavage of eIF4G
We next turned to the question of whether p100 can reverse the inhibition of translation caused by perturbation of endogenous eIF4F holoenzyme complex activity, rather than simple depletion of eIF4G. Inhibition of translation of capped BMV RNAs in the parent lysate was effected by one of three methods: addition of FMDV L-protease, which cleaves eIF4G (Kirchweger et al., 1994; Ohlmann et al., 1995, 1996); addition of m7GpppG cap analogue, which prevents eIF4E (and thus eIF4F) interaction with the 5′-cap; and addition of 4E-BP1, which sequesters eIF4E and thus indirectly prevents any interaction of eIF4G–4A complex with the 5′-cap itself (Pause et al., 1994). In all three cases, addition of p100 reversed the inhibition (Figure 6) with a similar concentration dependence to that seen with the depleted lysate. Interestingly, irrespective of which of the three inhibitors was used, translation of BMV RNA-3 was rescued much more effectively than BMV RNA-4, just as in the case of the depleted lysate supplemented with p100. Also of interest is the fact that addition of a high concentration of p100 to the parent, uninhibited lysate had at most only a marginal effect, a small increase in BMV RNA-3 translation and a small decrease in the yield of the RNA-4 translation product (Figure 6).
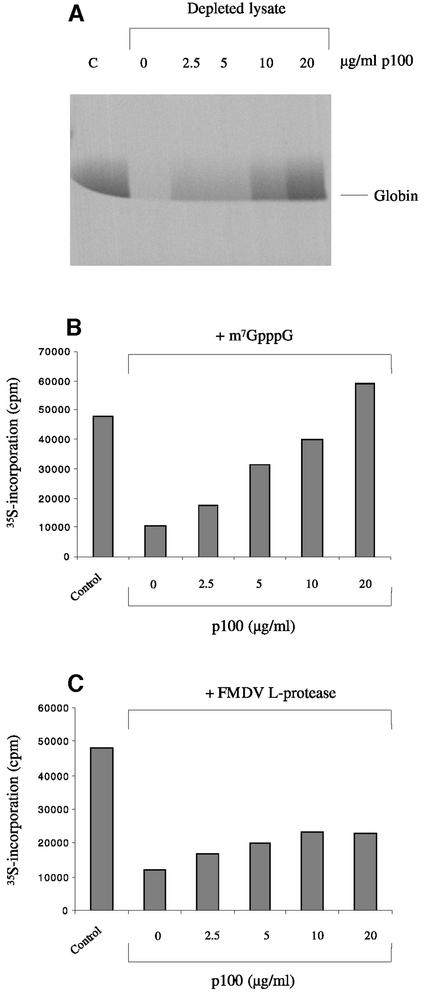
p100 can support the translation of capped globin mRNAs
One possible cause for concern about the general validity of the results described above is that they were obtained using a capped but non-polyadenylated plant viral RNA. Although there is absolutely no evidence that the translation of BMV RNAs in mammalian systems is atypical of capped mRNA translation, we thought it important to verify the more critical findings with capped and polyadenylated mRNA of mammalian origin, and for this purpose we chose globin mRNA extracted from reticulocytes. As shown in Figure 7, translation of globin mRNA was severely compromised by depletion of eIF4G, and it could be rescued completely by supplementing the system with p100, although complete rescue required rather more p100 (20 µg/ml) than was needed for maximum rescue of BMV RNA translation (∼10 µg/ml). As with BMV RNA, the translation of globin mRNA in the eIF4G-depleted system supplemented with p100 was completely refractory to inhibition by m7GpppG cap analogue (data not shown).
Fig. 7. p100 can rescue the translation of capped polyadenylated globin mRNA. (A) Globin mRNA was translated at a concentration of 15 µg/ml in either the control (non-depleted) lysate (C) or the eIF4G-depleted lysate supplemented with 0, 2.5, 5, 10 or 20 µg/ml recombinant p100, as indicated. Radiolabelled translation products were separated by gel electrophoresis, and the resulting autoradiograph is shown. (B) Globin mRNA was translated at a final concentration of 15 µg/ml in either control (untreated) lysate or lysate supplemented with 0.4 mM m7GpppG (with 0.32 mM additional MgCl2) and either 0, 2.5, 5, 10 or 20 µg/ml recombinant p100, as indicated. The incorporation of [35S]methionine into acid-precipitable protein was determined by scintillation counting and is plotted. (C) Globin mRNA was translated at a final concentration of 15 µg/ml in either control (untreated) lysate or lysate pre-incubated for 10 min at 30°C with recombinant FMDV L-protease and then supplemented with 0, 2.5, 5, 10 or 20 µg/ml recombinant p100 as indicated.
