Abstract
NEDD8/Rub1 is a ubiquitin (Ub)-like post-translational modifier that is covalently linked to cullin (Cul)-family proteins in a manner analogous to ubiquitylation. NEDD8 is known to enhance the ubiquitylating activity of the SCF complex (composed of Skp1, Cul-1, ROC1 and F-box protein), but the mechanistic role is largely unknown. Using an in vitro reconstituted system, we report here that NEDD8 modification of Cul-1 enhances recruitment of Ub-conjugating enzyme Ubc4 (E2) to the SCF complex (E3). This recruitment requires thioester linkage of Ub to Ubc4. Our findings indicate that the NEDD8-modifying system accelerates the formation of the E2–E3 complex, which stimulates protein polyubiquitylation.
Keywords: IκBα/NEDD8/ROC1–SCF/ubiquitylation/ubiquitin ligase
Introduction
Ubiquitin (Ub) is an 8.6 kDa highly conserved protein molecule. It forms a polyubiquitin chain on proteins, which becomes a degradation signal attacked by the 26S proteasome (Hershko and Ciechanover, 1998; Voges et al., 1999). Protein ubiquitylation is catalysed by a cascade reaction involving three enzymes, E1 (Ub-activating enzyme), E2 (Ub-conjugating enzyme) and E3 (Ub-ligating enzyme) (Hershko and Ciechanover, 1998). NEDD8 (a mammalian orthologue of the budding yeast Rub1, related to Ub1) has the highest homology to Ub (57% identity) among an expanding family of Ub-like proteins in eukaryotes (Hochstrasser, 1998; Jentsch and Pyrowolakis, 2000; Yeh et al., 2000). NEDD8 is covalently attached to target proteins via the C-terminal glycine residue in a manner analogous to ubiquitylation, which is catalysed by the APP-BP1/Uba3 heterodimer and Ubc12, similar to the E1- and E2-like enzymes, referred to here simply as ‘E1L’ and ‘E2L’, respectively (Osaka et al., 1998; Gong and Yeh, 1999). The NEDD8 conjugation system is conserved across species (Lammer et al., 1998; Liakopoulos et al., 1998) and, to date, its targets are unknown other than Cul-family proteins (Hochstrasser, 2000; Jentsch and Pyrowolakis, 2000). Cul-1 is a common subunit of Ub-protein ligases termed ‘SCF’, or the ‘ROC1–SCF’ complex, consisting of the core subunits Skp1, Cul-1/Cdc53, ROC1/Rbx1/Hrt1 and substrate recognition adaptors known as F-box proteins, which are involved in ubiquitylation of a multitude of proteins (Deshaies, 1999; Kipreos and Pagano, 2000). Recently, other Cul-family proteins have also been described to constitute a large family of distinct Ub–protein ligase complexes, since ROC1/Rbx1/Hrt1, a RING-finger protein, which is essential for the catalytic activity of SCF, interacts with all members of Cul-family proteins (Kamura et al., 1999a; Ohta et al., 1999; Seol et al., 1999). Intriguingly, it is also reported that the Rbx1 is involved in Rub1 modification of two cullins, Cdc53/Cul-1 and Cul-2, perhaps acting as an E3 for both ubiquitylation and NEDD8 modification (Kamura et al., 1999b). In addition, Furukawa et al. (2000) recently reported that ROC1 promotes nuclear accumulation of Cul-1 to facilitate its NEDD8 modification, enhancing ubiquitin ligase activity of Cul-1.
The multifunctional transcription factor, NF-κB, which consists of two distinct subunits, p50 and p65, is normally present in the cell cytoplasm as an inactive form due to association with IκB (Baldwin, 1996; Ghosh et al., 1998). IκB prevents the transportation of NF-κB into the nucleus by its nuclear localization signal. To date, it is clear that phosphorylation and subsequent proteolytic destruction of IκB result in the immediate removal of IκB (Finco and Baldwin, 1995; Maniatis, 1999). The multisubunit IκB kinase (abbreviated herein as IKK) phosphorylates two serine residues, Ser32 and Ser36, at the N-terminal region of IκBα (a major member of IκB family proteins) (May and Chosh, 1999). The ROC1–SCFβTrCP1 complex, identified as an IκBα–E3 ligase, binds phosphorylated IκBα (pIκBα) to ubiquitylate, generating a polyubiquitin chain, which is finally recognized by the 26S proteasome for ultimate degradation (Yaron et al., 1998; Winston et al., 1999).
We recently have reported that the putative IκBα–E3, homodimer of SCFβTrCP1 or SCFβTrCP2, is recruited rapidly to bind pIκBα in vivo and in vitro (Suzuki et al., 2000). During these studies, we noticed that Cul-1 is modified preferentially by NEDD8. More recently, it has been demonstrated that NEDD8 modification of Cul-1 activates ubiquitylation of IκBα (Furukawa et al., 2000; Read et al., 2000; Wu et al., 2000) and p27Kip1 (Morimoto et al., 2000; Podust et al., 2000), by SCFβTrCP1 and SCFSkp2, respectively. However, the mechanistic role of NEDD8 in promoting SCF Ub ligase activity is to a large extent unknown.
Using a fully in vitro reconstituted system, we examined in the present study the mechanism by which NEDD8 modification of Cul-1 augments the function of ROC1– SCFβTrCP1 as an E3–IκBα ligase. Our results showed that NEDD8-ylation of Cul-1 promotes recruitment of Ub-linked E2 (Ubc4) to the ROC1–SCFβTrCP1 complex, leading to efficient ubiquitylation of phosphorylated IκBα. Our results indicate that the NEDD8 system positively regulates SCF activity, possibly through a conformational change of Cul-1 that promotes the E2–E3 complex formation. These findings shed new light on the mechanism underlying the functional link between the Ub and NEDD8 systems that are highly conserved during evolution.
Results
NEDD8-ylation of Cul-1 stimulates ubiquitylation of IκBα by ROC1–SCFβTrCP1
To investigate the mechanistic role of NEDD8 modification of Cul-1, we devised a fully reconstituted system for NEDD8-ylation and IκBα ubiquitylation. To this end, we produced all components involved in these systems as recombinant proteins by Escherichia coli or the baculovirus system (for details, see Materials and methods). Figure 1A shows dye staining patterns after separation by SDS–PAGE, indicating that all recombinant proteins were purified to near homogeneity.
