Abstract
The BOB.1/OBF.1 coactivator is critically involved in mediating octamer-dependent transcriptional activity in B lymphocytes. Mice lacking this coactivator show various defects in B-cell development, most notably they completely lack germinal centers. Consistent with this phenotype, BOB.1/OBF.1 levels are massively upregulated in germinal center B cells as compared with resting B cells. We have addressed the mechanism of upregulation and found that only a minor part of this regulation can be attributed to increased levels of BOB.1/OBF.1-specific mRNA. Apparently, BOB.1/OBF.1 is also regulated at the protein level. In support of this suggestion we have been able to identify two related BOB.1/OBF.1 interacting proteins, SIAH1 and SIAH2, in a yeast two-hybrid screen. SIAH1 and SIAH2 are known regulators of protein stability. Cotransfection experiments revealed that coexpression of SIAH results in a destabilization of BOB.1/OBF.1 protein without affecting mRNA levels. Further more, proteasome inhibitors block the degradation of BOB.1/OBF.1 protein. Finally, B-cell receptor cross-linking also resulted in the degradation of BOB.1/OBF.1 and consequently reduced transcriptional activation of BOB.1/OBF.1-dependent reporters.
Keywords: B cell/BOB.1/OBF.1/coactivator/SIAH/ubiquitin ligase
Introduction
The octamer motif plays a crucial role in B-cell-specific transcription. It is conserved in virtually all immunoglobulin heavy chain and light chain gene promoters as well as in several immunoglobulin enhancer elements. It is essential for the B-cell-specific promoter function and contributes to the immunoglobulin enhancer activity in B cells (Matthias, 1998). Transcriptional activity at octamer-containing promoters in lymphoid cells requires a combination of specific transcription factors and coactivators. B cells express two octamer-binding transcription factors, Oct1 and Oct2. Furthermore, they express the B-cell-restricted coactivator BOB.1/OBF.1 (other names are OCA-B, Bob1, OBF-1). This coactivator interacts with the Oct proteins and is critical for B-cell-specific transcription of promoters containing the octamer motif (Gstaiger et al., 1995; Luo and Roeder, 1995; Pfisterer et al., 1995; Strubin et al., 1995).
BOB.1/OBF.1 is a transcription factor-specific coactivator and so far has only been found to interact functionally with a subgroup of Oct proteins (Sauter and Matthias, 1998). The BOB.1/OBF.1 protein is recruited to the transcription complex via its interaction with the POU-domains of the Oct proteins. In this complex, the N-terminal domain of BOB.1/OBF.1 contacts the DNA directly in the major groove (Cepek et al., 1996; Gstaiger et al., 1996; Chasman et al., 1999). By interaction with both subdomains of the POU-domain of the Oct proteins (the POU-homeo and the POU-specific domain) BOB.1/OBF.1 acts as a molecular clamp, fixing the Oct protein to the DNA (Sauter and Matthias, 1998). BOB.1/OBF.1 can be subdivided into two functional domains, the N-terminal domain is critical for the interaction with Oct1, Oct2 and the DNA. The C-terminal domain is mainly responsible for the transactivation function (Pfisterer et al., 1995; Gstaiger et al., 1996). Analysis of the crystal structure of a BOB.1/OBF.1 peptide bound to an Oct1 POU-domain/octamer DNA complex have shown that amino acids (aa) 16–38 form an ordered structure in the crystal. These amino acids are responsible for the interaction with the Oct POU-domains and in addition for the contact to an adenine at position five in the DNA octamer motif (Chasman et al., 1999). The C-terminal domain was shown to contact the basal transcription machinery as well as additional transcription cofactors like PC4 (Luo and Roeder, 1995).
BOB.1/OBF.1 is constitutively expressed in B cells whereas T cells show an inducible expression after costimulation (Zwilling et al., 1997). Similar expression levels were found in transformed cell lines representing the different stages of B-cell development (Schubart et al., 1996b). Disruption of the BOB.1/OBF.1 gene resulted in a reduction of transitional B cells in the bone marrow (Hess et al., 2001) and in a complete loss of germinal center formation in the spleen and in lymph nodes (Kim et al., 1996; Nielsen et al., 1996; Schubart et al., 1996b). Consistent with this phenotype it was shown that BOB.1/OBF.1 protein expression in primary B cells is highest in germinal center B cells (Qin et al., 1998; Greiner et al., 2000). In vitro, RNA expression of BOB.1/OBF.1 can be induced in resting splenic B cells by stimulation with lipopolysaccharide (LPS) or anti-CD40/interleukin-4 (IL-4), suggesting that the upregulation is a consequence of increased levels of transcript (Qin et al., 1998; Greiner et al., 2000). In line with these observations, the promoter of BOB.1/OBF.1 contains a cAMP regulatory element binding protein/activating transcription factor (CREB/ATF) binding site, crucial for induction by anti-CD40 and IL-4 (Stevens et al., 2000).
Several transcription factors such as p53 and c-Jun are regulated by protein stability. These proteins become ubiquitylated and subsequently degraded by the proteasome. Protein ubiquitylation depends on a ubiquitin activating enzyme (E1) and ubiquitin conjugating enzymes (Ubc or E2). In addition, E3 proteins (ubiquitin ligases) interact with E2 and the substrate and are critical for specificity. Many E3 proteins are characterized by the presence of a RING finger domain (Lorick et al., 1999; Joazeiro and Weissman, 2000). The p53 regulating protein Mdm2 is such an E3 RING finger protein (Honda et al., 1997; Fang et al., 2000). Another example is the Drosophila protein SINA (seven in absentia), which is necessary for R7 eye development (Carthew and Rubin, 1990). The human homologs of SINA, SIAH1 and SIAH2 (seven in absentia homolog), have been shown to interact with DCC (deleted in colon cancer) and NCoR (nuclear corepressor), and thereby induce their degradation (Hu et al., 1997b; Zhang et al., 1998).
Here we show that BOB.1/OBF.1 interacts with SIAH1 and SIAH2. The interaction with SIAH1 leads to a degradation of BOB.1/OBF.1 via the proteasome pathway. This degradation can be induced in Ramos B cells by triggering the B-cell receptor (BCR). The finding of an absence of a strong transcriptional regulation of BOB.1/OBF.1 in germinal center versus non-germinal center B cells suggests that the regulation of the BOB.1/OBF.1 protein stability plays a critical role and could be mediated by the interaction with the SIAH proteins.
Results
Stability of the BOB.1/OBF.1 protein is regulated
Previous analyses had shown that BOB.1/OBF.1 protein levels are much higher in CD38+ germinal center B cells compared with CD38– non-germinal center cells. To determine whether this upregulation is due to increased mRNA levels, primary CD38+ and CD38– B cells were isolated from human tonsils and mRNA expression was quantified by real time PCR analyses (LightCycler). As an internal control we used the housekeeping gene Porphobilinogen-Deaminase (PBGD), which shows low ubiquitous expression and has no known pseudogenes (Fink et al., 1998, 1999). As a positive control we measured CD38 mRNA expression in the two purified cell populations and saw a 15-fold upregulation in CD38+ selected cells, which was consistent with the purity of the cell populations determined by FACS analysis (Figure 1A and data not shown). We next determined the mRNA levels for BOB.1/OBF.1. To our surprise only a very weak induction (<2-fold) in CD38+ cells was observed (Figure 1A). When we analyzed the BOB.1/OBF.1 protein levels in these cell populations we found a >6-fold induction in CD38+ cells (Figure 1B). From these results we conclude that BOB.1/OBF.1 is not only regulated at the level of RNA transcription or RNA stability but also most likely at the protein level.