In addition, and likewise as seen with BMV RNA, p100 completely reversed the inhibition of globin mRNA translation caused by addition of m7GpppG cap analogue to standard (i.e. not eIF4G-depleted) reticulocyte lysate (Figure 7B). The dependence of this rescue on p100 concentration was very similar to that observed in the p100-dependent rescue of globin mRNA translation in the eIF4G-depleted lysate (Figure 7A), and again rather more p100 was needed than in the case of BMV RNA. In terms of the response to p100, globin mRNA is evidently intermediate between the extremes of BMV RNA-3 and RNA-4, and comparable to BMV RNA-2.
The inhibition of globin mRNA translation caused by addition of FMDV L-protease was also reversed by addition of p100 (Figure 7C), although the reversal was less complete than was seen with BMV RNAs -1, -2 and -3 (Figure 6). At present we do not have a viable explanation for this difference. As discussed later, what appeared at first sight to be the most obvious explanation turned out to be inadequate when subjected to a direct test.
Picornavirus type II IRESs outcompete scanning-dependent mRNAs for p100
In the experiment shown in Figure 2, addition of p100 to eIF4G-depleted lysate rescued translation of the IRES-dependent cistron very efficiently, but failed to resuscitate upstream cistron translation. We reasoned that this was likely to be due to competition between the two cistrons for p100, a supposition that is confirmed in Figure 8. A concentration of 2.5 µg/ml p100 was sufficient to effect near-maximum initiation on the EMCV IRES, but rescue of translation of the (capped) upstream NS cistron was barely detectable even at 10 µg/ml p100 (Figure 8A). However, with the same molar concentration of a transcript of the dicistronic cDNA construct linearized within the 5′-proximal part of the IRES, so as to generate a capped monocistronic mRNA coding for just NS, 10 µg/ml p100 effected a very efficient rescue (Figure 8B). Similarly, when the template was a mixture of the four BMV RNAs plus a monocistronic mRNA with the EMCV IRES, addition of 2.5 µg/ml p100 to the eIF4G-depleted lysate was sufficient to achieve maximal initiation on the EMCV IRES (Figure 8C), whereas rescue of BMV RNA translation, which was more than half-maximal at 5 µg/ml p100 in the absence of the EMCV IRES competitor (Figure 5), required appreciably higher p100 concentrations in the presence of the competitor. Whereas the EMCV IRES competed on roughly equal terms with BMV RNA-2 and RNA-4 in the parent control lysate (but strongly outcompeted translation of BMV RNA-1), in the depleted lysate supplemented with p100 it was completely dominant over all four scanning-dependent RNAs. Clearly, the EMCV IRES strongly outcompetes capped mRNAs for functional interaction with p100, a property that is shared by the FMDV IRES (data not shown). (We consider that this approach of assessing the relative affinities of p100 for different RNAs by using a strictly functional analysis of competition in translation assays is more meaningful than any data on physical interaction of p100 with mRNA, since p100 seems to bind RNA non-specifically, requiring arbitrary decisions as to how much rRNA and/or heparin competitor needs to be added to suppress non-specific interaction without affecting the specific binding.)