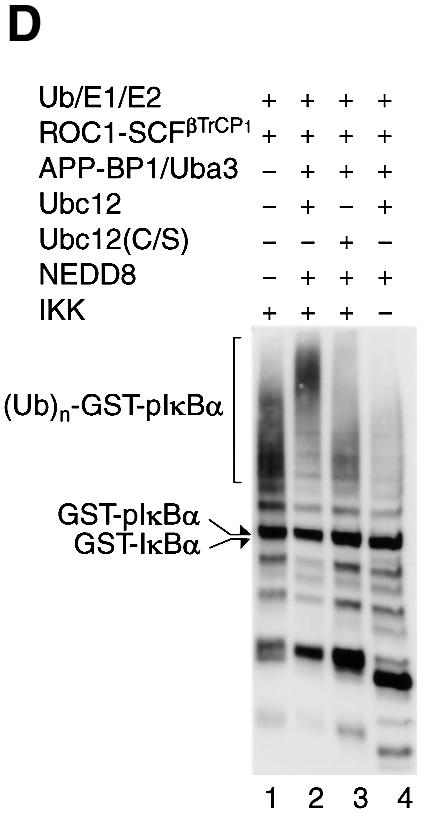
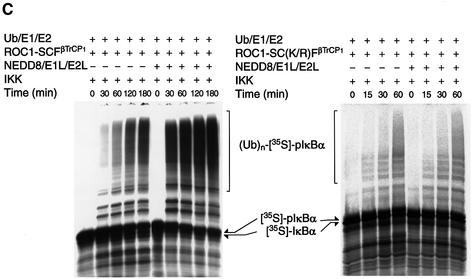
Fig. 1. NEDD8-ylation of Cul-1 stimulates ubiquitylation of IκBα by the ROC1–SCFβTrCP1 complex. (A) Electrophoretic patterns of recombinant proteins consisting of the IκBα-ubiquitylating [E1 (Uba1), E2 (Ubc4) and (E3) ROC1–SCFβTrCP1] and NEDD8-modifying [E1L (APP-BP1/Uba3) and E2L (Ubc12)] systems. A 5 µg aliquot of each affinity-purified protein, except ROC1–SCFβTrCP1 (10 µg), was subjected to SDS–PAGE followed by staining with Coomassie blue R-250. (B) Modification of Cul-1 by a reconstituted NEDD8 conjugation system. The ROC1–SCFβTrCP1 complex or ROC1–SC(K/R)FβTrCP1 was incubated at 25°C for 15–30 min in the presence or absence of the NEDD8 system containing His-NEDD8 or GST–NEDD8 as described in Materials and methods. After incubation, the reaction was terminated by the addition of 20 µl of SDS–PAGE sample buffer. One-quarter volume of the boiled supernatant was subjected to SDS–PAGE followed by western blotting with anti-HA antibodies to detect Cul-1. Arrows indicate Cul-1, His-NEDD8-Cul-1 and GST–NEDD8-Cul-1. (C) Time course of [35S]IκBα ubiquitylation by ROC1–SCFβTrCP1 (left) and ROC1–SC(K/R)FβTrCP1 (right). Ubiquitylation of [35S]IκBα by ROC1–SCFβTrCP1 was performed in the presence or absence of the NEDD8 system. IκBα pre-phosphorylated by IKK was used as a substrate, unless otherwise specified. The high molecular mass ubiquitylated [35S]IκBα is shown as (Ub)n-[35S]-pIκBα. (D) Effect of Ubc12(C/S) on the in vitro ubiquitylation of phosphorylated GST–IκBα. Experiments were similar to those of (B), except that GST–IκBα and Ubc12(C/S) were used for the assay. The high molecular mass ubiquitylated GST–IκBα detected by western blotting is shown as (Ub)n-GST–pIκBα.
First, we tested whether the reconstituted NEDD8 system is active for modification of Cul-1. As shown in Figure 1B, when ROC1–SCFβTrCP1 was incubated with E1L and E2L in the presence of His-NEDD8, two anti-HA (Cul-1)-reactive bands were detected, whereas only a single species was detected in the absence of the NEDD8 system. The fast migrating major band corresponded to unmodified Cul-1 and the species exhibiting reduced mobility on SDS–PAGE were thought to be Cul-1 conjugated to His-NEDD8, because this slow migrating band did not appear when we used ROC1–SC(K/R)FβTrCP1, whose NEDD8-targeted lysine of Cul-1 was replaced by arginine. To confirm further that the slow migrating band corresponds to the NEDD8-modified form of Cul-1, we used GST–NEDD8. When GST–NEDD8 was used instead of His-NEDD8, the high molecular mass band corresponding to Cul-1-conjugated with GST–NEDD8 was observed in a time-dependent fashion. After 30 min incubation, 10–20% of Cul-1 was conjugated to NEDD8, but longer incubation, e.g. >2 h, resulted in modification of ∼60–70% of Cul-1 (data not shown).
In the next step, we examined the role of the NEDD8 system in ubiquitylation of pIκBα mediated by ROC1–SCFβTrCP1. As shown in Figure 1C (left), ROC1–SCFβTrCP1 time-dependently catalysed polyubiquitylation of cell-free translated [35S]IκBα that had been phosphorylated by purified IκB kinase (IKK). The addition of NEED8 and NEDD8-ligating enzymes resulted in marked augmentation of IκBα ubiquitylation, particularly during the initial period of incubation, e.g. 30–60 min. However, less pIκBα ubiquitylation was observed in the absence of the NEDD8 system. Because reticulocyte extracts used for the synthesis of [35S]IκBα contained a significant amount of the NEDD8 system (Osaka et al., 1998; Hori et al., 1999), we next used GST–IκBα produced by E.coli as a substrate. As shown in Figure 1D (lane 2), the NEDD8 system markedly enhanced polyubiquitylation of GST–pIκBα in a phosphorylation-dependent manner (Figure 1D, lane 4). However, the ubiquitylating activity was still detected in the absence of the NEDD8 system, even if GST–pIκBα was used. Thus, NEDD8-ylation of Cul-1 was not essential for the ubiquitylating reaction at least in the present in vitro system. Nonetheless, because the NEDD8 system could augment the ROC1–SCFβTrCP1 activity in our in vitro system, we analysed its mechanistic reaction further.
We next examined whether this augmentation of ubiquitylating activity of ROC1–SCFβTrCP1 is mediated solely by modification of Cul-1, but not by co-operation of the NEDD8 and Ub system. When mutant Cul-1(K/R) whose NEDD8-targeted lysine was replaced by arginine was used instead of wild-type Cul-1, the NEDD8 system did not enhance pIκBα ubiquitylation (Figure 1C, right), strongly indicating that the modification of Cul-1 by NEDD8 plays a critical role in the enhancement of ROC1–SCFβTrCP1-dependent pIκBα ubiquitylation. This inability of Cul-1(K/R) was not due to loss of ROC1–SC(K/R)FβTrCP1 complex formation nor substrate (pIκBα) binding (see below). Moreover, when Ubc12(C/S) defective for thioester linkage of NEDD8 was added instead of wild-type Ubc12 in the NEDD8 system, no obvious stimulatory effect on the ROC1–SCFβTrCP1-dependent GST–pIκBα ubiquitylating activity was observed (Figure 1D, lane 3), again suggesting the importance of NEDD8-ylation of Cul-1 for activation of IκBα–E3 ligase.
It is worth noting that the ubiquitylating activity of pIκBα by ROC1–SCFβTrCP1 persisted for incubation up to 2 h even when the NEDD8 system was not supplemented (Figure 1C) or ROC1–SC(K/R)FβTrCP1 was used, suggesting that the ROC1–SCFβTrCP1 complex is fairly stable. To confirm this finding more clearly, we tested the effect of the NEDD8 system on heat inactivation of ROC1– SCFβTrCP1. Pre-incubation of ROC1–SCFβTrCP1 for 15 min at various temperatures did not alter the mode of loss of [35S]pIκBα ubiquitylating activity at high temperatures, irrespective of the presence or absence of the NEDD8 system (data not shown). Taken together, it is clear that the NEDD8 system does not function to protect the instability of the ROC1–SCFβTrCP1 complex.
