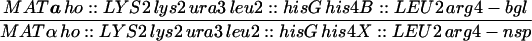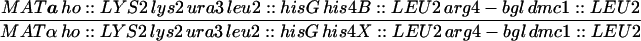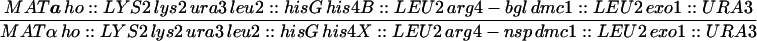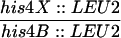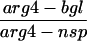Abstract
The MRE11, RAD50, and XRS2 genes of Saccharomyces cerevisiae are involved in the repair of DNA double-strand breaks (DSBs) produced by ionizing radiation and by radiomimetic chemicals such as methyl methanesulfonate (MMS). In these mutants, single-strand DNA degradation in a 5′ to 3′ direction from DSB ends is reduced. Multiple copies of the EXO1 gene, encoding a 5′ to 3′ double-strand DNA exonuclease, were found to suppress the high MMS sensitivity of these mutants. The exo1 single mutant shows weak MMS sensitivity. When an exo1 mutation is combined with an mre11 mutation, both repair of MMS-induced damage and processing of DSBs are more severely reduced than in either single mutant, suggesting that Exo1 and Mre11 function independently in DSB processing. During meiosis, transcription of the EXO1 gene is highly induced. In meiotic cells, the exo1 mutation reduces the processing of DSBs and the frequency of crossing over, but not the frequency of gene conversion. These results suggest that Exo1 functions in the processing of DSB ends and in meiotic crossing over.
INTRODUCTION
In mating-type switching and meiotic recombination in budding yeast, the formation of double-strand breaks (DSBs) is the initiating event. The DSB ends are subjected to 5′ to 3′ directed exonucleolytic processing to leave 3′-tailed single-strand (ss) DNA (Stahl, 1996). These ssDNA tails can be detected in vivo as intermediates during mating-type switching (White and Haber, 1990) and during meiotic recombination (Sun et al., 1989, 1991; Cao et al., 1990). Intermediates with ssDNA tails accumulate if the strand invasion step is blocked by a mutation in the RAD51, -52, -54, -55, or -57 gene during mating-type switching and also in DMC1 during meiotic recombination (Bishop et al., 1992; Shinohara et al., 1992; Sugawara and Haber, 1992; Ogawa et al., 1993; Sugawara et al., 1995). Several of these genes, RAD51, -55, -57, and DMC1, encode homologues of the Escherichia coli RecA protein.
One candidate for the enzyme involved in the production of 3′-tailed ssDNA is the Mre11/Rad50/Xrs2 protein complex. A lack of any one of these proteins makes cells highly sensitive to DSBs (Game and Mortimer, 1974; Ivanov et al., 1992; Ajimura et al., 1993) and reduces processing of DSB ends produced by the HO endonuclease during mating-type switching (Ivanov et al., 1994; Lee et al., 1998; Tsubouchi and Ogawa, 1998). As a consequence, the kinetics of recombinant formation is slow, but the amount of recombinant DNA ultimately formed is the same as in wild type (Ivanov et al., 1994; Tsubouchi and Ogawa, 1998).
The Mre11/Rad50/Xrs2 complex is implicated in several aspects of DNA metabolism, including mitotic recombination, telomere maintenance, and nonhomologous end joining (Haber, 1998). Recent biochemical studies show that Mre11 and its mammalian homologues have double-strand (ds) DNA 3′ to 5′ exonuclease activity and ssDNA endonuclease activity (Furuse et al., 1998; Paull and Gellert, 1998; Trujillo et al., 1998; Usui et al., 1998; Moreau et al., 1999; reviewed by Haber, 1998). These activities are not directly involved in the formation of 3′-tailed ssDNA at DSB ends, because mre11 mutants that lack them still show the wild-type level of DSB processing in vivo (Moreau et al., 1999).
The Exo1 protein of fission and budding yeasts has been isolated as a 5′ to 3′ dsDNA exonuclease (Szankasi and Smith, 1995; Fiorentini et al., 1997). This protein is conserved through higher eukaryotes and is implicated in recombination and mismatch correction in vivo (Digilio et al., 1996; Tishkoff et al., 1997, 1998; Qiu et al., 1999). In accord with the mismatch correction function, budding yeast Exo1 interacts with the Msh2 protein (Tishkoff et al., 1997). A homology search of the complete genome of budding yeast reveals four predicted proteins that share sequence similarity with Exo1: Rad2, Rad27, Din7, and Yen1 (Mieczkowski et al., 1997; Tishkoff et al., 1997). There could be functional redundancy among these proteins.
Proper segregation of homologous chromosomes at the reductional division of meiosis requires crossing over to establish chiasmata, which physically connect homologues after disassembly of the synaptonemal complex. Gene conversion has no significant role in segregation (Roeder, 1997). A mutation in any one of a group of genes (ZIP1, ZIP2, MSH4, MSH5, MLH1, MLH3, and MER3) reduces crossing over (Ross and Roeder, 1994; Sym and Roeder, 1994; Hollingsworth et al., 1995; Hunter and Borts, 1997; Chua and Roeder, 1998; Nakagawa and Ogawa, 1999; Wang et al., 1999). Among these, MSH4, MSH5, MLH1, and MLH3 are homologues of the mismatch repair proteins of E. coli, suggesting a mechanistic relationship between mismatch repair and crossing over (Nakagawa et al., 1999).
In this study, we isolated the EXO1 gene as a high-copy suppressor of the methyl methanesulfonate (MMS) sensitivity of mre11 mutants. The exo1 mutant itself is moderately sensitive to MMS. The exo1 mre11 double mutant, however, is more sensitive to MMS and shows a greater reduction in DSB processing efficiency during mating-type switching than either single mutant. During meiosis, the exo1 mutation reduces both DSB processing and crossover frequency and causes nondisjunction of homologous chromosomes. We suggest that Exo1, as well as the Mre11 complex, is involved in the processing of DSB ends and that Exo1 is also involved in crossing over to promote the accurate segregation of chromosomes during meiosis.