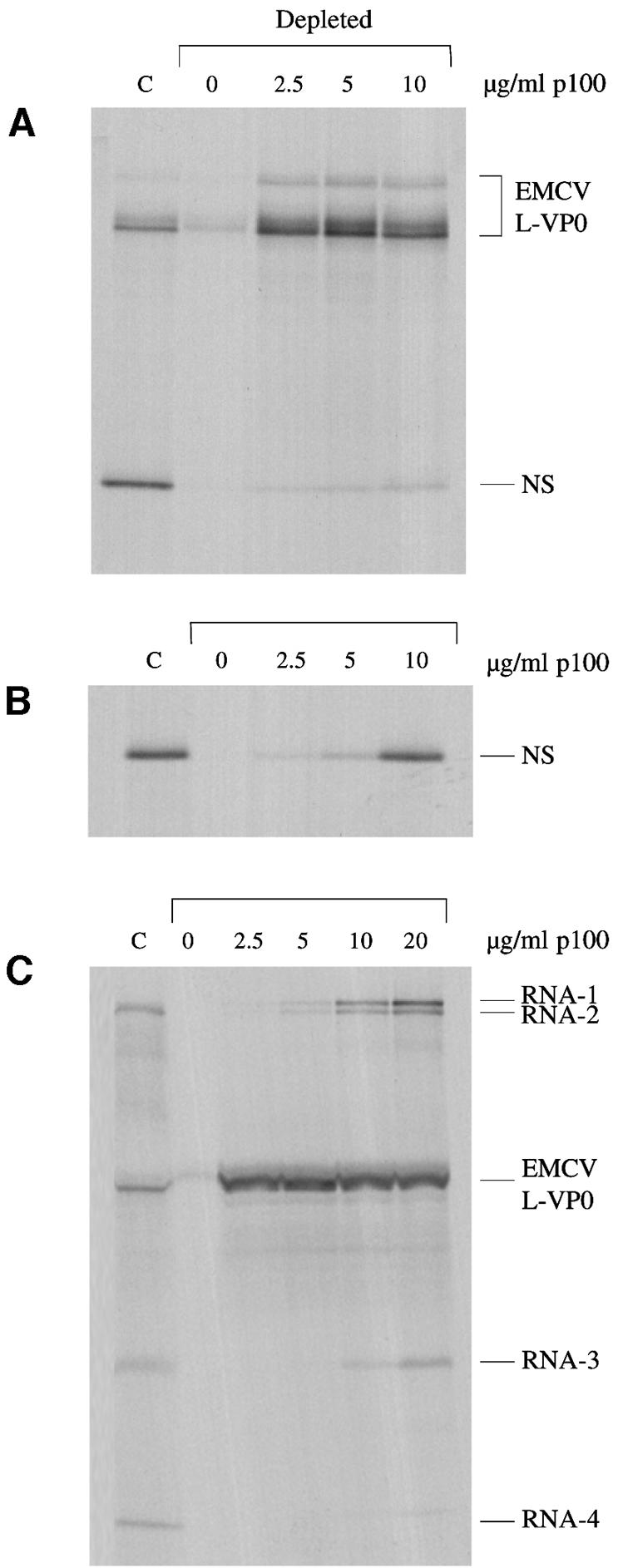
Fig. 8. The EMCV IRES strongly outcompetes capped scanning-dependent mRNAs for functional interaction with p100. (A) Capped dicistronic mRNA with an upstream cistron coding for influenzavirus NS1, an EMCV IRES, and EMCV coding sequences for L-VP0 (∼55 kDa) as downstream cistron, was translated at a final concentration of 20 µg/ml in the control (non-depleted) lysate (C), and in the eIF4G-depleted lysate supplemented with 0, 2.5, 5 or 10 µg/ml p100, as indicated. Radiolabelled translation products were separated by gel electrophoresis, and the resulting autoradiograph is shown. (B) A capped monocistronic mRNA encoding NS, transcribed from the same cDNA construct as the dicistronic mRNA in (A) but with linearization in the 5′-proximal part of the IRES, was translated at a final concentration of 8 µg/ml (same molar concentration as the dicistronic mRNA assayed in A), under the same conditions as in (A). (C) An equimolar mixture of BMV RNAs (13 µg/ml final concentration) and an uncapped monocistronic mRNA with the EMCV IRES linked to viral L-VP0 coding sequences (7 µg/ml) were translated under the same conditions as for (A) and (B).
In a similar vein we have noted that whilst translation in eIF4G-depleted lysate of a monocistronic mRNA with the HAV IRES is efficiently rescued by p100 with a dose– response not unlike that seen with BMV RNAs (Figure 5), globin mRNA (Figure 7A) or the capped monocistronic NS mRNA (Figure 8B), in a dicistronic mRNA background the HAV IRES-dependent translation is quite strongly outcompeted by the upstream scanning-dependent cistron (Ali et al., 2001). Thus, the hierarchy of functional affinities of p100 for the different RNAs is: EMCV (and FMDV) IRES >> capped scanning-dependent mRNAs > HAV IRES. Unfortunately, we are unable to place the picornavirus type I IRESs (poliovirus and rhinovirus) in this hierarchy, since translation dependent on these IRESs is not efficiently rescued by addition of p100 to the eIF4G-depleted lysate, most probably because the procedure also depletes another factor essential for the function of these IRESs.