The NEDD8 system has no effect on the assembly of ROC1–SCFβTrCP1
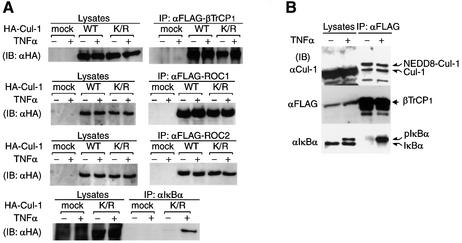
In an in vitro reconstituted system, we found that Cul-1(K/R) was complexed with Skp1, βTrCP1 and ROC1, as is wild-type Cul-1 (see Figure 1C, right panel and data not shown), indicating that NEDD8-ylation is not required for the assembling of the ROC1–SCFβTrCP1 complex at least in vitro. To confirm this in in vivo, we examined whether NEDD8-ylation of Cul-1 is essential for the assembly of the SCF complex. To this end, HA-Cul-1 or the HA-Cul-1(K/R) mutant was co-expressed with FLAG-βTrCP1 in HEK 293 cells, and immunoprecipitated by anti-FLAG antibody, followed by immunoblotting with anti-HA antibody. As shown in Figure 2A (top), Cul-1(K/R) was incorporated into the SCFβTrCP1 complex, as was the wild-type Cul-1. Similar results were obtained by immunoprecipitation with FLAG-ROC1 or its isoform FLAG-ROC2 (Figure 2A, second and third rows). Thus, it was clear that NEDD8 modification is not required for the assembly of the SCFβTrCP1 complex. Furthermore, Cul-1(K/R) could bind to phosphorylated IκBα (pIκBα) in tumour necrosis factor-α (TNF-α)-stimulated cells (Figure 2A, bottom), indicating that NEDD8 modification of ROC1–SCFβTrCP1 is not required for substrate binding.
Fig. 2. Modification of Cul-1 by NEDD8 in vivo. (A) Association of Cul-1(K/R) with the SCF complex and IκBα in living cells. pcDNA3-HA-Cul-1 and HA-Cul-1(K/R) were co-expressed with FLAG-βTrCP1, FLAG-ROC1 or FLAG-ROC2 in 293 cells and either left untreated or treated with TNF-α. After immunoprecipitation (IP) with anti-FLAG (upper three rows) and anti-IκBα (lower row) antibodies, immunoblotting (IB) was carried out with anti-HA antibody for cell lysates and the immunoprecipitates. (B) Co-immunoprecipitation of NEDD8-modified Cul-1 with βTrCP1 in vivo. pcDNA3-βTrCP1-FLAG was transfected into 293 cells and treated or not with TNF-α. The extracts or anti-FLAG antibody immunoprecipitates were analysed by western blotting using anti-FLAG, anti-Cul-1 and anti-IκBα antibodies. (C) NEDD8 modification of Cul-1 associated with phosphoryl ated IκBα. HEK-293 cells were either left untreated or treated with TNF-α. The anti-IκBα antibody immunoprecipitates were analysed by western blotting using anti-Cul-1 and anti-NEDD8 antibodies. Experiments were conducted in duplicate. (D) βTrCP1 augments modification of Cul-1 by NEDD8 in reticulocyte lysates. Following the synthesis of [35S]Cul-1 in the presence or absence of [35S]βTrCP1 or GST–NEDD8 as indicated for 60 min at 30°C in a reticulocyte lysate transcription/translation system, samples of the resultant translational products were subjected directly to SDS–PAGE in the presence of dithiothreitol and then autoradiographed. Arrows indicate [35S]βTrCP1, [35S]-Cul1, endogenous NEDD8-modified [35S]Cul-1 and GST–NEDD8-modified [35S]Cul-1. (E) Co-immunoprecipitation of NEDD8-modified Cul-1 with βTrCP1 or βTrCP1 lacking the WD40-repeat region. Thirty-six hours after pcDNA3-FLAG-βTrCP1 or the cDNAs of FLAG-tagged βTrCP1 lacking the WD40-domains (designated FLAG-βTrCP1ΔW1–7) was transfected into 293 cells, crude extracts were prepared as described in Materials and methods. After immunopre cipitation by anti-FLAG antibody, the resulting immunoprecipitates were analysed by western blotting using anti-FLAG, anti-Skp1, anti-Cul-1 and anti-NEDD8 antibodies.
Curiously, we noticed very little NEDD8-ylation of overexpressed HA-tagged wild-type Cul-1. Therefore, we tested whether endogenous Cul-1 is modified by NEDD8 in living cells. After FLAG-tagged βTrCP1 was expressed in 293 cells, endogenous Cul-1 in the SCFβTrCP1 complex was immunoprecipitated by anti-FLAG antibody. Western blotting showed that Cul-1 was observed mainly as a single species in the 293 cell extracts, whereas two bands were evident in immunoprecipitates with anti-FLAG antibody (Figure 2B). This pattern was unaffected upon TNF-α stimulation, which caused association of SCFβTrCP1 with pIκBα. The upper band reacted with anti-NEDD8 antibody (data not shown), indicating that it was NEDD8-ligated Cul-1. Thus, most Cul-1 present in cell extracts is not modified by NEDD8, while nearly half of Cul-1 in the SCFβTrCP1 complex seems to be modified by NEDD8. Nonetheless, Cul-1 co-immunoprecipitated with anti-IκBα antibody in TNF-α-treated 293 cell extracts was a single species, which was identified as a NEDD8-modified form (Figure 2C), indicating that Cul-1 in the SCFβTrCP1 complex associated with pIκBα is ligated preferentially by NEDD8. However, the mechanisms involved in the efficient modification of Cul-1 by NEDD8 in the SCF complex bound to target substrate, such as pIκBα, remain unclear.
Subsequently, we investigated the influence of SCFβTrCP1 complex formation on in vitro NEDD8 ligation of Cul-1. For this purpose, we added F-box protein, i.e. βTrCP1, in rabbit reticulocyte lysates, because we have already reported that NEDD8 efficiently modified all human Cul-family proteins in this in vitro system (Osaka et al., 1998; Hori et al., 1999). As shown in Figure 2D, when the cDNAs of Cul-1 were co-translated with those of βTrCP1 or none, the upper band of synthesized [35S]Cul-1 increased in a manner dependent on co-translation of [35S]βTrCP1 (lane 2). The reduced mobility band was assumed to be [35S]Cul-1 conjugated by endogenous NEDD8. To confirm this assumption, we carried out the same experiment in the presence of GST–NEDD8. The GST–NEDD8-conjugated [35S]Cul-1 increased proportionately with elevated levels of translated [35S]βTrCP1 (Figure 2B, lanes 3–5). Intriguingly, no obvious augmentation of the ligation of GST–NEDD8 to [35S]Cul-1 was observed following the addition of βTrCP1ΔF lacking the F-box domain that cannot associate with Skp1 and thus Cul-1 (data not shown), strongly indicating that the formation of the SCFβTrCP1 complex promotes NEDD8 modification of Cul-1.
