Abstract
A group of specialized genes has been defined to govern the molecular mechanisms controlling the circadian clock in mammals. Their expression and the interactions among their products dictate circadian rhythmicity. Three genes homologous to Drosophila period exist in the mouse and are thought to be major players in the biological clock. Here we present the generation of mice in which the founding member of the family, Per1, has been inactivated by homologous recombination. These mice present rhythmicity in locomotor activity, but with a period almost 1 h shorter than wild-type littermates. Moreover, the expression of clock genes in peripheral tissues appears to be delayed in Per1 mutant animals. Importantly, light-induced phase shifting appears conserved. The oscillatory expression of clock genes and the induction of immediate-early genes in response to light in the master clock structure, the suprachiasmatic nucleus, are unaffected. Altogether, these data demonstrate that Per1 plays a distinct role within the Per family, as it may be involved predominantly in peripheral clocks and/or in the output pathways of the circadian clock.
Keywords: circadian rhythm/clock gene/knock-out mouse/light response/Per1
Introduction
Most organisms display rhythms in various aspects of their physiology (Dunlap, 1999; Cermakian and Sassone-Corsi, 2000). Circadian rhythms, i.e. rhythms with a period close to 24 h, in animals are controlled by biological clocks located in the central nervous system, but also in peripheral non-neuronal tissues (Brown and Schibler, 1999). In mammals, a central clock controlling a large number of rhythms is located in neurons of the suprachiasmatic nucleus (SCN) of the anterior hypothalamus (Klein et al., 1991). Circadian clocks can function autonomously, independently of any external time cues, but can be reset by environmental cues, such as day–night cycles. The circadian system can be divided into three conceptual components (Eskin, 1979; Brown and Schibler, 1999): the clock itself, or pacemaker, which generates rhythmicity autonomously; the input pathways, through which the clock responds to external (e.g. environmental) signals; and the output pathways, which allow rhythmic information to be spread throughout the body and thereby regulate the animal’s physiology.
A number of genes involved in the clock mechanism have been isolated in recent years, and models have emerged to explain how their interconnected functions are able to sustain circadian rhythms (Dunlap, 1999; Cermakian and Sassone-Corsi, 2000). A basic molecular feature of all circadian clocks is the use of feedback regulatory loops, generally operating at the level of transcription. In Drosophila, the transcription factors encoded by the clock (clk) and cycle (cyc) genes dimerize and activate the expression of the timeless (tim) and period (per) genes (Williams and Sehgal, 2001). The protein products of tim and per accumulate in the cytoplasm and then dimerize, enter the nucleus and repress their own transcription through inhibition of the binding of the CLK–CYC dimer to specific regulatory sites. In addition to this negative feedback loop, a positive regulatory loop appears to involve the stimulation by PER and TIM of clk expression by relieving the negative regulation of this gene by CLK itself (Glossop et al., 1999). Mutations in any of these genes can produce flies that are either arrhythmic or display rhythms with an abnormal period (Williams and Sehgal, 2001).
Three homologs of per were found in mammals (Albrecht et al., 1997; Shearman et al., 1997; Zylka et al., 1998), termed Per1, Per2 and Per3. These are also thought to be part of a negative feedback loop, but the dimerization partners are rather the products of the Cryptochrome (Cry)1 and Cry2 genes (Griffin et al., 1999; Kume et al., 1999; Shearman et al., 2000b). Furthermore, similarly to what is observed in Drosophila, Per2 positively regulates the Bmal1 gene (Shearman et al., 2000b), the mammalian homolog of Drosophila cyc. Per1 and Per2 also appear to be part of the input pathways as their expression is induced in the SCN after light stimulation of the mouse (Albrecht et al., 1997; Shearman et al., 1997), whereas it is reduced by non-photic stimuli (Maywood et al., 1999; Horikawa et al., 2000) such as arousal by presentation of a new wheel. Targeted disruption of the mPer2 gene produces mice with strongly abnormal behavioral rhythms in constant conditions (Zheng et al., 1999): these mutant animals display short period rhythms and eventually become arrhythmic. Moreover, the amplitude of expression of clock genes in the SCN is severely blunted. mPer3 knock-out mice have a milder phenotype, as the only circadian phenotype uncovered to date is a period of activity rhythms slightly shorter than normal (Shearman et al., 2000a). The targeted mutation of the mouse Per1 gene has not been reported yet.
Here we describe the generation of Per1-null mice. The analysis of the circadian phenotype of these animals reveals that they display a shorter period of activity–rest cycles in constant conditions compared with wild-type littermates. Moreover, while the phase of clock gene expression in peripheral tissues is affected, this is not the case in the SCN. Finally, light-induced c-fos expression in the SCN and phase shifting of the clock are unaffected in the mutant animals. Altogether, these results suggest that Per1 function lies at the level of peripheral clocks and/or in the output pathways originating from the circadian pacemaker.