MATERIALS AND METHODS
Plasmids
Plasmids were constructed by standard procedures. A 4.4-kilobase (kb) SphI fragment containing EXO1 was cloned into the SphI site of YEplac195 (Gietz and Sugino, 1988) to make pHT131. An EcoRI fragment in the EXO1 coding sequence was replaced with a 1.1-kb EcoRI fragment containing URA3 from YEp24 to make pHT133 or with a 2.2-kb HpaI fragment containing LEU2 from YEp13 to make pHT256. Plasmids pHT133 and pHT254 were used for EXO1 disruption experiments. pHT59 is a pUC118-based plasmid carrying a 3.4-kb NsiI–BamHI fragment of XRS2 on which a XbaI–BglII fragment was replaced with a 3.8-kb hisGURA3hisG fragment from pNKY51 (Alani et al., 1987). pHT127 carries a 4.3-kb BamHI fragment containing MRE11 on YEplac195. pNKY291 contains sequences downstream of HIS4. pYtel is based on pBluescriptII SK− and carries a 1.0-kb XhoI fragment of a yeast chromosomal end, which contains 120-base pair telomere repeats and part of the Y′ subtelomeric repeat (a gift from F. Ishikawa, Tokyo Institute of Technology, Tokyo, Japan). pYA301 carries a 3.4-kb BamHI–EcoRI fragment that includes the yeast actin gene cloned into the BamHI–EcoRI sites of pBR322 (Gallwitz and Seidel, 1980). pBTM116 (Chien et al., 1991) was used to measure plasmid end joining (Boulton and Jackson, 1996b)
Yeast Strains and Media
Strains used in this study are listed in Table 1. All gene targeting was carried out by the one-step gene disruption method (Rothstein, 1983). KJC010 contains an mre11-1 allele and is MMS sensitive at 34°C (Ajimura et al., 1993). HTY836 was made by replacing a BglII–BglII region of the THR4 coding sequence of the R167 strain (a gift from J.E. Haber, Brandeis University, Waltham, MA) with a hisGlacZ insertion. HTY525, -1076, -1104, -1106, -1214, -1215, -1330, and -1336 are SK1 derivatives that enter the meiotic cell cycle in a highly synchronous manner (Fast, 1973). HTY1212, -1213, -1326, -1328, -1434, -1436, -1442, and -1444 are congenic to SK1. HTY1212, -1213, -1214, and -1215 are identical to a pair of haploids consisting of MY257 and a pair of haploids consisting of MY261, respectively (Sym and Roeder, 1994), and are gifts from G.S. Roeder (Yale University, New Haven, CT). The EXO1 gene was disrupted by transforming strains with pHT133 or pHT256 after SphI digestion. The XRS2 gene was disrupted by transforming strains with pHT59 after SphI and BamHI double digestion. Disruption of MRE11, RAD50, RAD52, and DMC1 was carried out as described previously (Alani et al., 1989; Bishop et al., 1992; Johzuka and Ogawa, 1995; Tsubouchi and Ogawa, 1998).
Table 1.
Strains used
| Strains | Genotypes | |
|---|---|---|
| CG379 | MATα ura3-52 leu2-3, 112 trp1-289 ade5-1 his7-2 | |
| KJC010 | MATα can1 ura3 ade2 ade6 cyh2 trp1 mre11-1 | |
| HTY525 |
|
|
| HTY836 | matΔ::LEU2 hmr-3Δ mal2 trp1 leu2 ura3 thr4::hisGlacZ | |
| HTY837 | HTY836 but mre11::hisG | |
| HTY838 | HTY836 but rad50::hisG | |
| HTY839 | HTY836 but xrs2::hisG | |
| HTY975 | HTY836 but rad52::hisG | |
| HTY987 | HTY837 carrying YEplac195 | |
| HTY989 | HTY837 carrying pHT131 | |
| HTY991 | HTY837 carrying pHT127 | |
| HTY1013 | HTY836 but exo1::URA3 | |
| HTY1015 | HTY836 but mre11::hisG and exo1::URA3 | |
| HTY1017 | HTY836 but rad50::hisG and exo1::URA3 | |
| HTY1076 | HTY525 but homozygous for exo1::URA3 | |
| HTY1104 |
|
|
| HTY1106 |
|
|
| HTY1212 | MATa ho::LYS2 lys2 URA3 HOM3 TRP2 leu2::hisG can1 | |
| HTY1213 | MATα ho::LYS2 lys2 ura3 hom3-10 trp2 leu2::hisG CAN1 | |
| HTY1214 | MATa ho::LYS2 lys2 ura3 leu2::hisG trp1-H3 CEN3-URA3 | |
| HTY1215 | MATα ho::LYS2 lys2 ura3 leu2::hisG trp1-H3 CEN3-TRP1 his4B::LEU2 | |
| HTY1326 | HTY1212 but exo1::LEU2 | |
| HTY1328 | HTY1213 but exo1::LEU2 | |
| HTY1330 | HTY1214 but exo1::LEU2 | |
| HTY1332 | CG379 with pHT51 | |
| HTY1333 | HTY1332 but mre11::hisGURA3hisG | |
| HTY1334 | HTY1332 but exo1::URA3 | |
| HTY1335 | HTY1332 but mre11::hisG exo1::URA3 | |
| HTY1336 | HTY1215 but exo1::LEU2 | |
| HTY1434 | HTY1213 but msh4::URA3 | |
| HTY1436 | HTY1212 but msh4::URA3 | |
| HTY1442 | HTY1213 but exo1::LEU2 msh4::URA3 | |
| HTY1444 | HTY1212 but exo1::LEU2 msh4::URA3 |
All media used in this study were detailed previously (Tsubouchi and Ogawa, 1998). YPLac contains 1% yeast extract, 2% bacto peptone, and 2% sodium lactate (pH 5.5), and YPGal is identical except that it contains 2% galactose instead of lactate.
Isolation of High-Copy Suppressors for MMS Sensitivity of mre11
KJC010 was transformed with a yeast genomic library constructed in YEp24 (Botstein et al., 1979), plated on SD-URA (synthetic complete medium lacking uracil) plates, and incubated at 34°C for 3 d. Transformants were replica-plated onto YPD plates containing 0.01% MMS and incubated for 3 d at 34°C. MMS-resistant Ura+ clones were streaked on 5-FOA plates to isolate plasmid segregants, and MMS sensitivity of mre11-1 strains with and without the suppressor candidate plasmids was compared. The plasmids that make the mre11-1 strain resistant to MMS were isolated and sequenced from both sides of the insert fragment. The primers used are Pri 1 (cagtcctgctcgcttcgc) and Pri 2 (atgtcggcgatataggcg).
Plasmid End-joining Assay
This assay was performed as described previously (Boulton and Jackson, 1996b). In brief, HTY987, -989, and -991 were transformed with supercoiled, or linear, pBTM116 (Chien et al., 1991) digested with BamHI. One hundred nanograms of each DNA was used to transform each strain. The ratio of Ura+ Trp+ transformants obtained with cut or uncut DNA was determined for each strain. The average value of two independent assays for each strain is shown.
Detection of Mating-type Switching
Induction of HO break and detection of mating-type switch process by Southern blot analysis have been described (Tsubouchi and Ogawa, 1998). Genomic DNA samples obtained at indicated times were cut with StyI (Eco130I), separated on 0.7% agarose gels, and subjected to Southern blot analysis with a 1.0-kb NdeI–HindIII fragment of pHT46 as a probe.
Detection of Telomeres
Genomic DNA was isolated from overnight cultures of the strains indicated (see Figure 7), cut with XhoI, separated on 0.7% agarose gels, and subjected to Southern blot analysis with a 1-kb XhoI–BamHI fragment of pYtel as a probe.