Antagonism between the inhibitory effects of m7GpppG cap analogue and FMDV L-protease
The ability of p100 to support capped mRNA translation was surprising in view of the well known fact that cleavage of the eIF4G component of eIF4F by picornavirus proteases, which generates p100 and an eIF4E–eIF4G (N-terminal domain) complex, inhibits translation of capped mRNAs. One difference between a lysate treated with picornavirus proteases and an eIF4G-depleted lysate supplemented with p100 is that the latter has no eIF4E–eIF4G (N-terminal domain) complex. This consideration prompted us to question whether the binding of this complex to 5′-caps might inhibit, by steric hindrance, the interaction of p100 with the cap-proximal part of the RNA, which is presumably necessary for p100-dependent initiation. If this were the case, we would expect that the inhibition of translation of capped mRNAs caused by addition of picornavirus proteases should be at least partly reversed by addition of m7GpppG cap analogue, which would prevent the putative inhibitory interaction of eIF4E–eIF4G (N-terminal domain) complex with the cap. We have indeed observed such a relief of inhibition, particularly with globin mRNA rather than with BMV RNAs, possibly because the N-terminal cleavage product can interact with PABP (Figure 1), and thus would remain tethered to a polyadenylated mRNA via PABP bound to the 3′-poly(A) tail. At high concentrations of FMDV L-protease, addition of m7GpppG cap analogue stimulated globin mRNA translation by up to 2-fold (data not shown). However, in terms of relief of protease-induced inhibition, this is trivial, typically a decrease from 98% inhibition to 96%. We conclude that interaction of the eIF4E–eIF4G (N-terminal domain) cleavage product with 5′-caps makes a very small, and effectively negligible contribution to the inhibition caused by the protease.
On the other hand, with BMV RNA rather than globin mRNA, we consistently observed the reciprocal outcome, namely addition of FMDV L-protease effecting a partial reversal of m7GpppG-dependent inhibition (Figure 9). This partial relief of inhibition is almost certainly effected by the p100 fragment, since the rescue of translation is slightly more efficient for BMV RNA-1 than RNA-2, and better for RNA-3 than RNA-4 (Figure 9), precisely the same hierarchy as when translation is driven by p100 addition to the eIF4G-depleted lysate system. This result is interesting because of the obvious parallels with the observation that FMDV L-protease stimulates the translation of uncapped versions of mRNAs that are normally capped (Ohlmann et al., 1995, 1996). Thus, in any situation where eIF4F cannot interact with the capped 5′-end of an mRNA via eIF4E–cap interactions, either because m7GpppG has been added or because the RNA is uncapped, translation is stimulated by the picornavirus protease. It would appear that the conformation of intact eIF4F holoenzyme complex is not conducive to direct (cap- or eIF4E-independent) interaction of the factor with the 5′-proximal region of the RNA, and that this conformational constraint is alleviated when the eIF4G component is cleaved.
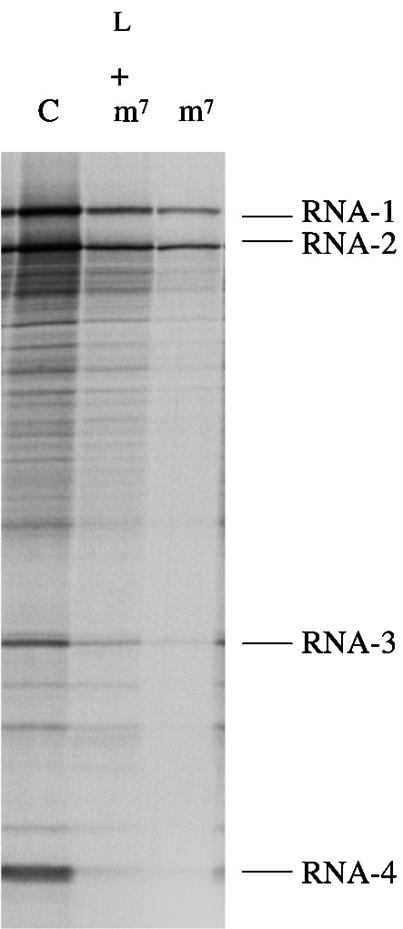
Fig. 9. FMDV L-protease can partially relieve inhibition of capped BMV RNA translation caused by m7GpppG cap analogue. BMV RNA was translated at a final concentration of 20 µg/ml in either control (untreated) lysate (C) or lysate supplemented with 0.4 mM m7GpppG cap analogue (with an additional 0.32 mM MgCl2), or supplemented with both recombinant FMDV L-protease and 0.4 mM m7GpppG cap analogue (with an additional 0.32 mM MgCl2), as indicated. Radiolabelled translation products were separated by gel electrophoresis, and the resulting autoradiograph is shown. The autoradiograph was exposed for longer than in the case of Figure 6 in order to facilitate visualization of the relative yield of products synthesized in the assay inhibited by cap analogue.