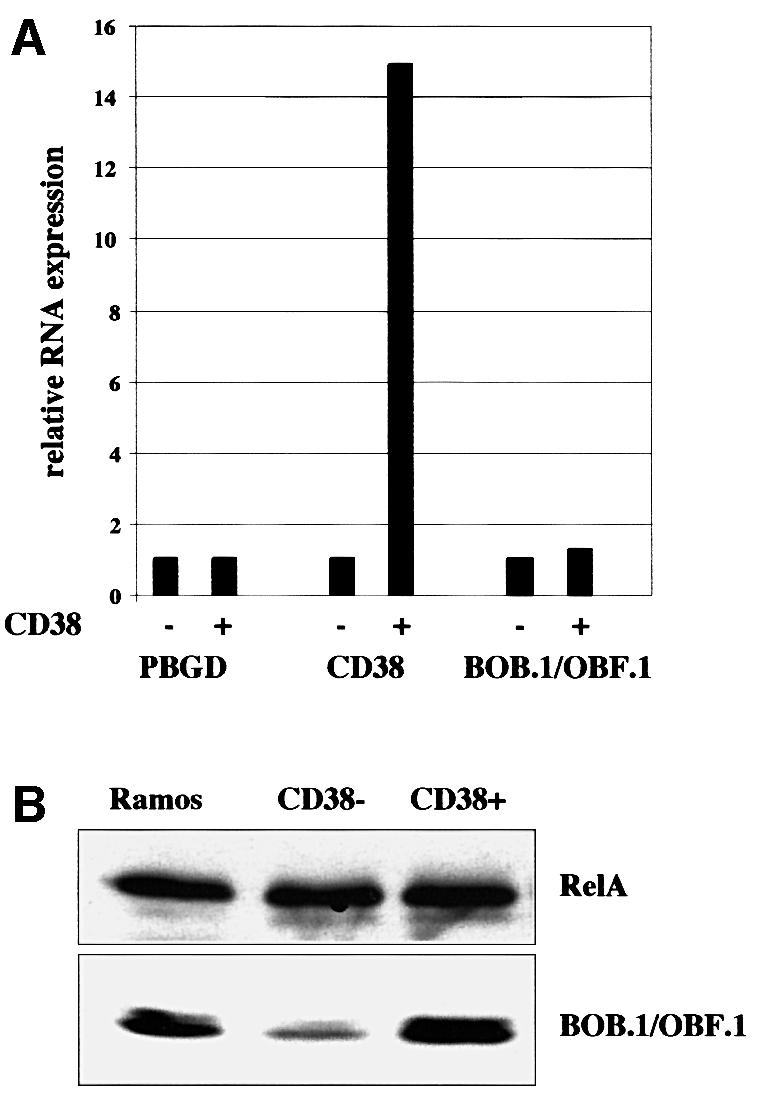
Fig. 1. BOB.1/OBF.1 protein levels are regulated in germinal center B cells. RNA and protein extracts were prepared from purified CD38– and CD38+ human tonsillar B cells. (A) BOB.1/OBF.1 expression levels were determined by quantitative real time PCR. CD38 was used as a positive control for the transcriptional regulation, PBGD was used for standardization. (B) Detection of BOB.1/OBF.1 protein levels by immunoblotting. Ramos B cells were used as positive control, RelA was used as a loading control.
BOB.1/OBF.1 interacts with SIAH proteins
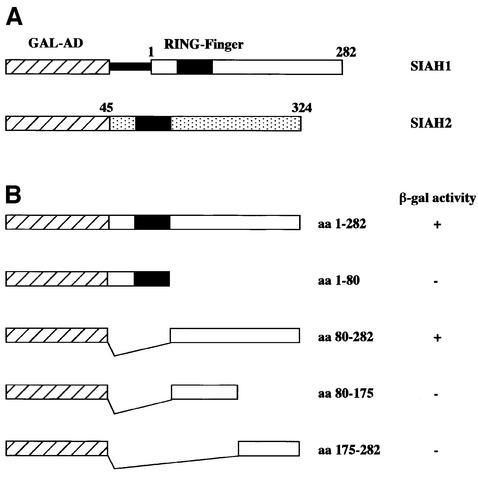
In order to identify proteins that could influence BOB.1/OBF.1 protein stability, we searched for interacting proteins in a yeast two-hybrid screen. The N-terminal domain of BOB.1/OBF.1 (aa 1–72) was cloned into a bait vector and a total of 9.3 × 106 independent transformants of a Jurkat library cloned in pGAD10 were analyzed. Only five positive clones were observed and their identity was established by sequencing. Four of the clones contained the same complete coding sequence of the human SIAH1 cDNA including 173 nt of 5′ untranslated sequence. The fifth clone contained most of the coding sequence of the SIAH2 cDNA, lacking only the first 45 aa (Figure 2A). The human SIAH1 and SIAH2 proteins are highly homologous (77% overall sequence identity) and differ predominantly by the presence of an extension in the N-terminus of the SIAH2 protein (Hu et al., 1997a).
Fig. 2. BOB.1/OBF.1 interacts with SIAH proteins in a yeast two-hybrid assay. (A) Clones of SIAH1 and SIAH2 obtained in the yeast two-hybrid assay. The SIAH1 clones contained the untranslated 5′-region, indicated by the thin black bar. The SIAH2 clone was truncated, missing the first 45 aa. GAL-AD, activation domain of the GAL4 transcription factor. (B) Mapping of the region of SIAH1 needed for the interaction with BOB.1/OBF.1. HF7c yeast cells were transformed with full-length SIAH1, the N-terminal part, the C-terminal part or truncated forms of the C-terminal part of SIAH1 together with the BOB.1/OBF.1 bait plasmid. Positive two-hybrid interaction was indicated by growth on -His +3AT plates and by β-gal activity. The ‘+’ for β-gal activity indicates that colonies turned blue within 1 h. The ‘–’ refers to no color change within 8 h.
There was no stop codon present in the leader sequence of the SIAH1 cDNA and the fusion retained the correct open reading frame of the SIAH1 protein. Nevertheless, we wanted to ensure that the interaction with BOB.1/OBF.1 was mediated by the SIAH1 open reading frame and generated an exact fusion between the GAL4 activation domain and the full-length SIAH1 protein eliminating the 5′ non-coding region. In addition, we generated fusion proteins with smaller fragments from the SIAH1 protein in order to identify the interaction domain with BOB.1/OBF.1. The results of these analyses are summarized in Figure 2B. Both the full-length SIAH1 protein as well as the C-terminal domain of SIAH1 interacted with BOB.1/OBF.1. In contrast, the N-terminal domain containing the ring finger domain (aa 39–76; Hu et al., 1997a) and sub-fragments of the C-terminal domain failed to show interaction. These results were consistent with the observation that in the case of the SIAH2 clone identified in the two-hybrid screen, the N-terminal domain was partially deleted. All of the subsequent experiments were performed with SIAH1.
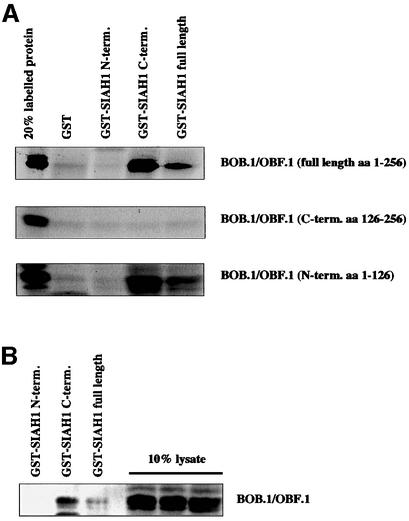
In order to verify the interaction of BOB.1/OBF.1 with SIAH1 in an independent assay system, glutathione S-transferase (GST) pull-down experiments were performed with recombinant proteins. GST fusion proteins containing either full-length SIAH1, or the N-terminal (aa 1–80) or C-terminal (aa 80–282) domains, were generated and purified. These fusion proteins and, as a control, GST were incubated with in vitro translated BOB.1/OBF.1 protein. Consistent with the results obtained in the yeast two-hybrid analyses, both the full-length SIAH1 and the C-terminal domain of SIAH1 interacted with BOB.1/OBF.1, whereas the N-terminal domain and GST by itself failed to show an interaction (Figure 3A, upper panel). This experimental approach was then used to determine the domains in the BOB.1/OBF.1 protein interacting with SIAH1. The fact that the N-terminal domain of BOB.1/OBF.1 was used as bait in the two-hybrid screen already indicated that it contains an interaction domain. Nevertheless, it did not rule out the possibility that the C-terminus of BOB.1/OBF.1 could also interact with SIAH1. However, when the BOB.1/OBF.1 subdomains were expressed independently, only the N-terminus showed interaction with full-length SIAH1 or the SIAH1 C-terminal domain (Figure 3A). Furthermore, when the GST–SIAH1 fusion proteins were incubated with whole-cell extracts from S194 murine B cells, native BOB.1/OBF.1 could be precipitated with full-length SIAH1 and the C-terminus, but not with the N-terminus of SIAH1 (Figure 3B).