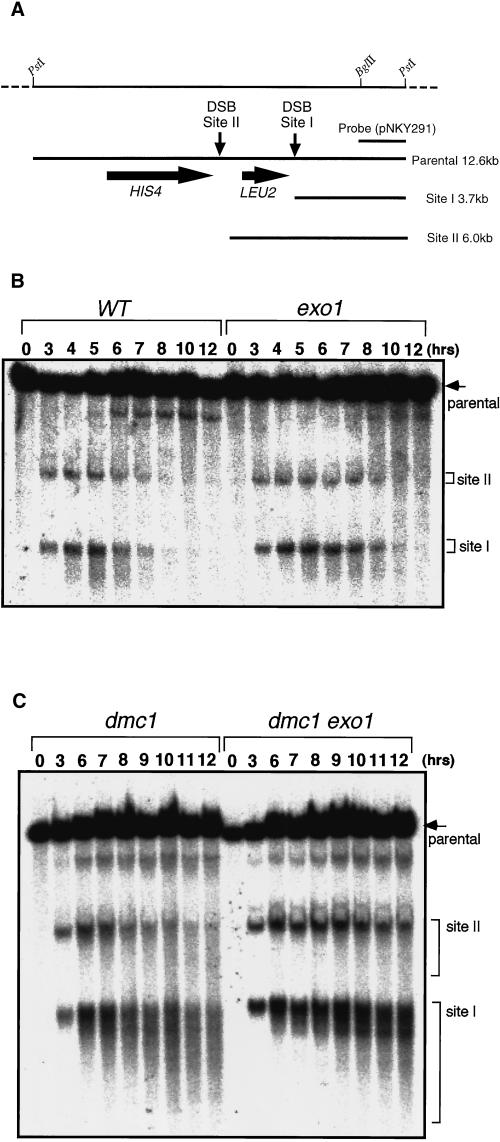
Figure 7.
Processing of meiotic DSBs is reduced in exo1. (A) Restriction map of the HIS4::LEU2 construct. DSBs are formed at sites I and II. Fragments diagnostic of DSBs at sites I and II and the unbroken parental fragment (parental) are shown. (B) Detection of meiotic DSBs in wild type (HTY525) and exo1 (HTY1076). (C) Detection of meiotic DSBs in dmc1 (HTY1104) and dmc1 exo1 (HTY1106). DNA was extracted from cultures of each strain at the indicated times during meiosis. After digestion with PstI and electrophoresis, DSBs were detected by Southern blot analysis with an appropriate probe (1.5-kb PstI–EcoRI band from pNKY291).
Return-to-Growth Experiment and Meiotic DSB Detection
Synchronous entry of cells into meiosis, return-to-growth experiments, and detection of meiosis-specific DSBs were performed as described previously (Cao et al., 1990; Johzuka and Ogawa, 1995). Genomic DNA samples at different times during meiosis were cut with PstI, separated on 0.7% agarose gels, and subjected to Southern blot analysis with a 1.5-kb PstI–BglII fragment of pNKY291 as a probe (Cao et al., 1990).
Detection of the EXO1 Transcript during Meiosis
Total RNA was extracted from meiotically dividing cells of HTY525, a diploid SK1 derivative, as reported previously (Johzuka and Ogawa, 1995), and subjected to Northern blot analysis. The amounts of RNA loaded to keep the amount of the ACT1 transcript nearly constant through meiosis are as follows: 10 μg for 0 h, 20 μg for 2 and 4 h, 40 μg for 6 h, and 60 μg for 8 and 10 h. As probes, a 0.9-kb EcoRI fragment of pHT130 and a 0.6-kb ClaI fragment of pYA301 were used to detect transcripts of EXO1 and ACT1, respectively.
RESULTS
The EXO1 Gene on a Multicopy Plasmid Suppresses the Repair Defect of an mre11 Mutant
In an mre11, rad50, or xrs2 null mutant, the kinetics of recombination during mating-type switching is slow because of inefficient DSB processing. However, the wild-type amount of recombinant is formed eventually, suggesting that there are additional proteins involved in this process. To identify a gene involved in the process, we screened for high-copy suppressors of the high MMS sensitivity of the mre11-1 missense mutant, which shows temperature-dependent MMS sensitivity (Ajimura et al., 1993). The mutant was transformed with a genomic DNA library constructed in a 2-μm vector, YEp24. From transformants that grew on YPD plates containing 0.01% MMS at 34°C, plasmids were retrieved and DNA sequences from both ends of the inserts were determined. Seventeen clones were found to contain the MRE11 gene itself, and eight contained the EXO1 gene. The 4.4-kb SphI–SphI fragment containing the EXO1 gene was cloned into YEplac195, a 2-μm vector, and this plasmid (pHT131) was used for further analysis.
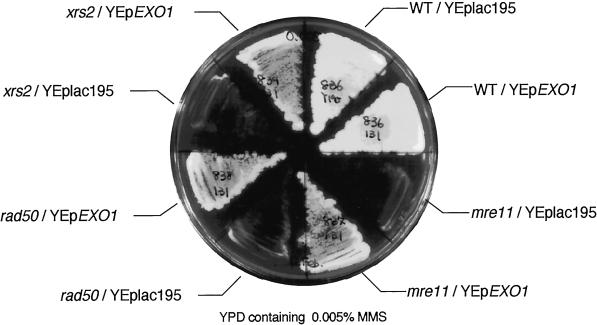
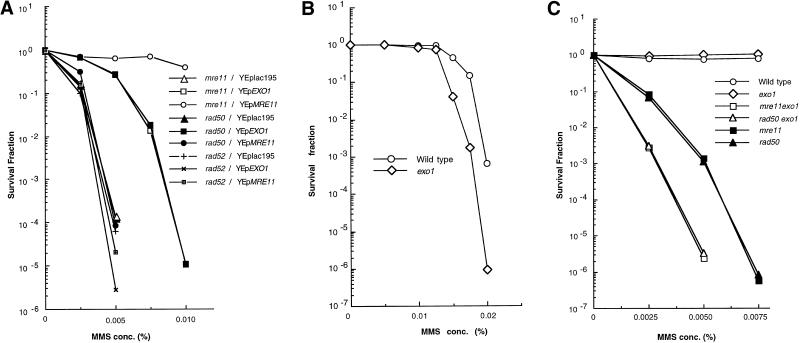
To determine if multiple copies of the EXO1 gene can suppress the defect of the mre11 null mutant, the mutant was transformed with vector alone (YEplac195), a plasmid containing EXO1 (pHT131), or a plasmid containing MRE11 (pHT127). Suppression of MMS sensitivity was clearly observed for the mutant with a high dosage of the EXO1 gene (Figure 1). Because the phenotypes of mre11, rad50, and xrs2 mutants are indistinguishable (Alani et al., 1990; Ivanov et al., 1992; Johzuka and Ogawa, 1995) and the Mre11 protein forms a complex with the Rad50 and Xrs2 proteins (Johzuka and Ogawa, 1995; Usui et al., 1998), the EXO1 gene was tested for its ability to suppress the MMS sensitivity of rad50 and xrs2 mutants as well. As expected, suppression was observed in these null mutants to the same extent as in the mre11 mutant (Figure 1). High dosage of EXO1 increased resistance to MMS >1000-fold at 0.005% MMS (Figure 2 A). However, EXO1 overexpression did not suppress the repair defect of the rad52 mutant (Figure 2 A), indicating that the suppression effect is specific to the mre11, rad50, and xrs2 mutants.
Figure 1.