Discussion
Through the use of an eIF4G-depleted reticulocyte lysate system, we have shown here that the p100 fragment of eIF4G (the C-terminal two-thirds fragment), which lacks an eIF4E binding site, can support efficient translation of capped mRNAs by a mechanism that is operationally cap independent as witnessed by the fact that it is totally refractory to inhibition by m7GpppG cap analogue (Figure 4). Entirely consistent with this observation, we also find that p100 can reverse the inhibition of capped mRNA translation in non-depleted lysates resulting from addition of either m7GpppG cap analogue, 4E-BP1, which sequesters eIF4E and thus blocks its association with eIF4G, or picornavirus proteases, which cleave endogenous eIF4G (Figure 6).
These results were most unexpected since it has hitherto been assumed that p100 cannot drive capped mRNA translation, an assumption based on the well established observation that cleavage by picornavirus proteases of the eIF4G component of the eIF4F complex, to give p100 and an eIF4E–eIF4G (N-terminal fragment) complex, correlates with the shut-off of host cell protein synthesis during picornavirus infection. The apparent discrepancy could be explained if the other cleavage product, the eIF4E–eIF4G (N-terminal domain) complex bound to 5′-caps and inhibited, via steric hindrance, the functional interaction of the p100 fragment with the mRNA. However, our results suggest that this is likely to make at most only a very marginal contribution to the shut-off of host cell mRNA translation.
Considerations of factor concentrations and relative affinities may provide a partial answer to the paradox, since maximum BMV RNA translation in our depleted lysate system required a concentration of p100 ∼4-fold greater than the concentration of endogenous eIF4G in the non-depleted lysate (Figure 5). The other feature that helps to explain host cell shut-off despite the fact that p100 is intrinsically capable of supporting capped mRNA translation is that at least some picornaviral IRESs (but not the HAV IRES) strongly outcompete capped mRNAs for p100 (Figure 8). It has previously been noted that the shut-off of host cell mRNA translation in poliovirus infected cells is not necessarily complete at a time when all eIF4GI has been cleaved. Intriguingly, the discrepancy in the timing of shut-off and eIF4GI cleavage is particularly large under conditions in which viral RNA replication is inhibited (Bonneau and Sonenberg, 1987; Irurzon et al., 1995; Gradi et al., 1998b). In the presence of RNA replication inhibitors, host cell mRNA translation may persist at up to ∼50% of the control cell rate at the time when all eIF4GI had been cleaved (Bonneau and Sonenberg, 1987; Irurzon et al., 1995; Gradi et al., 1998b). The recent discovery of another eIF4G species, eIF4GII (Gradi et al., 1998a), appeared to provide an explanation for this discrepancy, since eIF4GII is cleaved more slowly during the infectious cycle than eIF4GI (Gradi et al., 1998b). However, because eIF4GII is much less abundant than eIF4GI, yet appears to be no more active than eIF4GI (Gradi et al., 1998a), there is some difficulty in seeing how this relatively small amount of endogenous eIF4GII could alone drive host cell mRNA translation at ∼50% of the control cell rate. On the basis of the results reported here, we propose that it is not just the residual uncleaved eIF4GII that is sustaining this high rate of host cell mRNA translation, but that the p100 cleavage product of eIF4GI also makes a substantial contribution to host cell protein synthesis under these conditions, in which there is relatively little viral RNA to compete against host cell mRNA, because viral RNA replication is inhibited.
Our results also offer an explanation for why the injection of picornavirus proteases into Xenopus oocytes, a scenario where there is no competing viral RNA, resulted in an overall decrease of only 35% in global protein synthesis rate, despite the fact that complete cleavage of eIF4G occurred (Keiper and Rhoads, 1997). Interestingly, the translation of some Xenopus mRNAs was strongly inhibited whilst others were scarcely affected, an observation that parallels our finding that the relative efficiency of translation of the four BMV RNAs is different when translation is driven by p100 than in the normal situation where it is dependent on intact eIF4F.