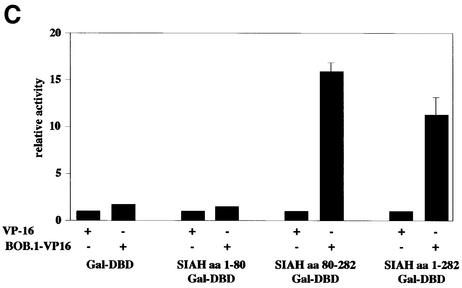
Fig. 3. In vitro and in vivo interaction between BOB.1/OBF.1 and SIAH1. (A) The SIAH1 C-terminus interacts with the N-terminus of BOB.1/OBF.1. Full-length SIAH1, the N-terminus or the C-terminus was fused to the GST protein. GST–SIAH1 fusion proteins were incubated with in vitro translated 35S-labeled BOB.1/OBF.1. The labeled N-terminal part, the C-terminal part and full-length BOB.1/OBF.1 were used for the pull-down assays. (B) In vitro interaction using GST–SIAH1 constructs. GST–SIAH1 fusion proteins were incubated with lysates from S194 B cells. BOB.1/OBF.1 was detected by immunoblotting. (C) Mammalian two-hybrid assay for BOB.1/OBF.1 and SIAH1. NIH 3T3 cells were cotransfected with the indicated Gal-SIAH1 constructs (N-, C-terminal part and full-length) together with the N-terminal part of BOB.1/OBF.1 fused to the VP16 transactivation domain. Interaction was detected using a luciferase reporter under the control of GAL binding sites. DBD, DNA binding domain.
We then addressed the question of whether BOB.1/OBF.1 and SIAH1 could interact in a mammalian cell. We therefore established a mammalian two-hybrid system, where either full-length SIAH1 or the subdomains were fused to the heterologous GAL4 DNA binding domain. These fusion proteins did not show any activity on their own when tested together with a luciferase reporter construct driven by four GAL4-binding sites in the promoter (Figure 3C). However, when an expression vector for a BOB.1/OBF.1–VP16 fusion protein was cotransfected, the activity of the GAL fusions with full-length SIAH1 or the C-terminal domain of SIAH1 was significantly induced. Taken together, these results demonstrate that SIAH1 is a bona fide BOB.1/OBF.1 interacting protein.
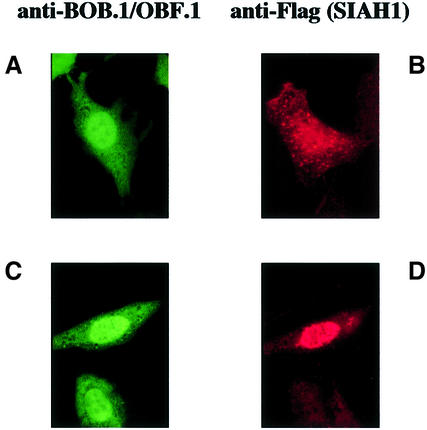
We reasoned that if there is an intricate relationship between BOB.1/OBF.1 and SIAH1, the two proteins should colocalize in cells. To test this hypothesis, we performed immunofluorescence analyses of transiently transfected cells expressing only BOB.1/OBF.1, only SIAH1 or both proteins together. When a Flag-tagged version of SIAH1 was transfected into NIH 3T3 cells, we saw a speckled staining all over the cell with some accumulation of SIAH1 in the nucleus (Figure 4). Expression of wild-type BOB.1/OBF.1 also showed both nuclear and cytoplasmic staining. This is consistent with the finding that BOB.1/OBF.1 is localized in both the nucleus and cytoplasm of B cells (Yu et al., 2001). Cotransfection of BOB.1/OBF.1 together with Flag-SIAH1 resulted in the appearence of an additional speckled staining for BOB.1/OBF.1, identical to that observed for SIAH1 (data not shown). However, even more convincing colocalization was observed when we used a version of the BOB.1/OBF.1 protein carrying an additional nuclear localization sequence (NLS-BOB.1/OBF.1). NLS-BOB.1/OBF.1 is almost exclusively localized in the nucleus in transfected NIH 3T3 cells (Figure 4). When both NLS-BOB.1/OBF.1 and Flag-SIAH1 were cotransfected into NIH 3T3 cells, immunofluorescence analysis revealed that the SIAH1 protein was redistributed to the nucleus (Figure 4). These results show that the proteins colocalize in transfected cells.
Fig. 4. Alteration of the subcellular localization of SIAH1 in cotransfection with BOB.1/OBF.1. NIH 3T3 cells were transfected with a BOB.1/OBF.1 construct containing an additional NLS sequence (A), Flag-SIAH (B) or both together (C and D). 18 h after transfection, the cells were fixed and stained with polyclonal anti-BOB.1/OBF.1 antibody and FITC-conjugated goat anti-rabbit antibody (A), monoclonal anti-FLAG antibody M2 and Cy3-conjugated goat anti-mouse antibody (B), or with both first and secondary antibodies together (C and D).
Interaction of BOB.1/OBF.1 with SIAH1 leads to a degradation of the BOB.1/OBF.1 protein
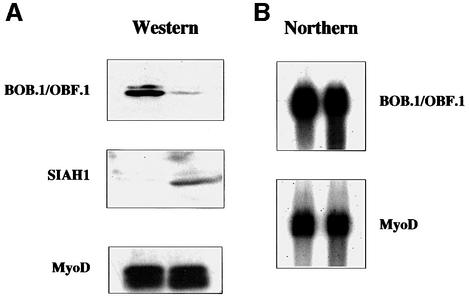
SINA, the Drosophila homolog of SIAH1, is involved in regulating protein stability by interacting with target proteins and mediating their ubiquitylation and degradation via the proteasome pathway. Similarly, some of the recently identified SIAH-interacting proteins such as DCC and NCoR were found to be degraded (Hu et al., 1997b; Zhang et al., 1998). We therefore investigated whether interaction with SIAH1 would result in BOB.1/OBF.1 degradation. After cotransfection of SIAH1 and BOB.1/OBF.1 into NIH 3T3 cells, expression of BOB.1/OBF.1 was analyzed both by protein and RNA analysis. As a control, an expression vector for myc-tagged MyoD was included. Whereas there was no reduction in the amount of BOB.1/OBF.1 RNA visible upon SIAH1 cotransfection (Figure 5B), a clear reduction in the amount of BOB.1/OBF.1 protein was observed (Figure 5A). The reduction in protein levels was specific, as the control protein was not affected.
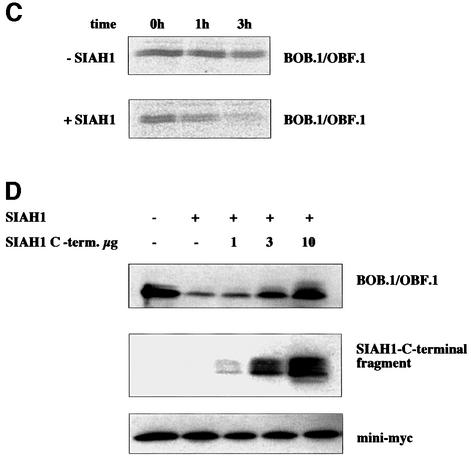
Fig. 5. SIAH1 mediates degradation of BOB.1/OBF.1. (A) Western blot of NIH 3T3 cells transfected with BOB.1/OBF.1 alone or together with SIAH1. Myc-tagged MyoD was used as a loading control. (B) Northern blot of the same transfected cells. (C) The BOB.1/OBF.1 half-life time becomes decreased by SIAH1. NIH 3T3 cells were transfected as described above. After pulse labeling the half-life time of BOB.1/OBF.1 was measured. (D) Western blot of NIH 3T3 cells transfected with vectors containing BOB.1/OBF.1, SIAH1 or increasing amounts of the myc-tagged SIAH1 C-terminal part. Mini-myc was used as a loading control.