Furthermore, we examined whether the substrate binding to the SCFβTrCP1 complex is necessary for Cul-1 modification by NEDD8. For this purpose, we deleted all seven WD40-repeat domains (termed FLAG-βTrCP1ΔW1–7) and transfected its cDNA into HEK 293 cells. As shown in Figure 2E, FLAG-βTrCP1ΔW1–7 formed complexes with Skp1 and Cul-1, because it retains the F-box domain. However, NEDD8-modified Cul-1 was not detected in anti-FLAG-immunoprecipitates with βTrCP1ΔW1–7, unlike βTrCP1. These results suggest that substrate binding to the SCFβTrCP1 complex may be necessary for Cul-1 modification by NEDD8, at least in vivo (see Discussion).
Stimulation of IκBα ubiquitylation by NEDD8-ylation is E2 dependent
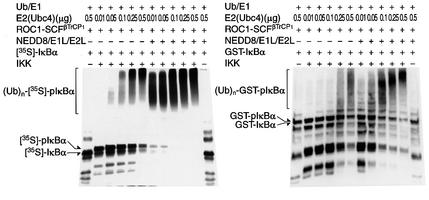
Considering the role of NEDD8-ylation in IκBα ubiquitylation, we next examined the effect of E2 (Ubc4). As shown in Figure 3 (left panel), augmentation of [35S]IκBα ubiquitylation by the NEDD8 system seemed to be dependent on the amount of E2 (Ubc4). Thus, in the presence of the NEDD8 system, very little E2 enzyme is required to exhibit the same effects as with high E2 concentration in the absence of the NEDD8 system. Similar dose-dependent effects of E2 on the ubiquitylation of GST–pIκBα were observed (Figure 3, right), suggesting that the role of the NEED8 system toward ROC1– SCFβTrCP1 may be related to the dose of E2.
Fig. 3. Dose-dependent effect of E2 (Ubc4) on the ubiquitylation of phosphorylated IκBα. The ubiquitylated assays of [35S]IκBα (left) and GST–IκBα (right) were performed as in Figure 1C and D, except for the use of various amounts of E2 (Ubc4) as indicated.
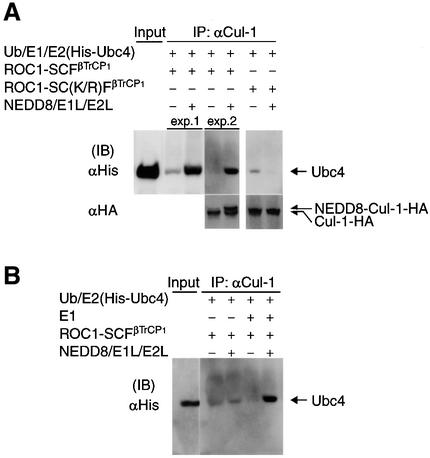
NEDD8-ylation enhances the recruitment of Ubc4 to ROC1–SCFβTrCP1 without affecting the association of ROC1–SCFβTrCP1 with pIκBα
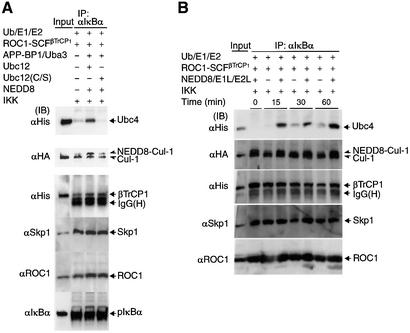
In Figure 2D, we showed that most of the endogenous SCFβTrCP1 bound to substrate (pIκBα) is modified by NEDD8, although this modification is not essential for the binding. To ascertain further that the NEDD8 system does not affect substrate binding of ROC1–SCFβTrCP1, the reaction was performed in the presence or absence of the NEDD8 system and followed by immunoprecipitation of IκBα and western blotting against SCF components. As shown in Figure 4A, almost the same amount of each component of the ROC1–SCFβTrCP1 complex was bound to pIκBα, irrespective of the presence or absence of the NEDD8 system, as expected.
Fig. 4. Effect of the NEDD8 system on recruitment of E2 (Ubc4) onto ROC1–SCFβTrCP1 bound to phosphorylated IκBα. The ROC1–SCFβTrCP1 complex was incubated at 25°C for 30 min in the presence or absence of the NEDD8 system together with Ub, E1 (Uba1) and E2 (Ubc4) for ubiquitylation, IκBα and IKK at the indicated combinations. After incubation, the reaction mixtures were treated with anti-IκBα antibody and the resulting immunoprecipitates were used for western blotting with various antibodies as indicated. ‘Input’ denotes various input controls (1/10 the amounts added in the assay mixtures). The effects of Ubc12(C/S) (A) and time course (B) on the binding of Ubc4 to ROC1–SCFβTrCP1 are shown.
Surprisingly, however, we found that the addition of the NEDD8 system caused a marked increase in the binding of His-tagged Ubc4 to pIκBα (Figure 4A), while little binding of E2 was observed in the absence of the NEDD8 system. This NEDD8-ylation-dependent binding of Ubc4 to pIκBα occurred in a time-dependent fashion (Figure 4B). Ubc12(C/S) had no profound effect on this Ubc4 recruitment. Moreover, UbcH7, which does not support pIκBα ubiquitylation, was not associated with pIκBα even when the NEDD8 system was also supplemented (data not shown).
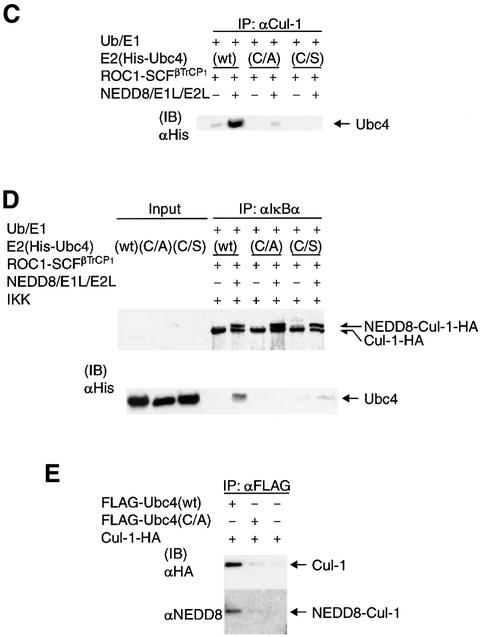
We next examined whether substrate binding of ROC1–SCFβTrCP1 is necessary for recruitment of E2. For this purpose, we performed immunoprecipitation analysis with anti-Cul-1 antibody in the absence of IκBα and IKK. As shown in Figure 5A, the NEDD8 system markedly promoted the binding of Ubc4 to ROC1–SCFβTrCP1, while no obvious binding of E2 was observed in the absence of the NEDD8 system. Furthermore, E2 was not significantly associated with the ROC1–SC(K/R)FβTrCP1 complex defective for NEDD8 modification (Figure 5A). These results suggest that Ubc4 is recruited directly to ROC1– SCFβTrCP1 in an NEDD8-ylation-dependent fashion without requiring the binding of the E3 (ROC1–SCFβTrCP1) to pIκBα. However, the purified ROC1–SCFβTrCP1 complex might have already associated with endogenous substrate(s) in insect cells, and substrate binding might be required for the assembly of ROC1–SCFβTrCP1. Thus, we believe that the substrate binding of ROC1–SCFβTrCP1 is required for its NEDD8 modification (shown in Figure 2E) and subsequent recruitment of E2.