In a recent report, p100 was found to support, albeit at low efficiency, the formation of 48S initiation complexes (40S ribosomal subunits associated with eIF3, eIF1, eIF1A and the eIF2–Met-tRNAi–GTP ternary complex, and bound to the mRNA at the correct initiation site according to toeprint assay results). The yield of such complexes in the presence of p100 was only ∼20% of the yield seen with a fragment of eIF4G equivalent to p100 extended 63 amino acid residues in the N-terminal direction in order to include the site on eIF4G to which eIF4E binds (Morino et al., 2000), but was decidedly above the ‘background’ observed in the complete absence of any eIF4G fragments, which was really zero (Figure 2B of Morino et al., 2000). With hindsight it seems likely that the 48S complex formation observed in these assays with p100 is a direct parallel with the observations reported here. However, it is not clear why the relative yield of such complexes was so low. A low yield would be explicable if the p100 concentration was at the lower end of the range that we studied (i.e. ∼2.5–5 µg/ml), but in fact these authors appear to have been using 50 µg/ml. Moreover, an additional complication is that in translation assays p100 failed to support translation of the 5′-proximal cistron of a capped dicistronic mRNA with the EMCV IRES (Figure 2C of Morino et al., 2000). No explanation was offered for the apparently conflicting results of the two assays. If the two results are both taken at face value, then the only self-consistent conclusion that can be drawn without referring to our own results is that p100 can support 48S initiation complex formation, albeit inefficiently, but cannot support the complete process of initiation. Reference to our results, however, suggests the alternative explanation that the apparent failure of p100 to support translation of the upstream cistron of the dicistronic mRNA was due to competition from the EMCV IRES (Figure 8).
What is rather surprising is that when translation is driven by p100, initiation still occurs overwhelmingly at the correct 5′-proximal AUG; the ‘noise’ level of incomplete products arising from incorrect initiation at internal sites seems no higher than when initiation is driven by the endogenous eIF4F holoenzyme complex in a strictly cap-dependent manner (Figure 4). Similarly, the strong stimulatory effect of p100 on the translation of uncapped versions of normally capped mRNAs is predominantly from the correct 5′-proximal initiation site (Ohlmann et al., 1995, 1996; De Gregorio et al., 1998). Thus, it appears that p100 has the property of delivering the 40S subunit to the mRNA almost exclusively in the vicinity of the 5′-end, irrespective of whether this end is capped or not.
Materials and methods
Materials
RNA from BMV was obtained from Promega, and globin mRNA from Gibco-BRL. Oligo (dT)25 Dynabeads (magnetic beads) were from Dynal.
eIF4G depletion procedure
A cDNA copy of EMCV Domain 3, nt 680–843 in the numbering system of Duke et al. (1992), was cloned between the HincII and BamHI sites of pSP64Poly(A) vector (Promega), which had previously been modified by inserting a T7 promoter between the HindIII and SalI sites, in order to facilitate large scale production of RNA. The resulting clone was linearized with EcoRI prior to in vitro transcription, which would generate an RNA with a poly(A) tail of 30 residues length. Large scale (0.2 ml) transcription reactions were carried out as described previously (Kaminski et al., 1998).
The standard eIF4G depletion procedure used 1 ml of suspension of oligo (dT)25 Dynabeads as supplied by the manufacturer (3.3 × 108 beads). The beads were washed once with 1 ml of 0.5× SSC buffer, and then resuspended in 200 µl of binding buffer (10 mM Tris–HCl pH 7.5, 100 mM KCl, 2 mM MgCl2). To this was added 10 µg of poly(A)+ EMCV Domain 3 RNA, and the material was incubated for 30 min at 4°C. The supernatant was then removed, and the beads were washed twice with 400 µl of binding buffer. All washes involved immobilizing the beads using the magnetic stand, and pipetting off the supernatant. The beads were then incubated on ice for 60 min with 200 µl of reticulocyte lysate translation assay mix as described below (complete except for the radiolabelled methionine). Following immobilization of the beads by use of the magnet, the reticulocyte lysate translation assay mix was removed, frozen in small aliquots and stored at –80°C.