Results
Generation of Per1-null mice
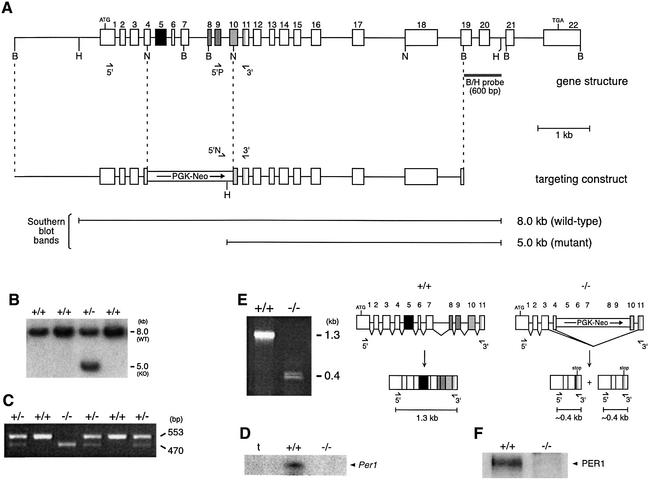
In order to elucidate the physiological function of the Per1 gene, we generated mutant mice with this gene disrupted by homologous recombination. We constructed a targeting vector in which a portion of the gene encompassing exons 4–10 was replaced by a phosphoglycerate kinase (PGK)– neomycin (Neo) cassette (Figure 1A). The targeted region would normally encode the whole PAS domain (PAS A and PAS B repeats) and part of the downstream PAC motif (Ponting and Aravind, 1997). The PAS domain is a structural and functional feature of PER proteins and of a number of clock and non-clock proteins that was shown to be essential for dimerization and regulatory functions (Huang et al., 1993; Crews, 1998). The construct was transfected into embryonic stem (ES) cells (129/Sv), and a clone that had undergone homologous recombination for one of the Per1 alleles (Figure 1B) was used to generate chimeric mice (C57BL/6 × 129/Sv). Intercrossing of heterozygous F0 offspring generated wild-type, heterozygous and homozygous mutant animals (Figure 1C) with a normal Mendelian ratio (234 +/+, 237 –/–, 386 +/–; thus 27, 28 and 46%, respectively). Both heterozygous and homozygous mutant animals, males and females, are fertile, and do not present any obvious anatomical defect (data not shown). RNase protection assay (RPA) using an RNA probe complementary to a part of the PAS domain confirmed that this region was absent in –/– mice (Figure 1D). However, Per1 transcripts were still present in these animals, as shown in RPAs and in situ hybridizations with a probe corresponding to an upstream region of the mPer1 mRNA (see Figures 3–5) and by RT–PCR. This latter technique was used to show that transcripts in which the Neo cassette and the rest of exon 4 and 10 are spliced out are expressed in –/– mice, introducing an in-frame stop codon (Figure 1E). Western analysis of protein extracts from these mice confirmed that the PER1 protein is absent (Figure 1F).
Fig. 1. Generation of Per1 knock-out mice. (A) Per1 gene structure. Numbered boxes are the exons. The position of BamHI (B), HindIII (H) and NcoI (N) restriction sites is shown, as well as the initiator and stop codons. Regions of boxes corresponding to the PAS A, PAS B and PAC regions are in black, gray and light gray, respectively. The targeting construct was generated by replacing an NcoI fragment by a PGK–Neo expression cassette and includes 2.5 and 4.5 kb of gene sequences upstream and downstream of the cassette, respectively. The regions complementary to the PCR or RT–PCR primers and to the Southern hybridization probe are indicated. Below is presented the size of the fragments hybridized with the ‘B/H probe’ in Southern hybridization on a HindIII digestion of the genomic DNA. (B) Isolation of an ES cell clone in which homologous recombination took place (third lane). (C) Genotyping of the mice by PCR. PCR is performed on DNA prepared from tail biopsies, using a cocktail of primers ‘5′P’, ‘5′N’ and ‘3′’ shown in (A). A 553 bp band is amplified for wild-type animals, a 470 bp band for homozygous mutants and both bands for heterozygous animals. (D) RNase protection assay on total RNA from wild-type and knock-out animals, using a riboprobe complementary to a part of the PAS domain. tRNA alone was used as a control (t). (E) RT–PCR on total RNA from wild-type and knock-out animals, using the primers 5′ and 3′ shown in (A). In wild-type animals, the expected 1.3 kb band is amplified. In knock-out animals, two smaller bands are obtained; sequence analysis reveals that they correspond to splicing products in which the Neo cassette (and exon 4 and 10; a piece of exon 4 remains in the larger product) was removed (see scheme). As a consequence, an in-frame stop codon arises just downstream of the splice site; so the only possible proteins could not extend beyond exon 3. (F) Western blot on embryonic fibroblasts derived from wild-type and knock-out embryos, using an antibody raised against a C-terminal peptide of PER1.
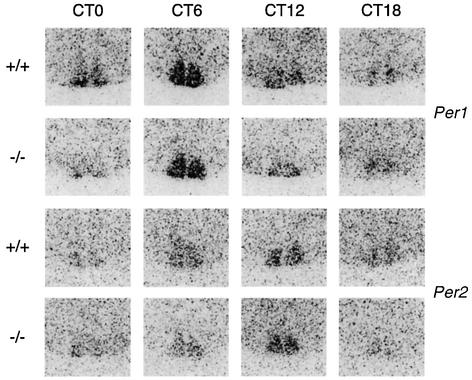
Fig. 3. Circadian expression of Per1 and Per2 is unaffected in the SCN of Per1 knock-out mice. In situ hybridization with Per1 and Per2 probes on brain cuts from animals entrained on a L12:D12 cycle and dissected at the indicated circadian times (CT) on the second day in DD. Only the region of the SCN is shown.