Suppression of the MMS sensitivity of the mre11 null mutant by high-copy EXO1. Wild-type (HTY836), mre11 (HTY837), rad50 (HTY838), and xrs2 (HTY839) null mutants containing YEplac195 or YEpEXO1 (pHT131) were streaked on YPD containing 0.005% MMS.
Figure 2.
(A) Quantitative analysis of the suppression efficiency of a high dosage of EXO1. Mutant mre11 (HTY837), rad50 (HTY838), and rad52 (HTY975) strains containing YEplac195, YEpEXO1 (pHT131), or YEpMRE11 (pHT127) were cultured and plated on SD-URA plates containing various concentrations of MMS. Colonies were counted after 5 d at 30°C and normalized to the number of colonies formed in the absence of MMS. (B) MMS sensitivity of the exo1 null mutant. (C) MMS sensitivity of double mutants. (B and C) Wild-type (HTY836), exo1 (HTY1013), mre11 (HTY837), rad50 (HTY838), mre11 exo1 (HTY1015), and rad50 exo1 (HTY1017) were cultured and plated on YPD plates containing various concentrations of MMS. Survival fractions were calculated as in A. Each experiment was performed at least twice for each strain; the results of single representative experiments are shown here.
Mating-type Switching Is Extremely Retarded in the Absence of Both Exo1 and Mre11
To test whether EXO1 is involved in DSB repair, the phenotype of an EXO1 deletion mutant was examined. The mutant is more sensitive to MMS than wild type, particularly in the presence of >0.0125% MMS (Figure 2B). Furthermore, exo1 mre11 and exo1 rad50 double mutants demonstrate more pronounced repair defects than the mre11 and rad50 single mutants (Figure 2C). Therefore, the EXO1 gene is involved in a damage repair pathway that is independent of MRE11, RAD50, and XRS2. However, its contribution to repair is not as great as that of MRE11/RAD50/XRS2.
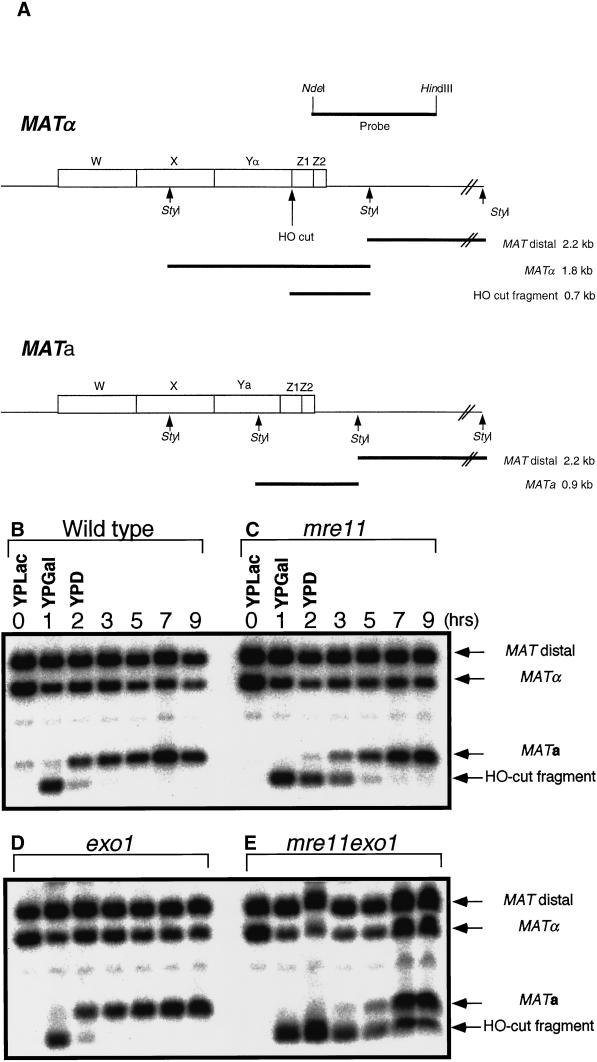
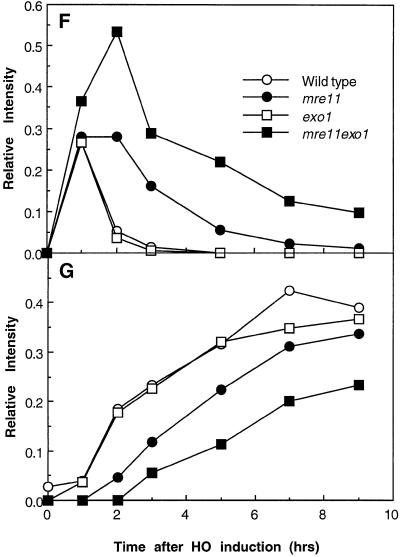
The process of mating-type switching is well characterized and provides a good model system for analyzing the effect of the exo1 mutation on homologous recombination. DSBs were formed at the MAT locus by the HO endonuclease, whose expression was controlled by a galactose-inducible promoter. When wild-type cells are incubated in medium containing galactose for 1 h, a 1.8-kb MATα band, which contains an HO target site, is reduced in amount and a 0.7-kb fragment corresponding to an HO-cut fragment appears (Figure 3, A, B, F, and G). When cells are shifted to glucose medium for 1 h, most of the 0.7-kb HO-cut fragment disappears and a 0.9-kb MATa band, the product of mating-type switching, appears. The 0.7-kb band completely disappears with an additional 1 h of incubation (Figure 3, B, F, and G). In the exo1 mutant, the kinetics of MAT switching is indistinguishable from that of wild type (Figure 3, D, F, and G). In the mre11 mutant, however, the 0.7-kb band persists after 3 h in glucose medium, and the appearance of the 0.9-kb MATa band is delayed about 2 h (Tsubouchi and Ogawa, 1998) (Figure 3, C, F, and G). In the exo1 mre11 double mutant, ∼10% of the 0.7-kb HO-cut fragment still remains at 9 h, and the appearance of the 0.9-kb MATa band is further delayed compared with the mre11 single mutant (Figure 3, E, F, and G). The relative intensity of HO-cut fragments in the mre11 exo1 double mutant increases even after shutting off the expression of HO, presumably because the remaining HO endonuclease is active while DSB processing is very slow, resulting in further accumulation of HO-cut fragments. These results indicate that recombination is retarded, but still occurs, even in the absence of both Exo1 and Mre11.
Figure 3.
Mating-type switching. (A) Physical map of the MAT locus. (B–E) Cells were preincubated in YPLac, then transferred into YPGal for 1 h, to induce a DSB at the Y/Z junction by HO endonuclease. Transcription was then repressed by changing the culture medium to YPD. Cells were harvested at the indicated times, and genomic DNA was extracted and subjected to Southern blot analysis with a probe shown in A. (B) Wild type (HTY1332); (C) mre11 (HTY1333); (D) exo1 (HTY1334); (E) mre11 exo1 (HTY1335). (F and G) Densitometric analyses of HO-cut fragments (F) and recombinant fragments (G) in Southern blot data from B–E. The band intensities of HO-cut fragments (in F) or recombinant fragments (in G) were divided by the combined intensities of bands corresponding to MAT distal, MATα, MATa, and HO cut fragment to obtain the relative intensities.