Fig. 5. Effect of the NEDD8 system on recruitment of E2 (Ubc4) to the ROC1–SCFβTrCP1 complex. (A) Effect of Ubc4 binding to ROC1–SCFβTrCP1 and ROC1–SC(K/R)FβTrCP1. Experiments and symbols are similar to those described in Figure 4, except that IκBα and IKK were not added and immunoprecipitation was carried out with anti-Cul-1 antibody. Some experiments were conducted in duplicate. (B) Only Ub-linked Ubc4 was recruited onto ROC1–SCFβTrCP1. The experiment was similar to that described in (A) except that E1 (Uba1) was omitted from the assay mixture. (C) The NEDD8 system does not support the binding of Ubc4(C/A) and Ubc4(C/S) to ROC1–SCFβTrCP1. The experiment was similar to that described in (A), except that Ubc4(C/A) and Ubc4(C/S) were used for wild-type Ubc4 (wt). (D) The NEDD8 system does not support the binding of Ubc4(C/A) and Ubc4(C/S) to pIκBα. The experiment was similar to that described in Figure 5 using Ubc4(C/A) and Ubc4(C/S). (E) Ubc4, but not Ubc4(C/A), co-immunoprecipitates Cul-1 in 293 cells. Thirty-six hours after pcDNA3-Cul-1-HA with unmodified FLAG-Ubc4(wt) or FLAG- Ubc4(C/A) were transfected into 293 cells, crude extracts were prepared as described in Materials and methods. After immuno precipitation by anti-FLAG antibody, the resulting immunoprecipitates were analysed by western blotting using anti-HA antibodies.
We then tested whether the thioester linkage of Ub is necessary for recruitment of E2 (Ubc4) to the ROC1– SCFβTrCP1 complex. As shown in Figure 5B, in the absence of E1 (Uba1), Ubc4 did not bind to ROC1–SCFβTrCP1, even in the presence of the NEDD8 system. This finding strongly indicates that thioester linkage of Ub to E2 is required for recruitment of E2 to ROC1–SCFβTrCP1. This conclusion was confirmed further using Ubc4(C/A) and Ubc4(C/S), which cannot form a thioester linkage with Ub. Neither Ubc4(C/A) nor Ubc4(C/S) bound to ROC1–SCFβTrCP1 immunoprecipitated by either anti-Cul-1 antibody (Figure 5C) or anti-IκBα antibody (Figure 5D) in the presence of the NEDD8 system. Thus, we concluded that the NEDD8 system enhances the formation of Ub-linked E2–E3 complex, leading to efficient ubiquitylation of pIκBα. Finally, we examined whether Ubc4 specifically interacts with Cul-1 in living cells. For this, Cul-1-HA was co-expressed with FLAG-Ubc4 or FLAG-Ubc4(C/A) and their interaction investigated by immunoprecipitation analysis. As shown in Figure 5E, FLAG-Ubc4, but not FLAG-Ubc4(C/A), immunoprecipitated Cul-1, the band of which reacted with anti-NEDD8 antibody, suggesting that formation of a thioester linkage between Ubc4 with Ub may be necessary for the interaction with SCF whose Cul-1 is modified by NEDD8.
While the presence of Uba1 (E1 for ubiquitylation) was required for recruitment of E2, Uba1 was not bound to pIκBα or ROC1–SCFβTrCP1 even in the presence of the NEDD8 system (data not shown). Furthermore, the binding of APP-BP1 (a regulatory subunit of E1 for NEDD8-ylation) or Ubc12 to ROC1–SCFβTrCP1 was undetectable, irrespective of the presence or absence of the NEDD8 system (data not shown), suggesting that the mechanistic actions for ubiquitylation and NEDD8-ylation are not equivalent. Taken together, these results indicate that NEDD8-ylation of Cul-1 specifically enhances recruitment of E2 (Ubc4) to a target substrate (pIκBα) through E3 (ROC1–SCFβTrCP1), which contributes to the efficient transfer of Ub for ultimate ubiquitylation.
Discussion
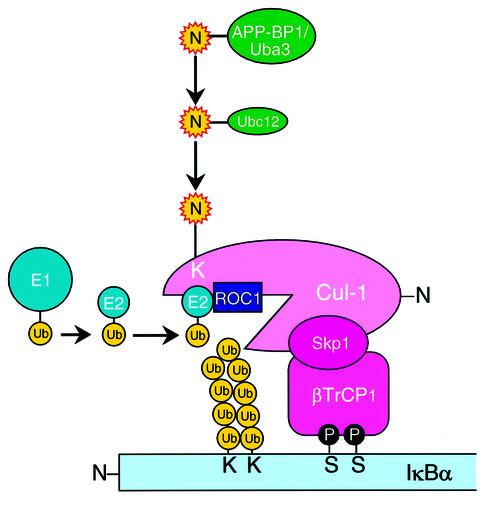
In the present study, we investigated the role of NEDD8 for Cul-1 in an in vitro reconstituted IκBα ubiquitylation system. We first showed that addition of the NEDD8 system resulted in the enhancement of polyubiquitylation of pIκBα phosphorylated by IKK. We also demonstrated that blockage of Cul-1 modification by NEDD8 with Cul-1(K/R) or Ubc12(C/S) did not enhance the ubiquitylation of pIκBα, indicating that NEDD8-ylation of Cul-1 truly activates the polyubiquitylation of pIκBα. Essentially similar results were reported recently by other groups for polyubiquitylation of pIκBα by SCFβTrCP1 (Furukawa et al., 2000; Read et al., 2000; Wu et al., 2000) and of p27Kip1 by SCFSkp2 (Morimoto et al., 2000; Podust et al., 2000). However, the molecular mechanism underlying the activation of SCF-Ub ligase by NEDD8 remains to be defined. To uncover the mechanistic role, we also demonstrated that the NEDD8 system enhanced the recruitment of Ub-conjugating enzyme (Ubc4) to the ROC1–SCFβTrCP1 E3 ligase complex to evoke efficient ubiquitylation of pIκBα. Indeed, the stimulatory effect of NEDD8 was evident more strikingly when there was a reduced supply of E2. Moreover, we found that a thioester-linkage between E2 and Ub is necessary for its binding to E3, because free E2 without supply of Ub from E1-Ub was dysfunctional for the binding to E3 and because Ubc4(C/S) and Ubc4(C/A) which are unable to trap Ub were not recruited to ROC1–SCFβTrCP1. In addition to these in vitro experiments, we showed that ectopically expressed Ubc4, but not Ubc4(C/A), interacted with endogenous NEDD8-linked Cul-1 in living cells. The results of our study allow us to design a hypothetical model for the role of the NEDD8 system in IκBα ubiquitylation (Figure 6). This model can solve for the first time the mechanistic role of the NEDD8 system in regulating the ROC1–SCFβTrCP1 E3 ligase complex.