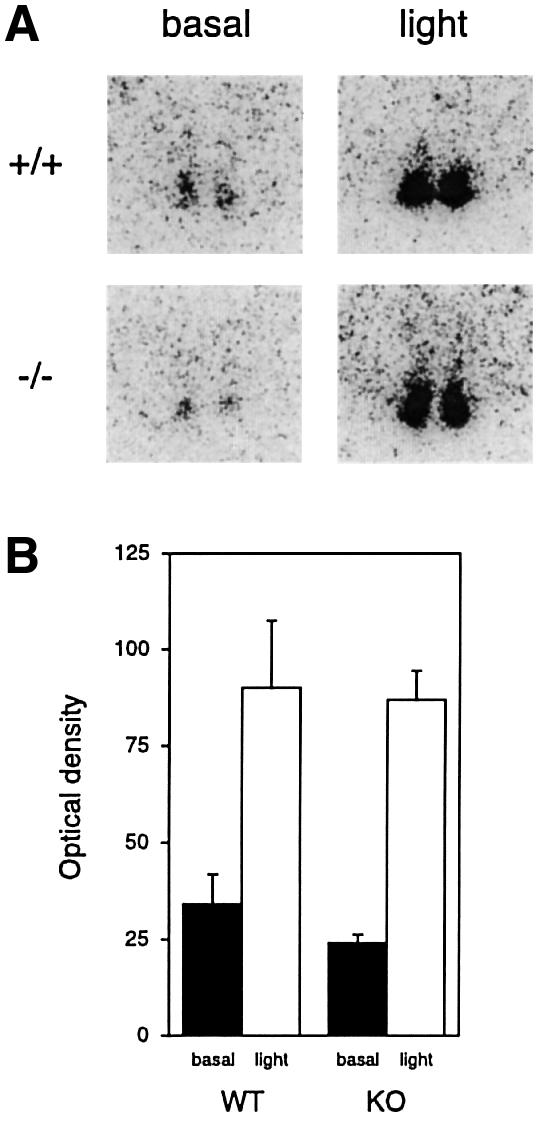
Fig. 5. Light-induced c-fos expression in Per1 knock-out mice. (A) In situ hybridization with a c-fos probe on brain cuts from animals entrained on a L12:D12 cycle. At CT14 on the third day in DD, the mice were either subjected to light for 15 min and kept another 15 min in the dark (light) or kept all this time in the dark (basal), before dissection. Only the region of the SCN is shown. (B) Quantification of the results (n = 3 for ‘basal’, n = 4 for ‘light’).
Short free-running period of behavioral rhythms in Per1 knock-out mice
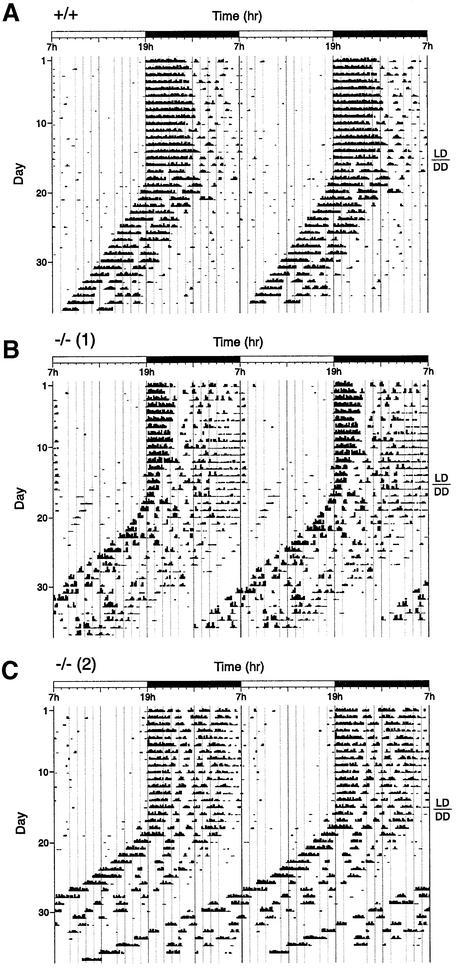
We used wheel-running measurements to assess the circadian rhythmicity of the activity of the Per1–/– mice, as compared with their wild-type littermates. Mice were housed individually in cages with running wheels and entrained on a regime of 12 h of light and 12 h of darkness (L12:D12 or LD), before being put in constant darkness (DD) in order to analyze their free-running behavior. Their activity was recorded and plotted in actograms (Figure 2). Three experiments with different groups of animals were conducted (Table I). In all three experiments, Per1–/– mice exhibited a shorter free-running period than their wild-type littermates (Table I). This difference ranged from 0.69 to 0.77 h and is statistically significant (Student’s t-test, p <0.05). In some cases (two mice in group 1, one mouse in group 2), the free-running behavior of the mutant animals could be divided into two phases, the period of the activity–rest cycle becoming suddenly shorter after an extended number of days in constant darkness (after 10–16 days; Figure 2C). Besides the difference in the free-running period, no other difference between wild-type and mutant animals could be uncovered in these three running wheel experiments. Mutant mice were entrained precisely and rapidly by LD cycles, and the average levels of wheel-running activity did not differ significantly between genotypes (data not shown).
Fig. 2. Per1 knock-out mice display a short free-running period of locomotor activity rhythms. Representative actograms for one wild-type (A) and two knock-out (B and C) animals. Animals were housed independently in cages with running wheels, and they were entrained on a light–dark cycle with 12 h of light and 12 h of darkness (LD). After 15 days, they were put in constant darkness (DD). Black bars represent the number of turns in 10 min. The plots were duplicated for clarity. Measurement of the free-running period was based on the onset of activity in DD (see Table I). In some mutant animals, the free-running period became even shorter after an extended time in DD [example in (C)].