Preparation of recombinant p100
Recombinant p100, amino acids 621–1560 in the revised numbering system of eIF4GI defined by Gradi et al. (1998a), was expressed with a C-terminal His-tag in Escherichia coli BL21 (DE3) plysS. The recombinant protein was initially isolated by chromatography using Ni2+-nitrilotriacetic acid-agarose (Qiagen), and was subsequently further purified by ion-exchange chromatography using heparin–Sepharose (Pharmacia) as described by Pestova et al. (1996b). The concentration of p100 was estimated by polyacrylamide gel electrophoresis of recombinant protein and bovine serum albumin standards, followed by Coomassie Blue staining.
Translation assays
In vitro translation assays were carried out as described by Jackson and Hunt (1983). Briefly, the reactions consisted of 70–75% (by vol.) micrococcal nuclease-treated rabbit reticulocyte lysate, and the following additional components at the final concentration stated: 100 mM KCl, 0.5 mM MgCl2, 10 mM creatine phosphate, 50 µg/ml creatine phosphokinase, 15 µM haemin, 0.1 mM each amino acid except methionine, 50 µg/ml calf liver tRNA (Boehringer), 0.5 mCi/ml [35S]methionine (SJ1515, Amersham Pharmacia Biotech). Assays supplemented with m7GpppG or GpppG also received additional MgCl2 at 0.8 mol/mol cap analogue, to counteract the chelating potential of the cap analogue; the ratio was chosen empirically on the basis of the observed shift in Mg2+ optimum caused by these cap analogues. Reactions were incubated at 30°C for 60 min, and then the translation products were either analysed by polyacrylamide gel electrophoresis followed by autoradiography, or quantitated by precipitation with trichloroacetic acid and scintillation counting of the filtered precipitates, as described previously (Jackson and Hunt, 1983). Hyperfilm βmax (Amersham Pharmacia Biotech) was used for autoradiography, and quantitative densitometry of the films was carried out using Phoretix software.
The generation of FMDV L-protease from plasmid pMM1 (Medina et al., 1993) by in vitro transcription and translation was carried out as described by Hunt et al. (1999). The protease preparation was diluted 100-fold into fresh lysate, which was then incubated for 10 min at 30°C to effect complete cleavage of the endogenous eIF4G.
Recombinant L-protease and 4E-BP1 were expressed and purified as described previously (Ohlmann et al., 1996). The protease was added to reticulocyte lysates at 5 µg/ml, and 4E-BP1 at 10 µg/ml.
Immunoblotting
Western blotting was carried out as described by Harlow and Lane (1988), with horseradish peroxidase-conjugated secondary antibodies. Detection was by ECL.
Acknowledgments
Acknowledgements
We thank Dr C.U.T.Hellen (SUNY Health Center, Brooklyn, NY) and Dr J.Lawrence, Jr (University of Virginia, VA) for providing reagents, Dr Graham Belsham for advice on the eIF4G depletion method, and Rosemary Farrell for providing technical assistance and infrastructure support to the R.J.J. group. This work was supported by grants from the Wellcome Trust to R.J.J. (051424 and 062348) and S.J.M. (040800, 045619, 056778, 057494 and 058915). S.J.M. is a Senior Research Fellow of the Wellcome Trust and I.K.A. was supported by a Medical Research Council postgraduate research studentship.
References
- Ahlquist P., Luckow,V. and Kaesberg,P. (1981) Complete nucleotide sequence of Brome Mosaic Virus RNA 3. J. Mol. Biol., 153, 23–38. [DOI] [PubMed] [Google Scholar]
- Ahlquist P., Dasgupta,R. and Kaesberg,P. (1984) Nucleotide sequence of the Brome Mosaic Virus genome and its implications for viral replication. J. Mol. Biol., 172, 369–383. [DOI] [PubMed] [Google Scholar]
- Ali I.K., McKendrick,L., Morley,S.J. and Jackson,R.J. (2001) The activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau A.-M. and Sonenberg,N. (1987) Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J. Virol., 61, 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta R. and Kaesberg,P. (1982) Complete nucleotide sequence of the coat protein messenger RNAs of Brome Mosaic Virus and Cowpea Chlorotic Mottle Virus. Nucleic Acids Res., 10, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M.C. and Jackson,R.J. (1989) On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res., 17, 3129–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Preiss,T. and Hentze,M.W. (1998) Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end dependent. RNA, 4, 828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke G.M., Hoffman,M.A. and Palmenberg,A.C. (1992) Sequence and structure elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol., 66, 1602–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught,B. and Sonenberg,N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Gradi A., Imataka,H., Svitkin,Y.V., Rom,E., Raught,B., Morino,S. and Sonenberg,N. (1998a) A novel functional human eukaryotic translation initiation factor eIF4G. Mol. Cell. Biol., 18, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A., Svitkin,Y.V., Imataka,H. and Sonenberg,N. (1998b) Proteolysis of human eukaryotic initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl Acad. Sci. USA, 95, 11089–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E. and Lane,D.P. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hershey J.W.B. and Merrick,W.C. (2000) The pathway and mechanism of initiation of protein synthesis. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (ed.), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–88.