To assess the effects of SIAH1 on the BOB.1/OBF.1 protein turnover more quantitatively, we measured half-life times of the BOB.1/OBF.1 protein in the presence or absence of SIAH1. NIH 3T3 cells were transfected with expression vectors for BOB.1/OBF.1 and SIAH1. BOB.1/OBF.1 protein levels were determined by metabolic labeling and pulse–chase experiments. In cells transfected with BOB.1/OBF.1 alone, the half-life time was ∼3 h. Cotransfection of SIAH1 leads to a reduction of the BOB.1/OBF.1 half-life time to 1.5 h (Figure 5C).
SIAH1 and SIAH2 as well as the Drosophila homolog SINA belong to a family of ring-zinc-finger proteins. These proteins act as E3 factors in the ubiquitylation pathway and thereby target their interaction partners for degradation (Lorick et al., 1999; Joazeiro and Weissman, 2000). Deletion in this ring-zinc-finger domain led to a block in ubiquitylation and to a stabilization of the SIAH1 protein (Hu and Fearon, 1999). The SIAH ring-zinc-finger is located in the N-terminal part whereas the BOB.1/OBF.1 interaction domain is C-terminally located. Cotransfection of increasing amounts of an N-terminally deleted SIAH1 construct together with SIAH1 full-length and BOB.1/OBF.1 led to a partial rescue of BOB.1/OBF.1 from degradation (Figure 5D).
SIAH1 and Oct1/2 contact different domains in BOB.1/OBF.1
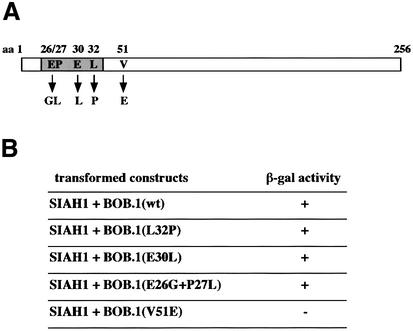
The N-terminal domain of BOB.1/OBF.1, which is involved in the interaction with SIAH1, has previously been shown also to be critical for mediating the contact with the POU-domains of Oct1 and Oct2. We were therefore interested to determine whether identical amino acids in the BOB.1/OBF.1 N-terminal domain would be critical for both contacts. Several point mutations of BOB.1/OBF.1 have been described that either did or did not affect the interaction with the POU-domain. Specifically, mutations in a short stretch of amino acids around position 30 were shown to interfere with POU-domain interaction (Gstaiger et al., 1996). This is also consistent with the X-ray crystal structure of the ternary complex (Chasman et al., 1999). In contrast, mutation of the amino acid at position 51 (V to E) did not affect BOB.1/OBF.1 interaction with the Oct1 POU-domain. We tested these mutants for their ability to interact with BOB.1/OBF.1 using the yeast interaction system. For this purpose, SIAH1 was fused to the GAL4 DNA binding domain and cotransformed with the GAL4 activation domain fused to BOB.1/OBF.1. This experiment revealed that all three single or double point mutants that were unable to interact with the POU-domain could still bind to SIAH1. In contrast, mutation of amino acid 51, which showed no effect on POU-domain interaction, failed to bind to the SIAH1 protein (Figure 6A and B). From this analysis we conclude that the interfaces of BOB.1/OBF.1 with either SIAH1 or the Oct1 POU-domain are in separate subdomains of the N-terminal region of BOB.1/OBF.1.
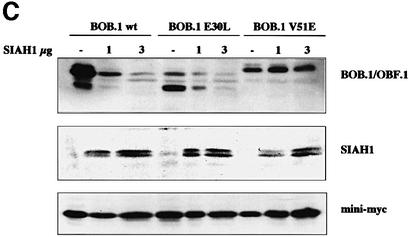
Fig. 6. Oct1/2 and SIAH1 do not bind to the same region in BOB.1/OBF.1. (A) Point mutations of BOB.1/OBF.1 (L32P, E30L, E26G+P27L) that are no longer able to interact with the Oct proteins are located in a short stretch of 7 aa, whereas the point mutation V51E, lying outside that stretch, is still able to interact with Oct1/2. (B) Yeast two-hybrid assay for the binding of SIAH1 to BOB.1/OBF.1 mutants. SIAH1 was cotransformed with the different point mutations (described in A) into HF7c yeast cells. The interaction of proteins was indicated by growth on -His +3AT plates and by β-gal activity. (C) The point mutants of BOB.1/OBF.1 (described in A) were cloned into pcDNA3 and cotransfected with different amounts of HA-tagged SIAH1 and an internal control vector called mini-myc into NIH 3T3 cells. Western blot analysis was performed using polyclonal anti-BOB.1/OBF.1 antibody, monoclonal anti-HA antibody 12CA5 or monoclonal anti-myc antibody 9E10.
We used the BOB.1/OBF.1 mutants to test whether the interaction with SIAH1 was necessary for degradation of BOB.1/OBF.1. Increasing amounts of cotransfected SIAH1 resulted in a decrease of wild-type BOB.1/OBF.1 protein levels in NIH 3T3 cells (Figure 6C). The E30L mutant, which could still interact with SIAH1, is also degraded in a SIAH1 concentration-dependent manner. In contrast, the V51E mutant, which does not interact with SIAH1, was also not degraded. A control protein was cotransfected in all experiments to confirm the specificity of the observed degradation effects (Figure 6C, lower panel). This result shows that interaction of BOB.1/OBF.1 with the SIAH1 protein is a prerequisite for degradation, whereas interaction with the Oct proteins is not required.
Both SIAH1 and the POU-domains of Oct1 and Oct2 interact with the N-terminal part of BOB.1/OBF.1. We considered the possibility that interaction of BOB.1/OBF.1 with the Oct POU-domain might protect the coactivator from SIAH-mediated degradation. We therefore cotransfected Oct2 together with SIAH1 and BOB.1/OBF.1 into NIH 3T3 cells. Even high amounts of Oct2 failed to interfere with the BOB.1/OBF.1 degradation (data not shown).
BOB.1/OBF.1 degradation occurs via the proteasome
Earlier studies had shown that SIAH1-mediated protein degradation involves the proteasome pathway. To investigate whether SIAH1-mediated degradation of BOB.1/OBF.1 also proceeds via this pathway, the proteasome inhibitor MG132 was included in the cotransfection experiments. Whereas BOB.1/OBF.1 was readily degraded in mock-treated cells, incubation of the transfected cells with MG132 for 6 h resulted in an efficient rescue of BOB.1/OBF.1 protein (Figure 7A). This suggests that BOB.1/OBF.1 is degraded by the proteasome in these experiments.