Table I. Mean values for the free-running period (τ) of wild-type and Per1 knock-out mice in running wheel experiments.
| Group no. | Genetic background and sex | τ (h) +/+ | τ (h) –/– | Student’s t-test |
|---|---|---|---|---|
| 1 | 50% C57BL/6 | 24.13 ± 0.23 | 23.40 ± 0.09a | p = 0.0002a |
| 50% 129/Sv females | (n = 5) | (n = 5) | ||
| 2 | 50% C57BL/6 | 23.47 ± 0.39 | 22.70 ± 0.46b | p = 0.011b |
| 50% 129/Sv males | (n = 6) | (n = 6) | ||
| 3 | 94% C57BL/6 | 23.74 ± 0.20 | 23.15 ± 0.37 | p = 0.007 |
| 6% 129/Sv males | (n = 6) | (n = 6) |
aThese values include the initial τ (23.31 and 23.51 h) of the two animals with a two-phase free-running behavior (see text). Using the τ observed after extended DD conditions (22.27 and 23.20 h, respectively), the mean τ for this group is 23.13 ± 0.50 h (p = 0.004).
bThese values include the initial τ (22.93 h) of the animal with a two-phase free-running behavior (see text). Using the τ observed after extended DD conditions (22.00 h), the mean τ for this group is 22.55 ± 0.51 h (p = 0.006).
It is known that the genetic background can profoundly affect the circadian behavior of mice (Oliverio and Malorni, 1979; Schwartz and Zimmerman, 1990). For this reason, we backcrossed the initial knock-out line (50% C57BL/6, 50% 129/Sv) with C57BL/6 mice. This latter strain is known to exhibit more clear-cut and less variable patterns of daily rhythmicity, and thus to be more suitable for circadian behavioral experiments (Oliverio and Malorni, 1979; Schwartz and Zimmerman, 1990). Our results show that quite similar values for the free-running period (τ) (Table I, compare groups 2 and 3) were obtained whether mice with 50% C57BL/6 or 94% C57BL/6 background were used. This is consistent with previous reports demonstrating that the C57BL/6 background is genetically dominant for circadian activity tests (Schwartz and Zimmerman, 1990), and strengthens our conclusion that Per1 knock-out mice have a shorter free-running period than normal.
Altered Per transcript expression in peripheral tissues
We next sought to determine whether the differences in behavioral activity could be paralleled to clock gene expression in different tissues. Figure 3 presents the expression of the Per1 and Per2 genes in the SCN in mice entrained under a L12:D12 cycle and then placed in DD for 2 days. As previously shown, both transcripts oscillate in the central clock structure, and peak during the subjective day (Albrecht et al., 1997; Shearman et al., 1997). No difference in the phase or the amplitude of these oscillations with respect to wild-type littermates could be observed in these experiments (Figure 3).
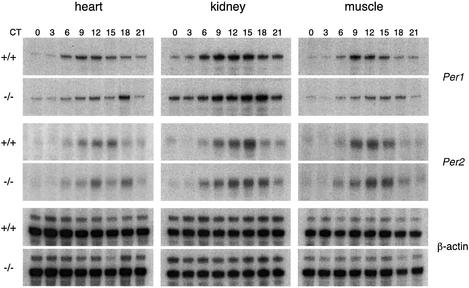
Per genes have been shown to oscillate in various non-neuronal peripheral tissues (Oishi et al., 1998; Zylka et al., 1998). The expression of Per1 and Per2 in kidney, heart and skeletal muscle of Per1-null mice and their wild-type littermates was analyzed by quantitative RPAs (Figure 4). As previously reported, the expression of Per genes in these tissues is delayed compared with that in the SCN, with a broad peak by the end of the subjective day or the first part of the subjective night (Zylka et al., 1998). Although Per1 and Per2 genes are expressed with circadian rhythmicity and with a similar amplitude between the two sets of mice, the peak of expression is delayed or broadened (declining later in the night) in all three tissues of the Per1-null mice (Figure 4).
Fig. 4. Delay in the expression of Per1 and Per2 in peripheral tissues of knock-out animals. RNase protection assays on total RNA from heart, kidney or skeletal muscle from wild-type or knock-out mice entrained on a L12:D12 cycle and dissected at the indicated circadian times (CT) on the third day in DD. A β-actin probe was used as a control for the amount of RNA. In all experiments, a tRNA control was used (not shown).
Light-induced c-fos expression and clock phase shifting in Per1-null mice
In order to investigate the possible involvement of Per1 in light response pathways in the SCN, mice were entrained on a L12:D12 cycle and, after being placed in DD, a 15 min light pulse was administered at circadian time (CT)14 on the second subjective night. After an additional 15 min in the dark, mice were killed and the brains processed for in situ hybridization. The c-fos gene was shown previously to be strongly induced in the SCN following light stimulation of the animal during the subjective night (Kornhauser et al., 1990), i.e. when this stimulation triggers phase shifting of the clock (Daan and Pittendrigh, 1976). As shown in Figure 5, c-fos induction in response to light is unaffected in Per1–/– mice.