EXO1 Does Not Suppress the Reduction in Nonhomologous End Joining in mre11
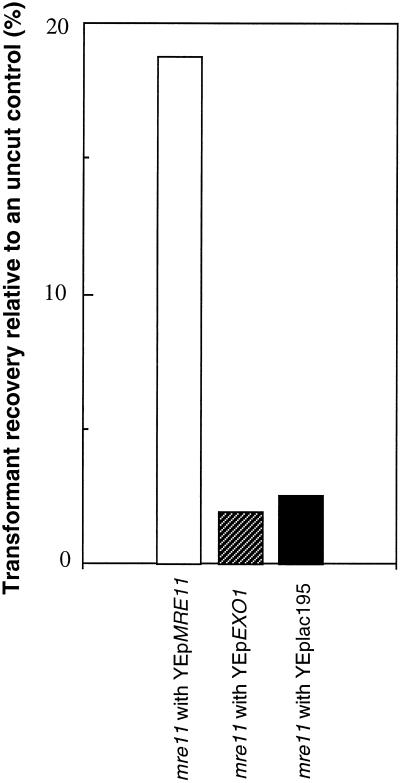
MRE11 is involved in the nonhomologous end joining (NHEJ) pathway as well as the homologous recombination pathway of DSB repair. To test whether EXO1 can suppress the mre11 defect in NHEJ, a plasmid recircularization assay was used (see MATERIALS AND METHODS). The mre11 null mutant containing vector alone (HTY987), a plasmid carrying EXO1 (HTY989), or a plasmid carrying MRE11 (HTY991) was transformed with pBTM116 linearized by BamHI or with a supercoiled plasmid control. Because the linearized plasmid must be recircularized to be propagated, the ratio of the number of transformants obtained with the linear plasmid tested relative to the number obtained with the supercoiled plasmid reflects the ability of the strains to repair DSBs. In pBTM116, there is no yeast-derived sequence around the restriction enzyme cleavage site; therefore, repair occurs predominantly by NHEJ. The proportion of transformants recovered with cut plasmid relative to uncut plasmid in mre11 carrying a vector alone is almost identical to that of mre11 carrying multicopy EXO1 (2.5 and 1.9%, respectively), and both of these values are ∼10-fold lower than the value obtained with mre11 carrying the MRE11 gene (18.8%) (Figure 4). We also tested the recovery of a plasmid linearized with a restriction enzyme that creates blunt ends; there was no difference in the plasmid recircularization ratio regardless of whether or not mre11 carried multicopy EXO1 (our unpublished result). These results indicate that EXO1 does not suppress the NHEJ defect of mre11.
Figure 4.
Multicopy EXO1 does not suppress the reduced NHEJ efficiency of mre11. The NHEJ assay is described in MATERIALS AND METHODS.
EXO1 Suppresses Neither Telomere Shortening nor Spore Inviability in mre11
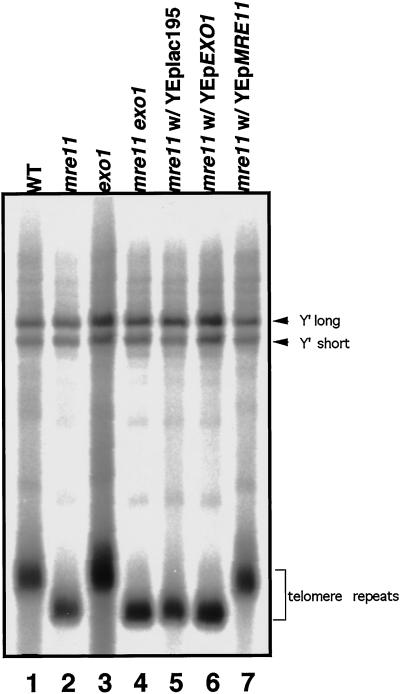
Telomeres become shortened in the mre11, rad50, and xrs2 mutants (Kironmai and Muniyappa, 1997; Boulton and Jackson, 1998; Nugent et al., 1998). Because a high dosage of the EXO1 gene suppresses the DSB repair defect of the mre11 mutant, we examined the effects of EXO1 on telomere shortening in mre11. In the exo1 mutant, telomeres are not shortened (Figure 5, lane 3), and shortening is not enhanced in the exo1 mre11 double mutant compared with mre11 alone (Figure 5, lane 4). Furthermore, high dosage of the EXO1 gene does not suppress the telomere shortening of mre11 (Figure 5, lane 6). Thus, unlike Mre11, Exo1 does not appear to be involved in telomere maintenance.
Figure 5.
Telomere length in mutant strains. Genomic DNAs were extracted, cut with XhoI, and subjected to electrophoresis, followed by Southern blot analysis to detect telomere sequences (see MATERIALS AND METHODS). Lane 1, wild-type (HTY836); lane 2, mre11 (HTY837); lane 3, exo1 (HTY1013); lane 4, mre11 exo1 (HTY1015); lane 5, mre11 with YEplac195 (HTY987); lane 6, mre11 with YEpEXO1 (HTY989); lane 7, mre11 with YEpMRE11 (HTY991).
mre11 produces inviable spores during meiosis. To determine if a high dosage of EXO1 suppresses this defect, spore viability was compared between an mre11 strain carrying multicopy EXO1 and a control mre11 strain carrying vector alone. In both cases, no viable spores were generated (44 tetrads dissected for each).
Meiotic Phenotypes of the exo1 Mutant
Transcription of EXO1 Is Induced during Meiosis
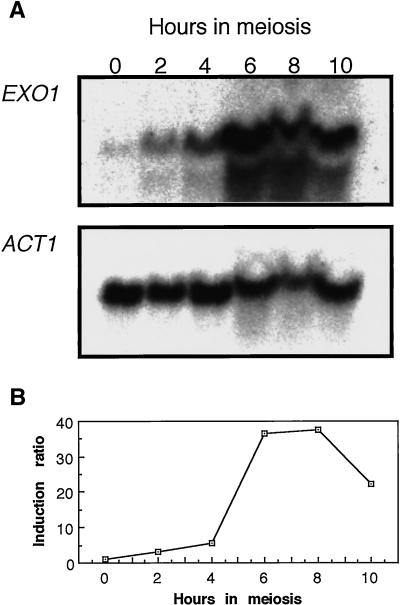
The EXO1 gene of the fission yeast was reported to be induced during meiotic prophase and implicated to have functions in mismatch correction (Szankasi and Smith, 1995). The Exo1 homologue of Drosophila, named Tosca, is also induced during meiosis (Digilio et al., 1996). We tried to determine if transcription of the EXO1 gene of budding yeast is induced during meiosis. Cells progressing synchronously through meiosis were harvested, and total RNA was extracted and subjected to Northern blot analysis. The amount of RNA loaded at each time point was normalized to the amount of ACT1 mRNA. In the strain used, premeiotic DNA synthesis is completed by 4 h, the DSB level reaches a maximum at around 5 h, and the fraction of cells that are binucleate is maximal at around 6 h. EXO1 transcripts are present at a low level during mitosis but increase drastically between 4 and 6 h after entry into meiosis, as has been shown before (Chu et al., 1998a,b) (Figure 6). The maximum increase of 36-fold is reached by 6 h; by 10 h, transcript levels begin to decline, suggesting its role by meiosis I.