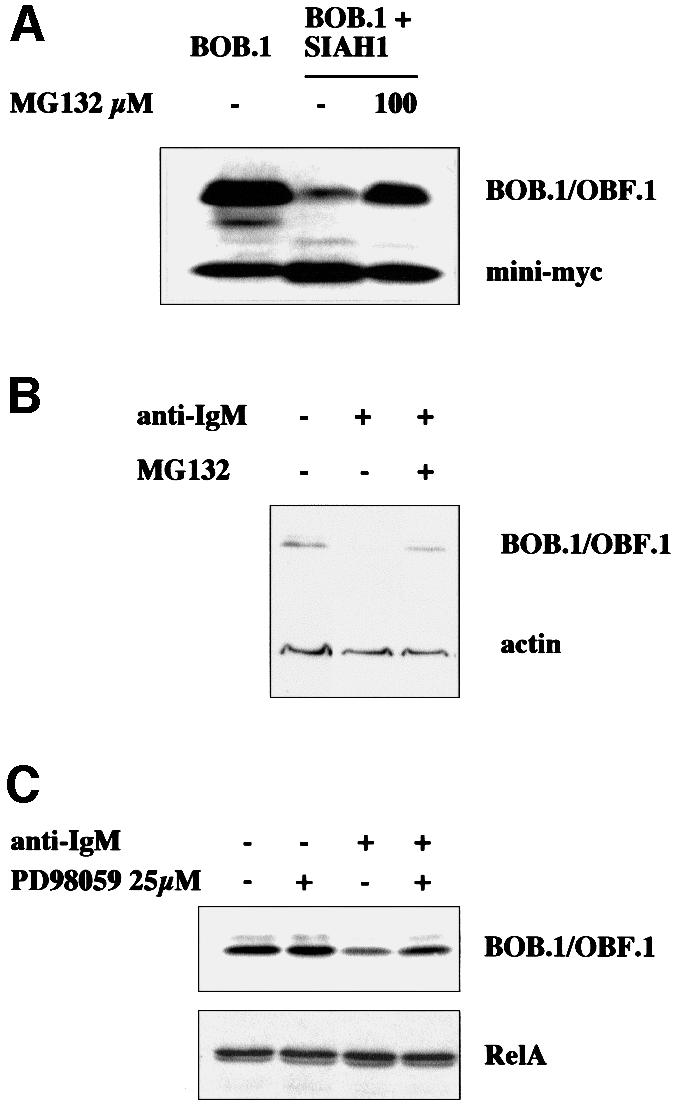
Fig. 7. BOB.1/OBF.1 becomes degraded in a proteasome-dependent manner. (A) NIH 3T3 cells were transfected with BOB.1/OBF.1 expression plasmid alone or together with SIAH1 expression plasmid. Eight hours later, the proteasome inhibitor MG132 was added. (B) Ramos B cells were incubated with anti-IgM antibodies and MG132 as indicated. After 12 h the cells were harvested and analyzed by western blotting with appropriate antibodies. (C) Thirty minutes before Ramos B cells were incubated with anti-IgM antibody, the MAPK inhibitor PD98059 was added (25 µM). Twelve hours later the cells were harvested and analyzed by western blotting.
Induction of BCR signaling in mature B cells in the absence of costimulation results in the downregulation of immunoglobulin gene transcription. We have used Ramos B cells and treated them with anti-IgM antibodies to mimic BCR activation. This treatment resulted in a specific reduction of BOB.1/OBF.1 protein levels. Furthermore, the degradation of BOB.1/OBF.1 could be blocked by addition of the proteasome inhibitor MG132 (Figure 7B). As mentioned before, Drosophila Sina is involved in regulating the protein stability of its target proteins. This regulation is dependent on the activation of the MAPK pathway. To test whether the MAPK pathway, which becomes activated by BCR cross-linking, is involved, we pre-incubated the cells with the MAPK inhibitor PD98059 (Figure 7C). As shown, the degradation of BOB.1/OBF.1 is partially blocked by inhibiting MAPK, indicating that as in Drosophila this pathway is critical in regulating protein stability by SIAH1.
SIAH1 is regulated at the transcription level
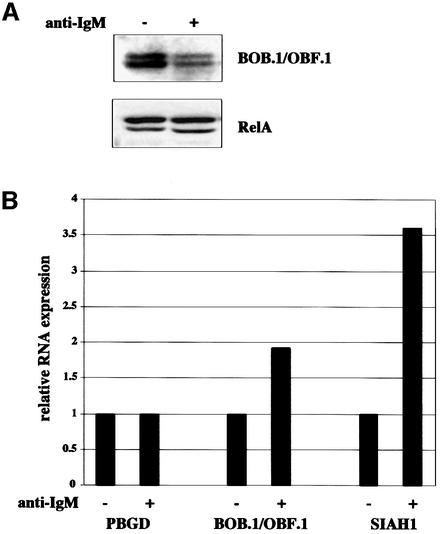
Two likely scenarios could explain the regulation of BOB.1/OBF.1 degradation by SIAH1. Either SIAH1 levels could be regulated, e.g. at the RNA expression level, or interaction of BOB.1/OBF.1 and SIAH1 might be controlled by protein modification. To investigate SIAH1 expression, we prepared protein and RNA from IgM cross-linked Ramos B cells. The BOB.1/OBF.1 protein degradation was analyzed as a control for the proper stimulation of the cells (Figure 8A). SIAH1 and BOB.1/OBF.1 RNA levels were analyzed by real time RT–PCR. In anti-IgM-treated cells, SIAH1 RNA levels were upregulated 3.6-fold (Figure 8B). Interestingly, although BOB.1/OBF.1 protein levels were reduced, BOB.1/OBF.1 RNA levels were also slightly induced (1.9-fold).
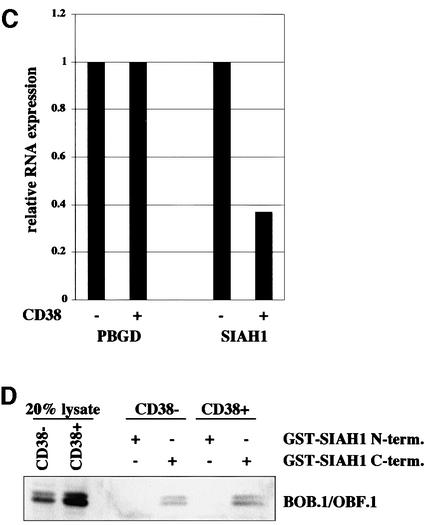
Fig. 8. SIAH1 is regulated at the transcriptional level. (A) BOB.1/OBF.1 protein levels from BCR cross-linked Ramos B cells were analyzed by western blotting. RelA was used as a loading control. (B) RNA from the same cells used in (A) was transcribed into cDNA and SIAH1 expression was analyzed by LightCycler PCR. (C) The cDNAs from CD38– and CD38+ tonsillar B cells, already used in Figure 1A, were analyzed for SIAH1 expression by LightCycler PCR. (D) Potential altered interaction of BOB.1/OBF.1 and SIAH1 in CD38– and CD38+ tonsillar B cells was analyzed by pull-down assays. Therefore, the GST–SIAH1 C-terminal and the GST–SIAH1 N-terminal part were mixed with lysates from CD38– and CD38+ tonsillar B cells.
We then analyzed SIAH1 expression in primary B cells. RNAs from sorted CD38– versus CD38+ B cells were analyzed by quantitative RT–PCR. Expression of SIAH1 was reduced 2.7-fold in CD38+ B cells as compared with the CD38– population (Figure 8C). This result is consistent with a model suggesting that regulation of SIAH1 levels is responsible for controlling BOB.1/OBF.1 protein levels. However, we also tested the hypothesis that a modification of BOB.1/OBF.1 could modulate the interaction with SIAH1 and, as a consequence, BOB.1/OBF.1 stability. We performed pull-down assays of extracts from CD38– and CD38+ cells with GST–SIAH1 fusion proteins. As shown in Figure 8D, the C-terminal domain of SIAH1 is able to precipitate BOB.1/OBF.1 both from CD38– and CD38+ B cells, whereas no interaction is seen with the N-terminal domain. This result indicates that the interaction of BOB.1/OBF.1 with SIAH1 is not altered by modification of BOB.1/OBF.1. However, it does not rule out a possible modification of SIAH1.