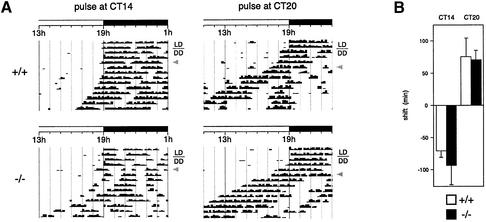
Per1 was suggested to be involved in the SCN response to environmental stimuli, given the observation that this gene is induced quickly in this clock structure after a light pulse administered during the subjective night (Albrecht et al., 1997; Shearman et al., 1997). Since a light stimulation during the night causes a phase shift of the clock (Daan and Pittendrigh, 1976), we analyzed the behavioral response of our Per1-null mice in response to this stimulus. Mice were housed in running-wheel cages, entrained on a L12:D12 cycle, placed in DD for 2 days and then subjected to a 30 min light pulse at either CT14 or CT20. Activity continued to be recorded and the extent of the phase shift (delay at CT14, advance at CT20) was calculated (Figure 6). Both wild-type and mutant animals displayed a phase shift in their activity rhythms (Figure 6A), the extent of phase shifting being the same in each case (Figure 6B). The phase shift occurred as quickly in the mutant mice as in the wild-type (data not shown).
Fig. 6. A light pulse causes a phase shift of locomotor activity rhythms in Per1 knock-out mice. (A) Representative actogram of mice entrained on a L12:D12 cycle and put for 2 or 3 days in constant darkness (DD), before a light pulse (gray arrowhead) was given at CT14 (left) or CT20 (right), causing a phase delay or advance, respectively, on the following days. Only part of the full actogram is shown. (B) Quantification of the extent of the phase shifts (n = 6 for each genotype, mice of group 2 in Table I). By convention, delays are negative and advances are positive.
Discussion
Current knowledge of molecular mechanisms of mammalian circadian clocks is based on a number of experimental approaches. A powerful tool to gain insight into the real physiological functions of clock genes and the way they contribute to circadian rhythmicity in vivo is the generation of mice carrying a targeted mutation for a given clock gene. We have generated a line of mice deficient for the founding member of the mPer gene family, mPer1. These mutant animals are rhythmic, but they present defects in activity rhythms, i.e. a short period, and in clock gene expression in peripheral tissues, i.e. a delay in the decline of their expression during the night.
The prevalent working model of molecular clocks implicates the products of Per genes, at least Per1 and Per2, in the transcriptional feedback loops generating circadian oscillations of cellular functions (Dunlap, 1999; Cermakian and Sassone-Corsi, 2000; Shearman et al., 2000b). PER1 and PER2 proteins are thought to associate with the CRY proteins (products of the Cry1 and Cry2 genes) and inhibit CLOCK/BMAL1-mediated transcription (Griffin et al., 1999; Kume et al., 1999). An additional role for PER2 in activating Bmal1 expression has also been proposed (Shearman et al., 2000b). It is also known that a light pulse induces an up-regulation of Per1 gene expression in the SCN (Albrecht et al., 1997; Shearman et al., 1997), an increase that is thought to change the balance of clock proteins within SCN neurons, and thereby affects the phase of the clock.
Our results question this simplistic model in two ways. As Per1 has been postulated to have a role in the inhibition of CLOCK/BMAL1-controlled transcription, then mutants of Per1 should impair nuclear entry of CRY and PER proteins, and thus diminish their negative action on transcription. Therefore, Per1-null mice should have longer periods. However, although the phase of Per1 and Per2 expression in peripheral tissues during the third day in DD is delayed in the mutant mice, we observe a shorter period of activity–rest cycles in running wheel experiments. It would seem as if the absence of PER1 protein leads to a better inhibition, i.e. a more efficient negative limb of the negative feedback loop. Our results suggest that additional elements would need to be added to the classical model for the function of Per1 in the circadian system. A possible scenario is that within the multisubunit complex of clock proteins considered as the ‘negative limb’ of the circadian loop, PER1 may decrease the stability of other PER proteins or of CRYs. As a consequence, the concentration of these proteins could reach higher levels in Per1 knock-out animals, thereby compensating for the lack of PER1.
Another important aspect of the results presented here deals with the response to light in the SCN. From these results, it appears that the phase shifting of the clock in response to light is unaffected in the Per1-null mice. This is unexpected, given that the Per1 gene responds quickly to the light stimulation in the SCN (Albrecht et al., 1997; Shearman et al., 1997), and also with respect to data suggesting that light-induced Per1 expression is involved in phase shift of locomotor activity cycles and SCN firing rhythms (Akiyama et al., 1999). Thus, the exact role of Per1 light-induced expression is still unclear. Alterna tively, the defect in Per1 may be compensated by another clock gene.
Among natural clock mutants and knock-out mice known, there are only two examples of animals that are completely arrhythmic in constant conditions: the mouse mutant for the gene encoding the BMAL1 factor (Bunger et al., 2000), and the double Cry1/Cry2 knock-out mouse (van der Horst et al., 1999) (Cry1 or Cry2 single mutants have a short and long period, respectively). Mice with a point mutation in the Clock gene (mutation that leads to the skipping of one exon) are almost arrhythmic in constant conditions, but the mice nevertheless display some circadian rhythmicity in running wheels for several days after switching to DD conditions (King et al., 1997). Per2 mutant mice (lacking part of the PAS B repeat) exhibit very unstable rhythms: their free-running period is very short, and the mice become arrhythmic after a variable number of days (Zheng et al., 1999). Finally, the Per3-null mice exhibit a mild phenotype: their free-running period of activity rhythms is slightly shorter than normal (Shearman et al., 2000a).