Figure 6.
Meiotic induction of EXO1 transcription. Total RNA from a wild-type diploid (HTY525) was isolated at the indicated times during meiosis, fractionated on formaldehyde agarose gels (0.7%), and subjected to Northern blot analysis. (A) Transcription of the EXO1 and ACT1 genes was monitored with EXO1 (0.9-kb EcoRI fragment from pHT130) and ACT1 (0.6-kb ClaI fragment from pYA301) coding sequences as probes. The ACT1 transcript, which is expressed at a constant level during meiosis, was used as an internal control (McKee and Kleckner, 1997). (B) Relative intensity of the EXO1 transcript compared with that of ACT1. At each time, the intensities of the EXO1 and ACT1 bands were measured and the amount of EXO1 transcript was divided by the amount of ACT1 transcript.
DSB Processing Is Impaired in the exo1 Mutant
Almost all meiotic recombination is initiated by DSBs formed at recombination hot spots (Haber, 1997). The effect of the exo1 mutation on meiotic recombination was monitored at one of these hot spots, the HIS4::LEU2 locus (Cao et al., 1990) (Figure 7A). DSB products appeared at 3 h and reached a maximal level at 5 h in both wild type and exo1, but their disappearance was delayed by 1–2 h in exo1 (Figure 7B). Progression through the meiotic cell cycle was monitored at the same time. In exo1, the fraction of cells that had passed the first meiotic division reached a maximum at 7 h after entry into meiosis, which is 1 h later than in wild type (our unpublished result). To understand the effect of the exo1 mutation on DSB processing more clearly, we took advantage of the dmc1 mutant, in which the recombination reaction is blocked at the strand-transfer stage and DSBs with ssDNA tails accumulate (Bishop et al., 1992). In the exo1 dmc1 double mutant, meiotic DSBs were formed as in a wild-type time course, but the DNA fragment appeared more discrete (Figure 7C), indicating a defect in exonucleolytic processing to expose ssDNA tails. This reduction in processing was first evident at 3 h and became more pronounced at later times (Figure 7C).
The exo1 Mutation Reduces Meiotic Crossing Over
To examine the effect of the exo1 mutation on recombination, gene conversion frequencies were measured with two sets of heteroalleles, his4X/his4B and arg4-bgl/arg4-nsp (Sherman and Roman, 1963; Esposito and Esposito, 1974). At both of these loci, recombination was induced almost to the same extent as in the wild type at 24 h after entry into meiosis (Table 2). Crossing over was measured in four intervals (CEN3-HIS4, CAN1-URA3, URA3-HOM3, and HOM3-TRP2) by tetrad analysis (Sym and Roeder, 1994). In each interval, map distance was reduced 1.5- to 2.0-fold in the exo1 mutant (Table 3); this reduction is statistically significant (CEN3-HIS4, p < 0.02; CAN1-URA3, p ≪ 0.0001; URA3-HOM3, p ≪ 0.0001; HOM3-TRP2, p < 0.004).
Table 2.
Gene conversion
| Locus | Wild type (HTY525) | exo1 (HTY1076) | |
|---|---|---|---|
|
0.013 ± 0.007 | 0.012 ± 0.009 | |
|
0.040 ± 0.012 | 0.034 ± 0.012 |
The meiotic prototroph frequencies at HIS4 and ARG4 loci at 24 h after entry into meiosis are shown. Values are the averages of four independent cultures for each strain (mean ± SD).
Table 3.
Crossing over
| Interval | PD | TT | NPD | Total | cM | Decrease (fold) |
|---|---|---|---|---|---|---|
| Wild type | ||||||
| CEN3-HIS4 | 47 | 87 | 2 | 136 | 36.4 | |
| CAN1-URA3 | 62 | 177 | 9 | 248 | 46.6 | |
| URA3-HOM3 | 80 | 160 | 7 | 247 | 40.9 | |
| HOM3-TRP2 | 183 | 58 | 2 | 243 | 14.4 | |
| exo1 | ||||||
| CEN3-HIS4 | 199 | 122 | 7 | 328 | 25 | 1.5 |
| CAN1-URA3 | 270 | 161 | 6 | 437 | 22.5 | 2.1 |
| URA3-HOM3 | 265 | 170 | 7 | 442 | 24.0 | 1.7 |
| CAN1-HOM3 | 155 | 254 | 31 | 440 | 44.2 | – |
| HOM3-TRP2 | 371 | 61 | 0 | 432 | 7.1 | 2.0 |
| msh4 | ||||||
| CAN1-HOM3 | 61 | 42 | 2 | 105 | 25.7 | |
| HOM3-TRP2 | 81 | 20 | 0 | 101 | 9.9 | |
| exo1 msh4 | ||||||
| CAN1-HOM3 | 52 | 39 | 3 | 94 | 30.3 | |
| HOM3-TRP2 | 73 | 19 | 0 | 92 | 10.3 |
To examine the CEN3-HIS4 interval, HTY1214 and HTY1215 (wild type) and HTY1330 and HTY1336 (exo1) were dissected after mating. In the CAN1-URA3, URA3-HOM3, CAN1-HOM3, and HOM3-TRP2 intervals, HTY1212 and HTY1213 (wild type), HTY1326 and HTY1328 (exo1), HTY1434 and HTY1436 (msh4), and HTY1442 and HTY1444 (msh4 exo1) were dissected after mating. Only four-spore–viable tetrads that did not show gene conversion of the markers indicated were used to calculate map distances. Three classes of tetrads were scored as follows: PD, parental ditype; TT, tetratype; NPD; nonparental ditype. Map distances are expressed in terms of centimorgans (cM), which were calculated from the following formula: cM = 100/2(TT + 6NPD)/(PD + TT + NPD).
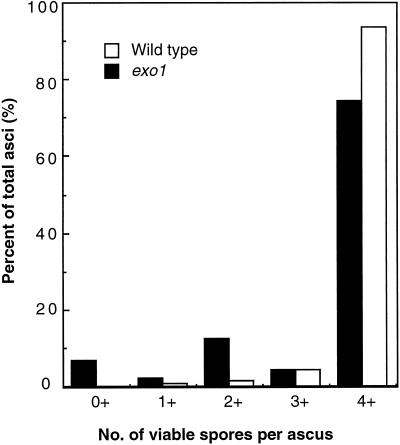
There is a modest but significant reduction of spore viability in the exo1 mutant (84%, 216 tetrads dissected, p ≪ 0.0001), as reported previously (Fiorentini et al., 1997). Furthermore, the number of viable spores per ascus is not random: fractions of asci that contain zero, two, or four viable spores per ascus (7, 12, and 74%, respectively) are more frequent than asci containing one or three viable spores per ascus (2 and 4%, respectively) (Figure 8). This suggests that a large fraction of the nondisjunction events is due to pairs of homologues segregating to the same pole at the first meiotic division. To examine the segregation of homologous chromosomes directly, a diploid strain was used in which the centromere on one chromosome III was marked with TRP1 and the other with URA3 (Sym and Roeder, 1994). In a cross between CENIII-TRP1 and CENIII-URA3 strains, spores that are disomic for chromosome III result from nondisjunction at the first meiotic division and can be identified as both Ura+ and Trp+. Among 63 two-spore–viable tetrads, 8 resulted from nondisjunction of chromosome III. Among them, no recombinant was found in CEN3-HIS4. In 45 of the remaining 55 tetrads containing two viable spores, the two spores were sisters (i.e., both spores were Ura+ or Trp+), suggesting that they resulted from meiosis nondisjunction of another pair of chromosomes. These results indicate that crossing over is reduced and nondisjunction is increased in exo1.