Fig. 6. Schematic model for the role of the NEDD8 system in IκBα ubiquitylation by a ROC1–SCFβTrCP1 Ub ligase complex. For details, see text. Ub, ubiquitin; E1 (Uba1) (Ub-activating); and E2 (Ubc4) (Ub-conjugating) enzymes; N, NEDD8; p, phosphorylation of serine. APP-BP1/Uba3 and Ubc12, as E1- and E2-like enzymes, respectively, for the NEDD8 system. S and K, serine and lysine residues, respectively.
However, it remains uncertain whether the effect of NEDD8 modification influences the length of the Ub chain or the efficiency of polyubiquitylation. Indeed, Wu et al. (2000) reported that the bacterially expressed ROC1– Cul-1 complex stimulates Ub polymerization in the absence of a substrate. This role of NEDD8-ylation resembles that mediated by a new ubiquitylation factor termed ‘E4’, which is involved in polyubiquitin chain assembly (Koegl et al., 1999). However, we favour the notion that the NEDD8-ylation of SCF ligase promotes the efficiency of polyubiquitylation, since the addition of the NEDD8 system resulted in a significant reduction in the added substrate, namely pIκBα. Further studies are required to clarify this issue.
Recently, Lisztwan et al. (1998) reported the association of Cul-1 and Cdc34/Ubc3(E2) with the F-box protein p45Skp2. Furthermore, it is well known that several RING-finger-type Ub-protein E3 ligases can bind specific classes of E2s (Deshaies, 1999). However, in our hands, we could not detect any obvious binding between Ubc4 and ROC1–SCFβTrCP1 in vitro in the absence of the NEDD8 system. Consequently, our results seem to be quite rational; the NEDD8 system apparently assists the formation of a stable complex between E2 and E3. We do not know the exact mechanism underlying this enhanced binding directed by NEDD8-ylation, but we favour the notion that NEDD8 modification of Cul-1 presumably induces conformational changes in Cul-1, which may open the binding pocket for Ub-E2 (see our model in Figure 6). However, it is still not clear whether Ubc4 binds to ROC1 and/or Cul-1, because they are hardly expressed as soluble recombinant proteins, and thus we are unable to conduct direct binding experiments in vitro. It is, however, worth noting that a significant activity of the ROC1–SCFβTrCP1 complex was detected without the NEDD8 system. In this case, E2 may be associated transiently with the E3 ligase complex during the ubiquitylating process, but the E2–E3 association was not observed appreciably in the present immunoprecipitation analysis due to the instability of the presumptive active complex.
Our results showed that the ubiquitylation reaction for pIκBα proceeded as a function of time in the absence of the NEDD8 system and that the ROC1–SCFβTrCP1 complex exhibited the same thermostability in the absence of the NEDD8 system. These results imply that NEDD8-ylation of Cul-1 is not involved in preventing the inactivation of ROC1–SCFβTrCP1 activity. Similar findings were reported recently by other groups (Podust et al., 2000; Read et al., 2000). Moreover, NEDD8-ylation is not involved in the formation of the ROC1–SCFβTrCP1 complex, because Cul-1(K/R), which is defective for NEDD8-ylation, could assemble into the complex similarly to Cul-1 in vitro and in vivo. Importantly, Read et al. (2000) came to the same conclusions and further found that NEDD8-conjugated SCFβTrCP and unmodified SCFβTrCP had similar affinity, namely apparently similar Km values, for binding to pIκBα. Consistent with these observations, we also demonstrated that the NEDD8 system is not required for the binding of the ROC1–SCFβTrCP1 complex to target pIκBα, because NEDD8-ylation-defective ROC1– SCFβTrCP1 with Cul-1(K/R) could bind normally to pIκBα in vitro and in vivo. Alternatively, for the stimulatory effect of NEDD8-ylation, it is possible that NEDD8-ylation of Cul-1 prevents ubiquitylation of E2 (Ubc4 in the present study), linked to the SCF ligase and consequently increases the availability of E2 for ubiquitylation. However, as in the present reconstituted system, no obvious ubiquitylation of Ubc4 was observed, making this possibility unlikely under our conditions. The above findings are not in conflict with our proposed model of the role of the NEDD8-modifying system (Figure 6). Previously, we reported that all members of the human Cul-family proteins are modified by NEDD8 (Hori et al., 1999), indicating that NEDD8 conjugation to Cul-family proteins represents a general mechanism of activation of the ROC–Cul-based Ub-protein ligase.
It is of note that only a small portion of the total cellular pool of Cul-1 was modified by NEDD8. However, nearly half of endogenous Cul-1 associated with ectopically expressed βTrCP1, and thus SCFβTrCP1, was modified by NEDD8, and NEDD8-modified Cul-1 was the only form detected in association with phosphorylated IκBα, the SCF substrate in vivo (Read et al., 2000; this study). These observations indicate the existence of a regulatory mechanism of the NEDD8-modifying system in vivo, but the mechanism that regulates NEDD8 conjugation to Cul-1 is unknown at present. We demonstrated in the present study that substrate binding of SCF might be required for NEDD8 modification of Cul-1, which, however, apparently is contradictory to the in vitro observation that NEDD8-ylation occurs in a substrate-independent manner (Wu et al., 2000). A similar in vivo/in vitro difference was described for the requirement of ROC1 for NEDD8 conjugation to CUL-1, i.e. ROC1 is necessary for in vivo, but not in vitro, NEDD8 modification of Cul-1 (Furukawa et al., 2000). In addition, Podust et al. (2000) reported that a continuous supply of the NEDD8 system is required for p27Kip1 ubiquitylation in crude cell lysates, indicating the existence of certain isopeptidase(s) capable of hydrolysing NEDD8 from the NEDD8–Cul-1 conjugate, leading to cessation of activation of SCFSkip2 ligase activity. Taken together, it seems that Cul modification by NEDD8 is a regulated process, which may be tightly linked to the SCF function, although the underlying mechanism(s) remains to be identified.