Among the Per mutant mouse lines (the published Per2 and Per3 mice and the Per1 mice described here), none is completely arrhythmic. This can be explained in two ways. First, there could be partial redundancy in the function of these genes, and the non-mutated genes could thus partially compensate for the loss of the third one. Secondly, it could be that PER genes are not involved in establishing and maintaining rhythms, but rather in fine-tuning the activity of the bona fide feedback loop components. Indeed, it has been suggested that the major negative element in the negative feedback loop is the CRY proteins (Shearman et al., 2000b), and not the PER proteins, as originally thought by analogy with the Drosophila system. It has also been suggested that Per3 may be part of the output pathways rather than a component of the pacemaker itself (Shearman et al., 2000a). The defects observed in running-wheel activity and in Per gene expression in peripheral tissues in Per1-null mice may indeed be due to dysfunction of the output pathways in these animals. Another attractive possibility is that Per1 is involved specifically in the physiological rhythmicity of peripheral clocks. A number of studies underscore a distinction between the biology of central ‘master’ clocks and of peripheral clocks. In Drosophila (Plautz et al., 1997; Giebultowicz et al., 2000), zebrafish (Whitmore et al., 1998) and the rat (Balsalobre et al., 1998; Yamazaki et al., 2000), autonomous oscillators have been shown to be present in non-neuronal organs. Moreover, these organ clocks can be uncoupled from the so-called master clock. For example, in Drosophila, transplanted excretory tubules maintain their own rhythms, out of phase with the host clocks (Giebultowicz et al., 2000). Uncoupling experiments in rodents have been possible by restricting food availability at specific times of the day (Damiola et al., 2000; Stokkan et al., 2001). By acting directly on the peripheral clock present in the liver, this approach has allowed SCN and liver rhythms to be uncoupled. Finally, in Clock mutant mice, Bmal1 gene expression is altered differently in the SCN and in other tissues (Oishi et al., 2000). Hence the molecular framework of circadian clocks in the SCN and peripheral tissues may differ, and Per1 may prove to have a more prominent role in the latter.
The phenotype of Per1-null mice is milder than that of Per2 mutants, but more impairing than in Per3-deficient animals. An obvious conclusion from the work presented here and elsewhere (Zheng et al., 1999; Shearman et al., 2000a) is that the three Per genes have different functions. A parallel can be drawn with the zebrafish system, in which the Per genes have acquired distinct functions: Per2 has a role in light response, whereas Per1 and Per3 are probably involved in core clock and/or output mechanisms (Pando et al., 2001). Intriguingly, in the mouse, Per1 and Per3 may have more similar function than Per2. Analysis of double Per1/Per3 mutant mice should help to clarify these issues. The Per1-null mice presented here will undoubtedly be a precious tool for the elucidation of the regulatory mechanisms involved in the circadian system.
Materials and methods
Generation of Per1 knock-out mice
Per1 genomic clones were isolated from an ES cell genomic SV129D3 library in λGEM12, screened with a Per1 cDNA clone obtained from H.Tei (Tei et al., 1997). BamHI fragments of 2, 3.2, 0.5, 5, 0.7 and 1.5 kb were subcloned into pBluescript SK– (pBS; Stratagene). A neomycin resistance gene under the control of the phosphoglycerate kinase promoter (PGK–Neo) was inserted between the NcoI sites located in exons 4 and 10. ES cells (129/SvPas line) were electroporated with the targeting construct and two positive clones were established. One of these was injected into C57BL/6 blastocysts. Male chimeras were bred with C57BL/6 female mice, and germline transmission was verified by Southern blotting on tail DNA, using a DNA probe external to the targeting construct (spanning exon 19 to intron 20). Heterozygous mice from this cross (founders, F0) were bred with 129/SvPas animals to obtain wild-type, heterozygous and homozygous mutant mice in normal Mendelian ratios (F1). All mice used for further analyses were from this generation (F1) or the following one (F2). The F0 mice (50% C57BL/6, 50% 129/SvPas) were also backcrossed for a number of generations with the C57BL/6 background. All the experiments were performed with 50:50 animals, except for the DD SCN in situ hybridization experiment (Figure 3) and the third group of mice in running wheels (94% C57BL/6). Seven- to 12-week old mice were used.
After verification of the germline transmission of the mutant allele and of the proper homologous recombination, a PCR test was set up to genotype further mouse progeny. Thirty cycles with an annealing temperature of 56°C were performed on DNA prepared from tail biopsies, using the following primers: 5′P (in exon 9), 5′TTTCTACATCCTGAGGACCGACC3′; 5′N (in the PGK–Neo cassette), 5′GGCCAGCTCATTCCTCCCACTCATGATC3′; and 3′ (in exon 11), 5′GGAATTCTGAGAGCTCCTGGATATCTGAG3′.
RT–PCR was carried out using the RNA PCR kit (Perkin Elmer) on total testis RNA, with a random hexamer for reverse transcription and the following primers for PCR: 5′ (in exon 1), 5′CAGCGGAGTTCTCATAGTTC3′; and 3′, as above. Amplified DNA fragments were cloned and sequenced. Similar results were obtained with other pairs of primers annealing upstream or downstream.