IgM cross-link in B cells leads to SIAH1-dependent reduction of BOB.1/OBF.1-driven transcription
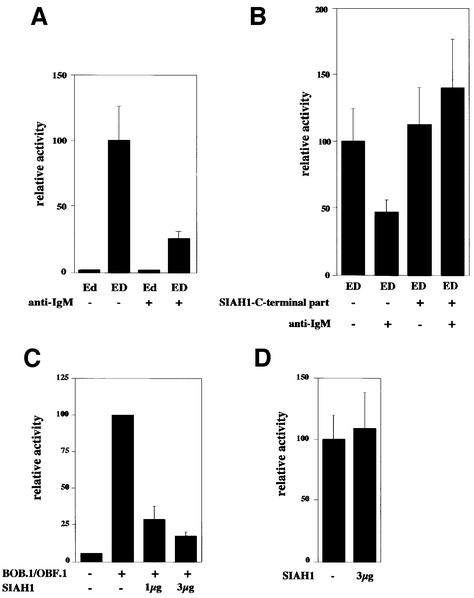
We then asked whether the activity of a reporter plasmid, which we had previously shown to be dependent on BOB.1/OBF.1, would be compromised in anti-IgM-treated Ramos B cells. This analysis revealed that indeed the activity was reduced upon anti-IgM treatment of Ramos B cells (Figure 9A). These experiments, however, do not prove that the observed inducible reduction was mediated by SIAH proteins nor do they definitely show that degradation of BOB.1/OBF.1 was the only critical event with respect to the activity of the reporter induced by anti-IgM treatment. To prove that SIAH1 is involved in the observed downregulation of the BOB.1/OBF.1-dependent reporter activity we cotransfected the C-terminal domain of SIAH1 (like in Figure 5D). Indeed, cotransfection fully restored the activity of the reporter, indicating that the BOB.1/OBF.1 degradation in cross-linked Ramos B cells is achieved via SIAH1 (Figure 9B). To test this reporter assay in an independent and more defined system, we transfected NIH 3T3 cells with the reporter together with a BOB.1/OBF.1 expression vector. This resulted in a strong induction of the reporter as had been shown before (Pfisterer et al., 1995). Cotransfection of SIAH1 strongly reduced the BOB.1/OBF.1-dependent reporter activity (Figure 9C). A control reporter independent of BOB.1/OBF.1 (the HSV thymidine kinase promoter) remained unaffected (Figure 9D).
Fig. 9. BOB.1/OBF.1 degradation results in downregulation of octamer-dependent transcription. (A) Ramos B cells were transfected with octamer-dependent luciferase reporter plasmids (ED contains the wild type, Ed contains a mutated octamer motif) and were incubated with anti-IgM antibodies for 20 h. (B) Downregulation of BOB.1/OBF.1 after IgM cross-linking is mediated by SIAH1. The same reporter plasmid described in (A) was cotransfected with the SIAH1 C-terminus followed by IgM cross-linking. (C) Octamer-dependent transcription is downregulated as a consequence of the BOB.1/OBF.1 degradation by SIAH1 in NIH 3T3 cells. NIH 3T3 cells were transfected with expression plasmids for BOB.1/OBF.1 and SIAH1. In addition, an octamer-dependent luciferase reporter plasmid (4× wt.TATA) was cotransfected. Eighteen hours after transfection the cells were harvested and luciferase activity measured. (D) No effect of SIAH1 was seen on the expression of thymidine kinase promoter-driven construct. Analysis as in (C).
Discussion
A significant part of the upregulation of BOB.1/OBF.1 in germinal center B cells is not due to increased mRNA expression levels. Rather we demonstrate that BOB.1/OBF.1 expression is regulated at the level of protein stability/protein degradation. As a potential regulator protein we identified the SIAH proteins that interact with BOB.1/OBF.1. As a consequence, BOB.1/OBF.1 is degraded in a proteasome-dependent manner. This degradation can be induced in Ramos B cells by IgM cross-linking.
So far only a few reports have addressed the regulatory mechanisms leading to the expression of BOB.1/OBF.1. Transcriptional induction was reported in Jurkat T cells after treatment with phorbol 12-myristate 13-acetate (PMA) and ionomycin (Sauter and Matthias, 1997; Zwilling et al., 1997). In addition, primary resting B cells were shown to upregulate BOB.1/OBF.1 transcripts after treatment with CD40-ligand and IL-4 as well as after treatment with LPS (Qin et al., 1998; Greiner et al., 2000). Analysis of the BOB.1/OBF.1 promoter identified a critical CREB/ATF site (Stevens et al., 2000). Interest ingly, when the time courses for the induction of BOB.1/OBF.1 transcript and protein were compared in B cells after LPS treatment, it was noted that protein induction was more enhanced than induction of the mRNA. In uninduced cells the protein was absent although the transcript was detectable. This had led to the hypothesis that BOB.1/OBF.1 regulation occurs at an additional translational or post-translational level (Qin et al., 1998). Consistent with these data and with the finding of a strong upregulation of BOB.1/OBF.1 in germinal center cells, we found that BOB.1/OBF.1 protein stability is regulated during development of germinal center B cells.
Potential regulator proteins for BOB.1/OBF.1 stability are SIAH1 and SIAH2. It has been demonstrated earlier that the interaction of proteins with SINA or SIAH1/2 results in a degradation of the targeted protein. In Drosophila, SINA acts together with Phyllopod (PHYL), a protein induced by Ras signaling, to target the transcriptional repressor Tramtrack. Tramtrack becomes ubiquitylated and degraded in a proteasome-dependent manner, allowing differentiation of the R7 cell (Li et al., 1997; Tang et al., 1997). In mammals, the proteins DCC and NCoR are both degraded in a SIAH-dependent manner (Hu et al., 1997b; Zhang et al., 1998). Interaction of SIAH2 with Vav1 (p95vav) does not lead to a degradation of Vav protein, but to a block in signal transduction via JNK activation (Germani et al., 1999). For NCoR, DCC and Vav1, it was shown that the interaction occurs with the C-terminus of SIAH. Consistent with these reports we found BOB.1/OBF.1 to interact with the C-terminus of SIAH1 and SIAH2. In contrast, BAG-1 was reported to require the full-length SIAH1 protein for its interaction (Matsuzawa et al., 1998). The C-terminal domain of SIAH is also responsible for the oligomerization of SIAH proteins. In line with these results we found that overexpression of this domain of SIAH1 can interfere with SIAH1 function and thereby block BOB.1/OBF.1 degradation. Interaction with SIAH led to a 4-fold reduction of the half-life of the DCC and NCoR proteins, and as shown here it reduced the half-life of BOB.1/OBF.1 ∼2-fold.
The observation that BOB.1/OBF.1 expression in germinal center B cells is at least in part regulated at a post-transcriptional level raises the question as to how protein stabilization might be achieved. We found that downregulation of BOB.1/OBF.1 via SIAH1 is downstream of BCR signaling in Ramos B cells. In germinal center B cell maturation, the following scenario for BOB.1/OBF.1 protein stabilization is likely. In the rapidly dividing cells of the dark zone in germinal centers, the centroblasts, expression of the BCR is downregulated (MacLennan, 1994). In addition, receptor signaling is modulated by CD22 or Fcγ (Cambier, 1997; Tedder et al., 1997). BCR downregulation and signal modification could lead to a block in SIAH1 expression and therefore to a block in BOB.1/OBF.1 ubiquitylation. This hypothesis is strengthened by the results shown in Figures 8 and 9. The enhancement of BCR signaling via cross-linking in Ramos B cells, which have a germinal center-like phenotype, leads to BOB.1/OBF.1 degradation via SIAH1. Therefore, the effect described in Figure 1 could be explained as followed: non-germinal center B cells do express the BOB.1/OBF.1 mRNA but the protein becomes rapidly degraded e.g. as a consequence of continuous BCR signaling. When the cells differentiate to rapidly dividing germinal center centroblasts they downregulate the BCR and as a consequence no longer degrade BOB.1/OBF.1 protein.
Alternative mechanisms might also be involved however. Due to a lack of a good SIAH-specific antibody, the potential modification of SIAH1 in germinal center B cells could not be addressed. In addition, enhanced protein stability could be achieved by blocking the degradation of the ubiquitylated protein. Recent data have shown a variation of proteasome subunit composition and of enzymatic activity in germinal center-derived Burkitt lymphoma and Hodgkin's lymphoma cell lines compared with non-germinal center-derived lymphoblastoid cell lines (Frisan et al., 2000). Therefore, a protection of ubiquitylated BOB.1/OBF.1 protein from degradation is conceivable.