It has been reported that deletion of the Rub1–NEDD8 ligation pathway in the budding yeast is viable and that modification of Cdc53 by Rub1 does not seem to be strictly essential for the function of SCFCDC4 in budding yeast (Lammer et al., 1998; Liakopoulos et al., 1998). In contrast, in fission yeast, we recently concluded that the NEDD8-modifying pathway is essential for cell viability and function of Pcu1 (Cul-1 orthologue), and thereby SCF. This conclusion was based on the finding that Pcu1K713R defective for NEDD8-ylation could not rescue growth arrest caused by deletion of pcu1+ and that forced expression of Pcu1K713R or depletion of NEDD8 in cells resulted in impaired cell proliferation and marked stabilization of the cyclin-dependent kinase (CDK) inhibitor Rum1, which is a substrate of the SCF complex (Osaka et al., 2000). In Arabidopsis thaliana, Axr1, an APP-BP1 homologue, plays a pivotal role in auxin response in synergy with SCF, but axr1 mutant cells remain viable; however, it is not clear at present whether its null mutation is lethal or not (Pozo et al., 1998; Gray and Estelle, 2000). In addition, the mutation of hamster SMC, encoding a protein almost identical to APP-BP1, is responsible for cell cycle defects in the hamster ts41 cell line (Chen et al., 2000). Recently, we also found that the NEDD8-conjugating pathway is essential for proliferation of mammalian cells, because deletion of the Uba3 gene encoding a catalytic subunit of NEDD8-activating enzyme in mice resulted in impaired cullin function and early embryonic death (K.Tateishi, M.Omata, K.Tanaka and T.Chiba, unpublished results). Taken together, these findings suggest that the NEDD8–Rub1-ligating pathway plays a critical role for the SCF complex in a variety of eukaryotic cells.
However, the reason why the NEDD8 system is essential in mammals and fission yeast but not in budding yeast is obscure (Lammer et al., 1998; Liakopoulos et al., 1998; Osaka et al., 2000). One possible explanation is that Cdc53 binds efficiently to E2 without NEDD8-ylation. This may be due to a structural difference between Cdc53 and Cul-1 or budding yeast may have other mechanism(s) to compensate for the loss of the NEDD8 system. Alternatively, it may be possible that the level of E2 in cells may differ among different organisms or, although unlikely, degradation of certain substrates by Cul-based E3 may be essential in some organisms but not in others. Genetic analyses have revealed an indispensable role for NEDD8-ylation for in vivo SCF function in various eukaryotes, except budding yeast, but biochemical analyses revealed that ROC1–SCFβTrCP1 or SCFSkp2 still retain significant activity without NEDD8-ylation (Morimoto et al., 2000; Podust et al., 2000; Read et al., 2000; Wu et al., 2000; this study). The reason is unknown at present, but it is possible that the level of thioester linkage of Ub to E2 is quite low in living cells, compared with an in vitro reconstitution system. Otherwise, it may be possible that another unknown protein(s), which is essential for cell viability, interacts with SCF in a NEDD8-dependent manner as E2-Ub does. For example, the essential gene named Sgt1 may be involved in the regulation of SCF activity in living cells (Kitagawa et al., 1999) although the addition of the human orthologue of Sgt1 had no effect on the IκBα ubiquitylation mediated by ROC1–SCFβTrCP1 (data not shown). Moreover, it was reported recently that Cks1/Suc1 is required for SCFSkp2-mediated ubiquitylation of p27Kip1 (Ganoth et al., 2001). Further studies nevertheless will help to clarify these unanswered issues.
Materials and methods
Construction of plasmids
Cul-1(K/R) (Lys696 in Cul-1 replaced by arginine), Ubc12(C/S) (Cys111 replaced by serine) and Ubc4(C/A) or (C/S) (Cys85 replaced by alanine or serine) mutants were synthesized by a PCR-assisted method using a site-directed mutagenesis kit (Stratagene, La Jolla, CA). For PCR amplification of human Ubc4, we used the sequence described by Rolfe et al. (1995). Note that there are two Cul-1 proteins; the longer one has a short stretch with an extra 24 amino acids near the N-terminal region of the shorter one, and, therefore, the NEDD8-ylated K696 of the latter Cul-1 corresponds to K720 of the former, as reported (see Morimoto et al., 2000; Read et al., 2000; Wu et al., 2000). The deletion mutant FLAG-βTrCP1ΔW1–7 (reisdues 260–569 deleted; see Suzuki et al., 2000) was amplified by PCR with appropriate primers. Plasmids for the expression of N- and C-terminally HA- or FLAG-tagged proteins were ligated into pcDNA3.1(+) vector (Invitrogen).
Cell cultures, transfections, immunoprecipitation and western blotting
The method used for culture of human HEK 239 cells, which subsequently were used for transfections, has been described previously (Suzuki et al., 2000). In brief, transfections were performed using the FuGENE 6 transfection reagent. MG132 (Z-Leu-Leu-Leu-H) at 50 µM and okadaic acid at 0.25 µM were pre-treated prior to TNF-α stimulation. Preparation of cell lysates and immunoprecipitation with various antibodies were performed as described before (Suzuki et al., 2000). For western blotting, the reaction mixtures were separated by SDS–PAGE and transferred to a PVDF membrane. The membranes were probed with various antibodies and visualized with horseradish peroxidase-conjugated second antibody using the enhanced chemiluminescence detection system (Amersham Pharmacia Biotech). The antibodies used this study were as described previously (Suzuki et al., 2000).
Preparation of recombinant proteins
Recombinant His-Ubc4, His-Ubc12, His-Ubc12(C/S), His-NEDD8, His-Ubc4(C/A), Ubc4(C/S) and GST–IκBα were produced in E.coli. Recombinant E1 (Uba1), His-APP-BP1/T7-Uba3, ROC1–SCFβTrCP1 and ROC1–SC(K/R)FβTrCP1 [T7-ROC1/FLAG-Skp1/HA-Cul-1 or HA-Cul-1(K/R)/His-βTrCP1] were produced from baculovirus-infected HiFive insect cells. These proteins, which are all human species except mouse E1, were affinity purified using a His Trap column (Pharmacia Biotech). Of these, APP-BP1/Uba3 acting as an E1 for NEDD8-ylation and the ROC1–SCFβTrCP1 complex functioning as an E3 for IκBα ubiquitylation were generated by simultaneously infecting two or four baculoviruses, respectively.
In vitro IκBα ubiquitylation and immunoprecipitation analysis
A 2 µl aliquot of in vitro translated 35S-labelled IκBα or 0.5 µg of GST–IκBα were pre-incubated at room temperature for 30 min with 25 ng of IKK in the presence of an ATP-regenerating system, followed by further incubation at 37°C for 0.5–3 h in a reaction volume of 20 µl containing 100 ng of recombinant mouse E1, 0.01–0.5 µg of Ubc4, 1 µg of ROC1–SCFβTrCP1 and 10 µg of bovine Ub. The reaction was carried out in the presence or absence of the NEDD8 system consisting of NEDD8 (10 µg), APP-BP1/Uba3 (0.5 µg) and Ubc12 (0.5 µg) that had been pre-incubated with ROC1–SCFβTrCP1 for 15 min at 25°C. After terminating the reaction by the addition of 8 µl of SDS–PAGE sample buffer, the boiled supernatant was separated by 10–20% SDS–PAGE and visualized by autoradiography or western blotting with anti-IκBα (mouse monoclonal IgG1, Santa Cruz Biotechnology, Santa Cruz, CA) (Chemiluminescence). The reaction mixtures were also used for immunoprecipitation with anti-IκBα antibodies (rabbit polyclonal IgG, Santa Cruz Biotechnology) or anti-Cul-1 serum raised in rabbits (Suzuki et al., 2000), followed by western blotting with various antibodies. IKK was purified from 293 cell extracts, as described by Lee et al. (1997).