Figure 8.
Distribution of tetrad types. The distribution of 216 asci with four (4+), three (3+), two (2+), one (1+), or no (0+) viable spores is shown for the wild type (HTY525) and the exo1 strain (HTY1076).
It was shown previously that the meiosis-specific mismatch repair homologues MSH4 and MSH5 are involved in meiotic crossing over (Ross and Roeder, 1994; Hollingsworth et al., 1995). These two gene products form a complex and act in the same pathway. In the msh4 mutant, crossing over is less than (in CAN1-HOM3, p ≪ 0.0001) or similar to (in HOM3-TRP2, the difference of the two values at this interval is statistically insignificant [p > 0.4]) that in the exo1 mutant (Table 3). To understand the relationship between the MSH4-dependent pathway and EXO1, a msh4 exo1 double mutant was constructed and crossing over was measured in the CAN1-HOM3 and HOM3-TRP2 intervals. The reduction of crossing over in msh4 was not much affected by the additional exo1 mutation (Table 3) (the differences of genetic distances in each interval are statistically insignificant [p > 0.5 and p > 0.9, respectively]). Unexpectedly, however, spore viability in msh4 exo1 (28%, 870 tetrads dissected) is lower than that in msh4 (43%, 570 tetrads dissected). The difference is statistically significant (p < 0.0001)
DISCUSSION
Exo1 and Mre11 Are Involved in DSB Processing
We isolated the EXO1 gene as a high-copy suppressor of the MMS sensitivity of the mre11-1 mutant. The EXO1 gene also suppresses the MMS sensitivity of the mre11, rad50, and xrs2 null mutants but not the rad52 mutant. These results suggest that the function of Exo1 can compensate for the defect of the mre11, rad50, and xrs2 mutants in MMS-induced DNA damage repair. The exo1 mutant shows moderate MMS sensitivity, and in combination with the mre11 mutation, MMS sensitivity is more pronounced. These results suggest that Exo1 and the Mre11/Rad50/Xrs2 complex act independently in MMS-induced DNA damage repair.
MMS, a radiomimetic agent, is assumed to cause DSBs, and the MMS sensitivity of the mre11 mutant is attributed to its inability to repair DSBs (Tavassoli et al., 1995; Bressan et al., 1998). DSBs are predominantly repaired by homologous recombination in yeast (Friedberg et al., 1995), and the process of mating-type switching serves as a model system for analyzing the recombination process (Haber, 1995). In this system, the mre11 and rad50 null mutations reduce 5′ to 3′ exonucleolytic processing from DSB ends, which delays the formation of mature recombinants (Sugawara and Haber, 1992; Lee et al., 1998; Tsubouchi and Ogawa, 1998). Although DSB processing at the MAT locus appears normal in the exo1 mutant, processing in the exo1 mre11 double mutant is delayed compared with that in the mre11 single mutant.
Because EXO1 encodes a dsDNA 5′ to 3′ exonuclease (Fiorentini et al., 1997; Tishkoff et al., 1997), promotion of ssDNA formation at DSB ends by EXO1 overexpression could explain the suppression of the repair defect of mre11. Although MRE11 is involved in the NHEJ pathway as well, EXO1 overproduction does not suppress the reduction of NHEJ, suggesting that EXO1 suppression is independent of the NHEJ pathway.
These results argue that Exo1 and Mre11 are involved in DSB processing to create 3′-tailed ssDNA. It is likely that Exo1 is involved directly in this process, whereas recent biochemical studies have demonstrated that Mre11 (and human Mre11 complex, consisting of hMre11, hRad50, and p95) displays 3′ to 5′ dsDNA exonuclease activity as well as ssDNA endonuclease activity in vitro, and that these activities are not important for the creation of 3′-tailed ssDNA in vivo (Furuse et al., 1998; Paull and Gellert, 1998; Trujillo et al., 1998; Usui et al., 1998; Moreau et al., 1999). Thus, it is not likely that Mre11 plays a major role in DSB processing as a nuclease. A second possibility is that the role of the Mre11 complex in DSB processing is to facilitate ssDNA formation indirectly, e.g., the complex may recruit other enzyme(s) and help them to process DSB ends.
DSB Processing and DSB Repair
We have observed a correlation between MMS sensitivity and the efficiency of DSB processing during mating-type switching: no reduction in DSB processing and moderate MMS sensitivity in exo1; reduced processing and more severe MMS sensitivity in mre11; and even greater reduction in processing and sensitivity to MMS in the mre11 exo1 double mutant. The fact that the exo1 mutation does not affect mating-type switching, although the mutant is slightly sensitive to MMS, might be due to a difference in the number of DSBs induced. In mating-type switching, only one DSB is created per cell, whereas multiple DSBs are produced at high doses of MMS.
Functional Redundancy in DSB Processing
Although delayed, recombination still occurs in the absence of both Mre11 and Exo1, suggesting the existence of other protein(s) with exonuclease activity. There are four genes (RAD2, RAD27, DIN7, and YEN1) that encode putative proteins sharing homology with Exo1. DIN7 was recently shown to function specifically in mitochondria (Fikus et al., 2000), so it is unlikely that DIN7 and EXO1 function redundantly. In particular, RAD27 may share common functions with EXO1, because exo1 rad27 double mutants exhibit a synthetic lethal phenotype, and the EXO1 gene is a high-copy suppressor of the temperature-sensitive and mutator phenotypes of a rad27 mutant (Tishkoff et al., 1997). There may be other enzymes with exonuclease activity in vivo.
Mre11 and Regulation of Telomere Length
Although MRE11/RAD50 are implicated in the telomerase-mediated pathway for telomere replication, it remains unclear how they function (Haber, 1998; Nugent et al., 1998). We have confirmed that telomere shortening occurs in the mre11 null mutant, as expected from the earlier report for the rad50 mutant (Kironmai and Muniyappa, 1997; Boulton and Jackson, 1998; Nugent et al., 1998). The exo1 null mutation, however, does not affect the telomere length, and high copies of the EXO1 gene do not affect the telomere shortening seen in the mre11 null mutant. Therefore, the reduction in DSB processing observed in the mre11 mutant is not likely to be related to telomere shortening. Mutations in yeast Ku homologues abolish NHEJ and reduce telomere length (Boulton and Jackson, 1996a,b, 1998; Milne et al., 1996; Porter et al., 1996). Both yeast Ku homologues and MRE11, RAD50, and XRS2 are implicated in the same pathway of NHEJ (Milne et al., 1996; Boulton and Jackson, 1998); therefore, the NHEJ activity of the Mre11 complex could contribute to telomere maintenance. This is consistent with our finding that high-copy EXO1 does not suppress the NHEJ defect of the mre11 mutant.