Many B-cell lymphomas are derived from germinal center B cells and we found high level expression of BOB.1/OBF.1 in germinal center-derived lymphomas (Greiner et al., 2000). Interestingly, the expression of BOB.1/OBF.1 is highly correlated with the expression of Bcl-6, a classical marker for germinal center-derived lymphomas. The parallels between Bcl-6 and BOB.1/OBF.1 are striking. Mutations in both genes result in a specific block of germinal center formation. Both proteins are preferentially expressed in germinal center cells. Finally, as shown in this paper for BOB.1/OBF.1 as well as previously for Bcl-6, both are regulated at the protein level (Allman et al., 1996; Kim et al., 1996; Nielsen et al., 1996; Schubart et al., 1996a; Dent et al., 1997; Ye et al., 1997). The cross-linking of Ramos B cells with anti-IgM leads in both cases to a proteasome-dependent degradation (Figure 7B and Niu et al., 1998). Bcl-6 belongs to a family of POZ/zinc finger transcription factors, like Tramtrack in Drosophila. The Tramtrack protein TTK88 is degraded by its interaction with Phyllopod and SINA, the Drosophila homolog of human SIAH. In both cases the activation of the MAPK pathway leads to downregulation of the POZ/zinc finger transcription factor (Li et al., 1997; Tang et al., 1997; Niu et al., 1998). After IgM cross-linking of Ramos B cells BOB.1/OBF.1 similarly to Bcl-6 degradation is blocked by treating the cells with a MAPK inhibitor (Figure 7C and Niu et al., 1998). We have found Bcl-6 as an interaction partner in a yeast two-hybrid screen using the SIAH1 C-terminus (aa 80–282) as a bait (data not shown). These obvious parallels in the regulation of BOB.1/OBF.1 and Bcl-6, two factors absolutely essential for germinal center formation, could hint towards a central role of SIAH in B-cell development, especially in germinal center formation.
Materials and methods
Plasmids
BOB.1/OBF.1 mutants as well as the wild type were cloned into pcDNA3. SIAH1 C-terminus containing aa 80–282 was cloned into pCS2+MT with a myc tag in front of SIAH1. SIAH1-HA was cloned into pCGN-M2, the Flag-tagged version into pCXN-Flag. For the mammalian two-hybrid system we used a BOB.1/OBF.1–VP16 fusion construct cloned into RC/CMV; SIAH1 was cloned as a GAL fusion construct into the GAL4 expression vector described before and read-out took place by using a reporter plasmid (pG4.TATA) containing the luciferase gene under the control of four GAL4-binding sites (Pfisterer et al., 1995). The mini-myc protein is a non-sense protein established by cloning of BOB.1/OBF.1 with a +1 frameshift into pCS2+MT, leading to a protein of 27 aa with a myc tag. For luciferase assays in Ramos cells the octamer-dependent luciferase reporter plasmids Ed-TATA and ED-TATA were used and in NIH 3T3 cells the plasmid 4× wt.TATA was used, as described before. For standardization of the transfections, CMV-LacZ or TK-Renilla-luciferase was cotransfected as described (Pfisterer et al., 1995; Laumen et al., 2000). All transfection experiments were performed at least three times.
LightCycler PCR
CD38 negative and positive cells from human tonsils were isolated as described before (Greiner et al., 1997, 2000). Total RNA was reverse transcribed using M-MLV reverse transcriptase (Gibco).
For PCR the following primers were used. PBGD forward: AGCTGCAGAGAAAGTTCCC; PBGD reverse: GTTACGAGCAGTGATGCC; CD38 forward: ATCAGCCACTAATGAAGTTGG; CD38 reverse: TCAGATGTGCAAGATGAATCC; BOB.1/OBF.1 forward: CACAGCTCCGGAGCAAGCC; BOB.1/OBF.1 reverse: CCTTCCACAGAGAGAGTGTGG; SIAH1 forward: GCCGTCAGACTGCTACAGC; SIAH1 reverse: AGCTGTACGATTGCGAAGAAC.
The PCR was performed in a Roche LightCycler using the LightCycler-FastStart DNA Master SYBR Green I system (Roche). The cDNAs were serially diluted (4:2:1) to check the efficiency of the PCR. Each PCR was performed for each dilution at least three times. Calculations were performed using the LightCycler software version 3.39. The quantitation was performed relative to the expression levels of CD38– cells. The efficiencies for any pair of PCRs did not differ by >0.05 and the overall efficiencies were between 1.8 and 1.98.
Cell culture and transfection methods
NIH 3T3 and S194 B cells were cultured in Dulbecco’s modified Eagle’s medium Glutamax medium (Gibco) supplemented with 10% fetal calf serum (FCS) and antibiotics. The NIH 3T3 cells were transfected by electroporation (Bio-Rad) with 450 V and 250 mA in phosphate-buffered saline (PBS). Ramos B cells were grown in RPMI medium (Gibco) supplemented with glutamine, 10% FCS and antibiotics. The cells were transfected by using DEAE as described previously (Pfisterer et al., 1995). The cross-linking was performed by adding goat F(ab′)2 anti-human IgM (Dianova and Biosource) to a final concentration of 25 µg/ml to the medium.
GST pull-down
The SIAH1 fragments containing aa 1–80, 80–282 and full-length SIAH1 were cloned into pGEX-4T-2 and transformed into Escherichia coli DH5α. Bacterial cells were grown, lysed and the protein purified via glutathione–Sepharose 4B beads as described (Zwilling et al., 1997). BOB.1/OBF.1 constructs were in vitro transcribed and translated (Pfisterer et al., 1995) using the TNT Coupled Reticulocyte Lysate System (Promega) in the presence of [35S]methionine and [35S]cysteine mix (Amersham). The in vitro translated proteins were mixed with the GST–SIAH1 constructs and precipitated by the addition of glutathione–Sepharose 4B beads. Separation took place on a 12.5% SDS–polyacrylamide gel, followed by autoradiography. The pull-down with S194 B-cell lysates and primary tonsillar B-cell lysates was performed in lysis buffer containing 25 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 10% glycerol and 1% Triton X-100.
Pulse–chase
NIH 3T3 fibroblasts were transfected with a BOB.1/OBF.1 expression plasmid and cultured for 12 h. After trypsination, the cells were seeded to 70% confluence and grown for 6 h in 6 cm dishes. The cells were starved for 60 min in an amino acid-free medium and then labeled by adding a 1 mCi/ml [35S]methionine and [35S]cysteine mix in medium (Amersham). After 30 min the cells were washed and incubated with medium containing cold methionine and cysteine (150 µg/ml). Cells were harvested at the indicated time points and the BOB.1/OBF.1 protein precipitated with the monoclonal anti-BOB.1/OBF.1 antibody. Separation took place on 12.5% SDS–polyacrylamide gels, followed by autoradiography.
Yeast two-hybrid assays
The N-terminal region of BOB.1/OBF.1 containing aa 1–72 was cloned into pPCH1 (Hagemann et al., 1997) and used as a ‘bait’. HF7c yeast cells expressing the bait vector were transformed (Gietz et al., 1995) with a Jurkat T cell library (Clontech). The triple dropout plates contained 5 mM 3-aminotriazole (3-AT), and putative clones were tested for β-gal activity. The different tested subdomains of SIAH1 were cloned into the library vector pGAD10 (Clontech) and cotransformed with the BOB.1/OBF.1 ‘bait’ vector in HF7c yeast cells.
For the interaction assay with the BOB.1/OBF.1 point mutants, HF7c yeast cells were cotransformed with the indicated vectors and plated onto double dropout plates (-Trp, -Leu). SIAH1 was cloned into pAS2.1 (Clontech) and the BOB.1/OBF.1 mutants were used as described before (Gstaiger et al., 1996). After 5 days growing clones were streaked onto triple dropout plates (-Trp, -Leu, -His) and 1–2 days later β-gal assays were performed.
Northern and western blot analysis
Total RNA was isolated from cells by using the High Pure RNA Isolation Kit (Roche) and separated on 1% formaldehyde agarose gels.