- Hunt S.L., Skern,T., Liebig,H.-D., Kuechler,E. and Jackson,R.J. (1999) Rhinovirus 2A proteinase mediated stimulation of rhinovirus RNA translation is additive to the stimulation effected by cellular RNA binding proteins. Virus Res., 62, 119–128. [DOI] [PubMed] [Google Scholar]
- Imataka H. and Sonenberg,N. (1997) Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol., 17, 6940–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Gradi,A. and Sonenberg,N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irurzon A., Sanchez-Palomino,S., Nonoa,I. and Carrasco,L. (1995) Monensin and nigericin prevent the inhibition of host translation by poliovirus without affecting p220 cleavage. J. Virol., 69, 7453–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. and Hunt,T. (1983) Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol., 96, 50–74. [DOI] [PubMed] [Google Scholar]
- Kaminski A., Ostareck,D.H., Standart,N.M. and Jackson,R.J. (1998) Affinity methods for isolating RNA binding proteins. In Smith,C.W.J. (ed.), RNA:Protein Interactions—A Practical Approach. Oxford University Press, Oxford, UK, pp. 137–160.
- Keiper B.D. and Rhoads,R.E. (1997) Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res., 25, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchweger R. et al. (1994) Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4γ. J. Virol., 68, 5677–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V.G., Pestova,T.V., Hellen,C.U.T. and Shatsky,I.N. (1998) Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem., 273, 18599–18604. [DOI] [PubMed] [Google Scholar]
- Korneeva N.L., Lamphear,B.J., Hennigan,F.L.C. and Rhoads,R.E. (2000) Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem., 275, 41369–41376. [DOI] [PubMed] [Google Scholar]
- Korneeva N.L., Lamphear,B.J., Hennigan,F.L.C., Merrick,W.C. and Rhoads,R.E. (2001) Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem., 276, 2872–2879. [DOI] [PubMed] [Google Scholar]
- Lamphear B.J., Kirchweger,R., Skern,T. and Rhoads,R.E. (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J. Biol. Chem., 270, 21975–21983. [DOI] [PubMed] [Google Scholar]
- Lomakin I.B., Hellen,C.U.T. and Pestova,T.V. (2000) Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosome entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol., 20, 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M., Domingo,E., Brangwyn,J.K. and Belsham,G.J. (1993) The two species of foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology, 194, 355–359. [DOI] [PubMed] [Google Scholar]
- Meyer K., Petersen,A., Niepmann,M. and Beck,E. (1995) Interaction of eukaryotic initiation factor eIF4B with a picornavirus internal translation site. J. Virol., 69, 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino S., Imataka,H., Svitkin,Y.V., Pestova,T.V. and Sonenberg,N. (2000) Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol., 20, 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T., Rau,M., Morley,S.J. and Pain,V.M. (1995) Proteolytic cleavage of initiation factor eIF-4γ in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res., 23, 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T., Rau,M., Pain,V.M. and Morley,S.J. (1996) The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J., 15, 1371–1382. [PMC free article] [PubMed] [Google Scholar]
- Pause A., Belsham,G.J., Gingras,A.C., Donze,O., Lin,T.A., Lawrence,J.C. and Sonenberg,N. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′ cap function. Nature, 371, 762–767. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Hellen,C.U.T. and Shatsky,I.N. (1996a) Canonical eukaryotic initiation factors determine translation by internal ribosome entry. Mol. Cell. Biol., 16, 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky,I.N. and Hellen,C.U.T. (1996b) Functional dissection of eukaryotic initiation factor eIF4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol., 16, 6870–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau M., Ohlmann,T., Morley,S.J. and Pain,V.M. (1996) A re-evaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J. Biol. Chem., 271, 8983–8990. [DOI] [PubMed] [Google Scholar]
- Stassinopoulos I.A. and Belsham,G.J. (2001) A novel protein–RNA binding assay: functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA, 7, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]