Standard molecular biology techniques (phage library screening, subcloning, sequencing, genomic DNA purification, Southern blotting) were conducted as described (Sambrook and Russell, 2001). Western blotting was performed according to the manufacturer’s instructions (Bio-Rad), on mouse embryo fibroblasts prepared by passing a 13-day-old embryo through an 18-gauge syringe and plating in Dulbecco’s modified Eagle’s medium + 10% fetal calf serum. Immunostaining was carried out as described (Crosio et al., 2000), with an antibody raised in rabbit against amino acids 1261–1278 of mPER1 (coupled with keyhole limpet hemocyanin; Pierce), and affinity purified (Sulfolink Coupling Gel; Pierce).
Locomotor activity in running wheels
Mice were housed in individual cages and entrained on a L12:D12 cycle for 2 weeks. They were then put in DD to study their free-running behavior. Wheel-running activity data were collected using the VitalView Data Acquisition System (Minimitter, Sunriver, OR) with a sampling interval of 10 min. Actograms were designed and data were analyzed with the Actiview Biological Rhythm Analysis software (Minimitter). The free-running period (τ) was calculated according to the onset of activity on days 4–20 in DD (except for the animals with two phases of free-running behavior: the calculation was then made independently for the two phases). The average activity per day was calculated for 8 days in LD and for 8 days (days 4–11) in DD. For the pulse-induced shift experiments, after entrainment on the L:D cycle, mice were put in DD for 2–5 days and then exposed to a 30 min light pulse at CT14 or 20. The phase shift (delay at CT14, advance at CT20) was assessed by measuring the interval between lines drawn on the onset of activity in free-running conditions before and after the light stimulation.
In situ hybridization and RNase protection assay
In situ hybridization (ISH) was performed as described (Crosio et al., 2000). Data were quantified by evaluating the optical density of the SCN region on autoradiograms. For each brain, five evenly spaced 10 µm cuts hybridized with the c-fos probe were quantified and their values summed. RPAs were performed as described (Whitmore et al., 1998). Total RNA extractions were performed using the RNAsolv reagent (Omega Biotek). RNAs were equilibrated on agarose gel by ethidium staining. The riboprobes were generated using an in vitro transcription kit (Promega). The probes used cover nucleotides 1–336 of the Per1 reading frame (for RPAs of Figure 4 and ISH), 1144–1488 of the Per1 reading frame (for RPAs of Figure 1D) and 456–585 of the Per2 sequence (DDBJ/EMBL/GenBank accession No. AF036893), cloned in pBS.
Acknowledgments
Acknowledgements
We thank Estelle Heitz and Maryam Rastegar for expert technical assistance, H.Tei for the mPer1 cDNA, and all the members of the Sassone-Corsi laboratory for helpful discussions. N.C. was supported by a Human Frontier Science Program long-term fellowship and a Canadian Institutes of Health Research postdoctoral fellowship. M.P.P. is supported by a postdoctoral fellowship of the Fondation de la Recherche Médicale. Work in our laboratory is supported by grants from CNRS, INSERM, CHUR, Human Frontier Science Program, Organon Akzo/Nobel, Fondation pour la Recherche Médicale and Association pour la Recherche sur le Cancer.
Note added in proof
While our manuscript was under review, two other groups have reported the generation of mice with a targeted mutation in the Per1 gene:
Bae,K., Jin,X., Maywood,E.S., Hastings,M.H., Reppert,S.M. and Weaver,D.R. (2001) Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron, 30, 525–536.
Zheng,B. et al. (2001) Non-redundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell, 105, 683–694.
Additional experiments in our laboratory show that—as in wild-type mice—a light pulse during the subjective day (CT6) does not phase shift activity rhythms of Per1 knock-out mice.
References
- Akiyama M. et al. (1999) Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J. Neurosci., 19, 1115–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U., Sun,Z.S., Eichele,G. and Lee,C.C. (1997) A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell, 91, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Damiola,F. and Schibler,U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell, 93, 929–937. [DOI] [PubMed] [Google Scholar]
- Brown S.A. and Schibler,U. (1999) The ins and outs of circadian timekeeping. Curr. Opin. Genet. Dev., 9, 588–594. [DOI] [PubMed] [Google Scholar]
- Bunger M.K., Wilsbacher,L.D., Moran,S.M., Clendenin,C., Radcliffe,L.A., Hogenesch,J.B., Simon,M.C., Takahashi,J.S. and Bradfield,C.A. (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell, 103, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N. and Sassone-Corsi,P. (2000) Multilevel regulation of the circadian clock. Nature Rev. Mol. Cell Biol., 1, 59–67. [DOI] [PubMed] [Google Scholar]
- Crews S.T. (1998) Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev., 12, 607–620. [DOI] [PubMed] [Google Scholar]
- Crosio C., Cermakian,N., Allis,C.D. and Sassone-Corsi,P. (2000) Light induces chromatin modification in cells of the mammalian circadian clock. Nature Neurosci., 3, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Daan S. and Pittendrigh,C.S. (1976) A functional analysis of circadian pacemakers in nocturnal rodents. II. The variability of phase response curves. J. Comp. Physiol., 106, 253–266. [Google Scholar]
- Damiola F., Le Minh,N., Preitner,N., Kornmann,B., Fleury-Olela,F. and Schibler,U. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev., 14, 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J.C. (1999) Molecular bases for circadian clocks. Cell, 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Eskin A. (1979) Circadian system of the Aplysia eye: properties of the pacemaker and mechanisms of its entrainment. Fed. Proc., 38, 2573–2579. [PubMed] [Google Scholar]
- Giebultowicz J.M., Stanewsky,R., Hall,J.C. and Hege,D.M. (2000) Transplanted Drosophila excretory tubules maintain circadian clock cycling out of phase with the host. Curr. Biol., 10, 107–110. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Lyons,L.C. and Hardin,P.E. (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science, 286, 766–768. [DOI] [PubMed] [Google Scholar]
- Griffin E.A. Jr, Staknis,D. and Weitz,C.J. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science, 286, 768–771. [DOI] [PubMed] [Google Scholar]
- Horikawa K., Yokota,S., Fuji,K., Akiyama,M., Moriya,T., Okamura,H. and Shibata,S. (2000) Nonphotic entrainment by 5-HT1A/7 receptor agonists accompanied by reduced Per1 and Per2 mRNA levels in the suprachiasmatic nuclei. J. Neurosci., 20, 5867–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.J., Edery,I. and Rosbash,M. (1993) PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature, 364, 259–262. [DOI] [PubMed] [Google Scholar]
- King D.P. et al. (1997) Positional cloning of the mouse circadian clock gene. Cell, 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D., Moore,R.Y. and Reppert,S.M. (1991) Suprachiasmatic Nucleus: The Mind’s Clock. Oxford University Press, New York, NY.