Acknowledgments
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas (Intracellular Proteolysis) from the Ministry of Education, Science, Sports and Culture of Japan.
References
- Baldwin A.S. (1996) The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol., 14, 649–681. [DOI] [PubMed] [Google Scholar]
- Chen Y., McPhie,D.L., Hirschberg,J. and Neve,R.L. (2000) The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S–M checkpoint and causes apoptosis in neurons. J. Biol. Chem., 275, 8929–8935. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and Cullin/RING-H2-based ubiquitin-ligases. Annu. Rev. Cell. Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Finco T.S. and Baldwin,A.S. (1995) Mechanistic aspects of NF-κB regulation: the emerging role of phosphorylation and proteolysis. Immunity, 3, 253–272. [DOI] [PubMed] [Google Scholar]
- Furukawa M., Zhang,Y., McCarville,J., Ohta,T. and Xiong,Y. (2000) The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification and ubiquitin ligase activity of CUL1. Mol. Cell. Biol., 20, 8185–8197.11027288 [Google Scholar]
- Ganoth D., Bornstein,G., Ko,T.K., Larsen,B., Tyers,M., Pagano,M. and Hershko,H. (2001) The cell-cycle regulatory protein Cks1 is required for SCFSkp2-mediated ubiquitinylation of p27. Nature Cell Biol., 3, 321–324. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May,M. and Kopp,E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- Gong L. and Yeh,E.T.H. (1999) Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem., 274, 12036–12042. [DOI] [PubMed] [Google Scholar]
- Gray W.M. and Estelle,M. (2000) Function of the ubiquitin–proteasome pathway in auxin response. Trends Biochem. Sci., 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1998) There’s the Rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev., 12, 901–907. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (2000) Evolution and function of ubiquitin-like protein-conjugation system. Nature Cell Biol., 2, E153–E157. [DOI] [PubMed] [Google Scholar]
- Hori T., Osaka,F., Chiba,T., Miyamoto,C., Okabayashi,K., Shimbara,N., Kato,S. and Tanaka,K. (1999) Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene, 18, 6829–6834. [DOI] [PubMed] [Google Scholar]
- Jentsch S. and Pyrowolakis,G. (2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol., 10, 335–342. [DOI] [PubMed] [Google Scholar]
- Kamura T. et al. (1999a) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science, 284, 657–661. [DOI] [PubMed] [Google Scholar]
- Kamura T., Conrad,M.N., Yan,Q., Conaway,R.C. and Conaway,J.W. (1999b) The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev., 13, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K., Skowyra,D., Elledge,S.J., Harper,J.W. and Hieter,P. (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell, 4, 21–33. [DOI] [PubMed] [Google Scholar]
- Kipreos E.T and Pagano,M. (2000) The F-box protein family. Genome Biol., 1, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M., Hoppe,T., Schlenker,S., Ulrich,H.D., Mayer,T.U. and Jentsch,S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Lammer D., Mathias,N., Laplaza,J.M., Jiang,W., Liu,Y., Callis,J., Goebl,M. and Estelle,M. (1998) Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev., 12, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F.S., Hagler,J., Chen,Z.J. and Maniatis,T. (1997) Activation of the IκBα kinase complex by MEKK1, a kinase of the JNK pathway. Cell, 88, 213–222. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D., Doenges,G., Matuschewski,K. and Jentsch,S. (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J., 17, 2208–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J.A., Marti,A., Sutterluty,H., Gstaiger,M., Wirbelauer,C. and Keck,W. (1998) Association of human Cul-1 and ubiquitin-conjugating enzyme Cdc34 with the F-box protein p45 (Skp-2): evidence for evolutionary conservation in the subunit composition of the Cdc34–SCF pathway. EMBO J., 17, 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T. (1999) A ubiquitin ligase complex for the NF-κB, Wt/Wingless and Hedgehog signaling pathways. Genes Dev., 13, 505–510. [DOI] [PubMed] [Google Scholar]
- May M.J. and Chosh,S. (1999) IκB kinases: kinsmen with different crafts. Science, 284, 271–273. [DOI] [PubMed] [Google Scholar]
- Morimoto M., Nishida,T., Honda,R. and Yasuda,H. (2000) Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCFskp2 toward p27kip1. Biochem. Biophys. Res. Commun., 270, 1093–1096. [DOI] [PubMed] [Google Scholar]
- Ohta T., Michel,J.J., Schottelius,A.J. and Xiong,Y. (1999) ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell, 3, 535–541. [DOI] [PubMed] [Google Scholar]
- Osaka F., Kawasaki,H., Aida,N., Saeki,M., Chiba,T., Kawashima,S., Tanaka,K. and Kato,S. (1998) A new NEDD8-ligating system for cullin-4A. Genes Dev., 12, 2263–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F. et al. (2000) Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J., 19, 3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust V.N., Brownell,J.E., Gladysheva,T.B., Luo,R.-S., Wang,C., Coggins,M.B., Pierce,J.W., Lightcap,E.S. and Chau,V. (2000) A Nedd8 conjugating pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl Acad. Sci. USA, 97, 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo J.C., Timpte,C., Tan,S., Callis,J. and Estelle,M. (1998) The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science, 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Read M. et al. (2000) Nedd8 modification of Cul-1 activates SCFβTrCP-dependent ubiquitination of IκBα. Mol. Cell. Biol., 20, 2326–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe M., Beer-Romero,P., Glass,S., Eckstein,J., Berdo,I., Theodoras,A., Pagano,M. and Draetta,G. (1995) Reconstitution of p53-ubiquitinylation reactions from purified components: the role of human ubiquitin-conjugating enzyme UBC4 and E6-associated protein (E6AP). Proc. Natl Acad. Sci. USA, 92, 3264–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol J.H. et al. (1999) Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev., 13, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Chiba,T., Suzuki,T., Fujita,T., Ikenoue,T., Omata,M., Furuichi,K., Shikama,H. and Tanaka,K. (2000) Homodimer of two F-box proteins βTrCP1 or βTrCP2 binds to IκBα for signal-dependent ubiquitination. J. Biol. Chem., 275, 2877–2884. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Strack,P., Beer-Romero,P., Chu,C.Y., Elledge,S.J. and Harper,J.W. (1999) The SCFβTRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev., 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Chen,A. and Pan,Z.-Q. (2000) Conjugation of Nedd8 to CUL1 enhances the activity of the ROC1–CUL1 complex. J. Biol. Chem., 275, 32317–32324. [DOI] [PubMed] [Google Scholar]
- Yaron A. et al. (1998) Identification of the receptor component of the IκBα–ubiquitin-ligase. Nature 396, 590–594. [DOI] [PubMed] [Google Scholar]
- Yeh E.T.H., Gong,L. and Kamitani,T. (2000) Ubiquitin-like proteins: new wines in new bottles. Gene, 248, 1–14. [DOI] [PubMed] [Google Scholar]