Exo1 and Crossing Over during Meiosis
In the present investigation, we observed increased fractions of tetrads exhibiting zero, two, and four viable spores in exo1 mutants, reminiscent of conditions that cause defects in meiosis I. Crossing over is reduced to ∼50% of the wild-type level and increased levels of chromosome nondisjunction are observed, suggesting that Exo1 is important for homologous chromosome segregation by ensuring crossing over. Consistent with this view, none of the eight chromosome III disomes was recombinant. In the absence of Mlh1, Mlh3, Msh4, Msh5, Zip1, Zip2, and Mer3, crossing over is reduced twofold to threefold during meiosis, leading to abnormal reductional division (Ross and Roeder, 1994; Sym and Roeder, 1994; Hollingsworth et al., 1995; Hunter and Borts, 1997; Chua and Roeder, 1998; Nakagawa and Ogawa, 1999; Wang et al., 1999). Mlh1, Mlh3, Msh4, and Msh5 are homologues of mismatch repair proteins, and both Mlh1 and Mlh3, like Exo1, are also required for mismatch correction; thus, a close mechanistic correlation between mismatch repair and crossing over is suggested. Our genetic data show that crossing over in the exo1 msh4 double mutant is no more reduced than in msh4, arguing that Exo1 and Msh4 work in a common pathway. There is a strong interaction between EXO1 and MSH2; MSH2 encodes a homologue of the MutS protein of E. coli and plays a central part in recognition of mismatch base pairing (Tishkoff et al., 1997), but it does not play a role in crossing over (Hunter and Borts, 1997). Exo1 physically interacts with Msh2, and EXO1 acts in the MSH2-dependent mismatch repair pathway. Because Msh2 and Msh4 are two homologues of the bacterial MutS protein, Exo1 function in mismatch repair may be mechanistically related to its role in crossing over.
Although the rate of meiotic crossing over in msh4 is not affected by the exo1 mutation, fewer spores are viable in the exo1 msh4 double mutant than in the msh4 single mutant, suggesting that Exo1 and Msh4 have additional roles during meiosis other than crossing over. Crossing over interference is severely reduced in msh4 strains, and the distribution of crossing over among chromosomes is random (Roeder, 1997). The greater spore viability of exo1 strains, despite a similar reduction in crossing over, suggests that interference is unaffected by the exo1 mutation. On the other hand, we have observed a reduction in the processing of meiosis DSBs in exo1. The greater reduction in spore viability in msh4 exo1 strains, compared with the corresponding single mutants, might be due to the combination of a defect in interference and a failure to repair some DSBs.
ACKNOWLEDGMENTS
We thank G.S. Roeder, L. Symington, J.E. Haber, and A. Shinohara for helpful discussions and critical reading of the manuscript, T. Tsubouchi for helpful discussions, R. Nobuki for technical assistance, and other members of the Ogawa laboratory for their stimulating comments. We also are indebted to J.E. Haber, F. Ishikawa, G.S. Roeder, and N. Kleckner for providing yeast strains and plasmids. This work was supported by Grants-in-Aid for Specially Promoted Research from the Ministry of Education, Science, Sports, and Culture of Japan, by the Howard Hughes Medical Institute, and by CREST of JST (Japan Science and Technology).
REFERENCES
- Ajimura M, Leem SH, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Alani E, Subbiah S, Kleckner N. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics. 1989;122:47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Botstein D, Falco SC, Stewart SE, Brennan M, Scherer S, Stinchomb DT, Struhl K, Davis RW. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996a;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996b;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998a;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998b;282:1421. doi: 10.1126/science.282.5389.699. (Errata). [DOI] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- Digilio FA, Pannuti A, Lucchesi JC, Furia M, Polito LC. Tosca: a Drosophila gene encoding a nuclease specifically expressed in the female germline. Dev Biol. 1996;178:90–100. doi: 10.1006/dbio.1996.0200. [DOI] [PubMed] [Google Scholar]
- Esposito RE, Esposito MS. Genetic recombination and commitment to meiosis in Saccharomyces. Proc Natl Acad Sci USA. 1974;71:3172–3176. doi: 10.1073/pnas.71.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast D. Sporulation synchrony of Saccharomyces cerevisiae grown in various carbon sources. J Bacteriol. 1973;116:925–930. doi: 10.1128/jb.116.2.925-930.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fikus MU, Mieczkowski PA, Koprowski P, Rytka J, Sledziewska GE, Ciesla Z. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics. 2000;154:73–81. doi: 10.1093/genetics/154.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: American Society of Microbiology Press; 1995. [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami MK, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D, Seidel R. Molecular cloning of the actin gene from yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1980;8:1043–1059. doi: 10.1093/nar/8.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC, Mortimer RK. A genetic study of X-ray sensitive mutants in yeast. Mutat Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Haber JE. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- Haber JE. A super new twist on the initiation of meiotic recombination. Cell. 1997;89:163–166. doi: 10.1016/s0092-8674(00)80194-4. [DOI] [PubMed] [Google Scholar]
- Haber JE. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- Hunter N, Borts RH. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Korolev VG, Fabre F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics. 1992;132:651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai KM, Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moor JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- McKee AH, Kleckner N. Mutations in Saccharomyces cerevisiae that block meiotic prophase chromosome metabolism and confer cell cycle arrest at pachytene identify two new meiosis-specific genes SAE1 and SAE3. Genetics. 1997;146:817–834. doi: 10.1093/genetics/146.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Fikus MU, Ciesla Z. Characterization of a novel DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, which is a structural homolog of the RAD2 and RAD27 DNA repair genes. Mol Gen Genet. 1997;253:655–665. doi: 10.1007/s004380050369. [DOI] [PubMed] [Google Scholar]
- Milne GT, Jin S, Shannon KB, Weaver DT. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Datta A, Kolodner RD. Multiple functions of MutS- and MutL-related heterocomplexes. Proc Natl Acad Sci USA. 1999;96:14186–14188. doi: 10.1073/pnas.96.25.14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Ogawa H. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 1999;18:5714–5723. doi: 10.1093/emboj/18.20.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman EH, Ogawa H. RecA-like recombination proteins in eukaryotes: functions and structures of RAD51 genes. Cold Spring Harbor Symp Quant Biol. 1993;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Porter SE, Greenwell PW, Ritchie KB, Petes TD. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Ross MP, Roeder GS. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sherman F, Roman H. Evidence for two types of allelic recombination in yeast. Genetics. 1963;48:255–261. doi: 10.1093/genetics/48.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Stahl F. Meiotic recombination in yeast: coronation of the double-strand-break repair model. Cell. 1996;87:965–968. doi: 10.1016/s0092-8674(00)81791-2. [DOI] [PubMed] [Google Scholar]
- Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Ivanov EL, Fishman LJ, Ray BL, Wu X, Haber JE. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- Tavassoli M, Shayeghi M, Nasim A, Watts FZ. Cloning and characterization of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 1995;23:383–388. doi: 10.1093/nar/23.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci USA. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]