- Kornhauser J.M., Nelson,D.E., Mayo,K.E. and Takahashi,J.S. (1990) Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron, 5, 127–134. [DOI] [PubMed] [Google Scholar]
- Kume K., Zylka,M.J., Sriram,S., Shearman,L.P., Weaver,D.R., Jin,X., Maywood,E.S., Hastings,M.H. and Reppert,S.M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell, 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Maywood E.S., Mrosovsky,N., Field,M.D. and Hastings,M.H. (1999) Rapid down-regulation of mammalian period genes during behavioral resetting of the circadian clock. Proc. Natl Acad. Sci. USA, 96, 15211–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Fukui,H. and Ishida,N. (2000) Rhythmic expression of BMAL1 mRNA is altered in Clock mutant mice: differential regulation in the suprachiasmatic nucleus and peripheral tissues. Biochem. Biophys. Res. Commun., 268, 164–171. [DOI] [PubMed] [Google Scholar]
- Oishi K., Sakamoto,K., Okada,T., Nagase,T. and Ishida,N. (1998) Antiphase circadian expression between BMAL1 and period homologue mRNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochem. Biophys. Res. Commun., 253, 199–203. [DOI] [PubMed] [Google Scholar]
- Oliverio A. and Malorni,W. (1979) Wheel running sleep in two strains of mice: plasticity and rigidity in the expression of circadian rhythmicity. Brain Res., 163, 121–133. [DOI] [PubMed] [Google Scholar]
- Pando M.P., Pinchak,A., Cermakian,N. and Sassone-Corsi,P. (2001) A cell based system that recapitulates the dynamic light-dependent regulation of the vertebrate clock. Proc. Natl Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz J.D., Kaneko,M., Hall,J.C. and Kay,S.A. (1997) Independent photoreceptive circadian clocks throughout Drosophila. Science, 278, 1632–1635. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. and Aravind,L. (1997) PAS: a multifunctional domain family comes to light. Curr. Biol., 7, R674–R677. [DOI] [PubMed] [Google Scholar]
- Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schwartz W.J. and Zimmerman,P. (1990) Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J. Neurosci., 10, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman L.P., Jin,X., Lee,C., Reppert,S.M. and Weaver,D.R. (2000a) Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol. Cell. Biol., 20, 6269–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman L.P. et al. (2000b) Interacting molecular loops in the mammalian circadian clock. Science, 288, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Shearman L.P., Zylka,M.J., Weaver,D.R., Kolakowski,L.F.,Jr and Reppert,S.M. (1997) Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron, 19, 1261–1269. [DOI] [PubMed] [Google Scholar]
- Stokkan K.A., Yamazaki,S., Tei,H., Sakaki,Y. and Menaker,M. (2001) Entrainment of the circadian clock in the liver by feeding. Science, 291, 490–493. [DOI] [PubMed] [Google Scholar]
- Tei H., Okamura,H., Shigeyoshi,Y., Fukuhara,C., Ozawa,R., Hirose,M. and Sakaki,Y. (1997) Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature, 389, 512–516. [DOI] [PubMed] [Google Scholar]
- van der Horst G.T. et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature, 398, 627–630. [DOI] [PubMed] [Google Scholar]
- Whitmore D., Foulkes,N.S., Strahle,U. and Sassone-Corsi,P. (1998) Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nature Neurosci., 1, 701–707. [DOI] [PubMed] [Google Scholar]
- Williams J. and Sehgal,A. (2001) Molecular components of the circadian system in Drosophila. Annu. Rev. Physiol., 63, 729–755. [DOI] [PubMed] [Google Scholar]
- Yamazaki S. et al. (2000) Resetting central and peripheral circadian oscillators in transgenic rats. Science, 288, 682–685. [DOI] [PubMed] [Google Scholar]
- Zheng B., Larkin,D.W., Albrecht,U., Sun,Z.S., Sage,M., Eichele,G., Lee,C.C. and Bradley,A. (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature, 400, 169–173. [DOI] [PubMed] [Google Scholar]
- Zylka M.J., Shearman,L.P., Weaver,D.R. and Reppert,S.M. (1998) Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron, 20, 1103–1110. [DOI] [PubMed] [Google Scholar]