Whole-cell protein lysates were performed by use of RIPA buffer and the proteins were separated on 12.5% SDS–polyacrylamide gels. The following antibodies were used for western blot analysis: polyclonal anti-BOB.1/OBF.1 (Pfisterer et al., 1995), our new monoclonal anti-BOB.1/OBF.1 (directed against the C-terminus), anti-HA 12CA5, anti-myc 9E10, polyclonal anti-RelA (Santa Cruz) and anti-actin antibody (Sigma).
Immunofluorescence
NIH 3T3 cells were transfected with the indicated vectors, seeded onto cover slips and cultured for 18 h. The cells were washed with PBS and fixed by using 4% paraformaldehyde. After washing with PBS, cells were blocked with 5% FCS, 0.5% Triton X-100 in PBS. Incubation with polyclonal rabbit anti- BOB.1/OBF.1 and/or monoclonal anti-Flag M2 (Kodak) was performed for 30 min at 37°C. Afterwards the cells were washed with PBS and incubated with FITC-conjugated goat anti-rabbit antibody (Dianova) and/or Cy3-conjugated anti-mouse antibody (Dianova). The nucleus was stained using a 1 µg/ml DAPI solution (Roche).
Acknowledgments
Acknowledgements
We thank Walter Schaffner and Matthias Gstaiger for the BOB.1/OBF.1 point mutations. We thank Maria Reichert for technical help. This work was supported by a grant to T.W. from the Deutsche Forschungsgemein schaft (SFB 497, C5).
Note added in proof
Independently, similar results are reported by Tiedt et al. on pages 4143–4152 of this issue.
References
- Allman D., Jain,A., Dent,A., Maile,R.R., Selvaggi,T., Kehry,M.R. and Staudt,L.M. (1996) BCL-6 expression during B-cell activation. Blood, 87, 5257–5268. [PubMed] [Google Scholar]
- Cambier J.C. (1997) Positive and negative signal co-operativity in the immune system: the BCR, Fcγ RIIB, CR2 paradigm. Biochem. Soc. Trans., 25, 441–445. [DOI] [PubMed] [Google Scholar]
- Carthew R.W. and Rubin,G.M. (1990) seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell, 63, 561–577. [DOI] [PubMed] [Google Scholar]
- Cepek K.L., Chasman,D.I. and Sharp,P.A. (1996) Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes Dev., 10, 2079–2088. [DOI] [PubMed] [Google Scholar]
- Chasman D., Cepek,K., Sharp,P.A. and Pabo,C.O. (1999) Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein–DNA interface. Genes Dev., 13, 2650–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent A.L., Shaffer,A.L., Yu,X., Allman,D. and Staudt,L.M. (1997) Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science, 276, 589–592. [DOI] [PubMed] [Google Scholar]
- Fang S., Jensen,J.P., Ludwig,R.L., Vousden,K.H. and Weissman,A.M. (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem., 275, 8945–8951. [DOI] [PubMed] [Google Scholar]
- Fink L., Seeger,W., Ermert,L., Hanze,J., Stahl,U., Grimminger,F., Kummer,W. and Bohle,R.M. (1998) Real-time quantitative RT–PCR after laser-assisted cell picking. Nature Med., 4, 1329–1333. [DOI] [PubMed] [Google Scholar]
- Fink L., Stahl,U., Ermert,L., Kummer,W., Seeger,W. and Bohle,R.M. (1999) Rat porphobilinogen deaminase gene: a pseudogene-free internal standard for laser-assisted cell picking. Biotechniques, 26, 510–516. [DOI] [PubMed] [Google Scholar]
- Frisan T., Levitsky,V. and Masucci,M.G. (2000) Variations in proteasome subunit composition and enzymatic activity in B-lymphoma lines and normal B cells. Int. J. Cancer, 88, 881–888. [DOI] [PubMed] [Google Scholar]
- Germani A., Romero,F., Houlard,M., Camonis,J., Gisselbrecht,S., Fischer,S. and Varin-Blank,N. (1999) hSiah2 is a new Vav binding protein which inhibits Vav-mediated signaling pathways. Mol. Cell. Biol., 19, 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl,R.H., Willems,A.R. and Woods,R.A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast, 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Greiner A., Knorr,C., Qin,Y., Sebald,W., Schimpl,A., Banchereau,J. and Muller-Hermelink,H.K. (1997) Low-grade B cell lymphomas of mucosa-associated lymphoid tissue (MALT-type) require CD40-mediated signaling and Th2-type cytokines for in vitro growth and differentiation. Am. J. Pathol., 150, 1583–1593. [PMC free article] [PubMed] [Google Scholar]
- Greiner A., Müller,K., Hess,J., Pfeffer,K., Müller-Hermelink,K.H. and Wirth,T. (2000) Up-regulation of BOB.1/OBF.1 expression in normal germinal center B cells and germinal center-derived lymphomas. Am. J. Pathol., 156, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M., Knoepfel,L., Georgiev,O., Schaffner,W. and Hovens,C.M. (1995) A B-cell coactivator of octamer-binding transcription factors. Nature, 373, 360–362. [DOI] [PubMed] [Google Scholar]
- Gstaiger M., Georgiev,O., van Leeuwen,H., van der Vliet,P. and Schaffner,W. (1996) The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J., 15, 2781–2790. [PMC free article] [PubMed] [Google Scholar]
- Hagemann C., Kalmes,A., Wixler,V., Wixler,L., Schuster,T. and Rapp,U.R. (1997) The regulatory subunit of protein kinase CK2 is a specific A-Raf activator. FEBS Lett., 403, 200–202. [DOI] [PubMed] [Google Scholar]
- Hess J., Nielsen,P.J., Fischer,K.-D., Bujard,H. and Wirth,T. (2001) The B lymphocyte-specific coactivator BOB.1 (OBF.1) is required at multiple stages of B-cell development. Mol. Cell. Biol., 21, 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Tanaka,H. and Yasuda,H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett., 420, 25–27. [DOI] [PubMed] [Google Scholar]
- Hu G. and Fearon,E.R. (1999) Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol., 19, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Chung,Y.L., Glover,T., Valentine,V., Look,A.T. and Fearon,E.R. (1997a) Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics, 46, 103–111. [DOI] [PubMed] [Google Scholar]
- Hu G., Zhang,S., Vidal,M., Baer,J.L., Xu,T. and Fearon,E.R. (1997b) Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev., 11, 2701–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C.A. and Weissman,A.M. (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell, 102, 549–552. [DOI] [PubMed] [Google Scholar]
- Kim U., Qin,X.-F., Gong,S., Stevens,S., Luo,Y., Nussenzweig,M. and Roeder,R.G. (1996) The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature, 383, 542–547. [DOI] [PubMed] [Google Scholar]
- Laumen H., Nielsen,P.J. and Wirth,T. (2000) The BOB.1/OBF.1 coactivator is essential for octamer-dependent transcription in B cells. Eur. J. Immunol., 30, 458–469. [DOI] [PubMed] [Google Scholar]
- Li S., Li,Y., Carthew,R.W. and Lai,Z.C. (1997) Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell, 90, 469–478. [DOI] [PubMed] [Google Scholar]
- Lorick K.L., Jensen,J.P., Fang,S., Ong,A.M., Hatakeyama,S. and Weissman,A.M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA, 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. and Roeder,R.G. (1995) Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol., 15, 4115–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan I.C. (1994) Germinal centers. Annu. Rev. Immunol., 12, 117–139. [DOI] [PubMed] [Google Scholar]
- Matsuzawa S., Takayama,S., Froesch,B.A., Zapata,J.M. and Reed,J.C. (1998) p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J., 17, 2736–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias P. (1998) Lymphoid-specific transcription mediated by the conserved octamer site: who is doing what? Semin. Immunol., 10, 155–163. [DOI] [PubMed] [Google Scholar]
- Nielsen P.J., Georgiev,O., Lorenz,B. and Schaffner,W. (1996) B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur. J. Immunol., 26, 3214–3218. [DOI] [PubMed] [Google Scholar]
- Niu H., Ye,B.H. and Dalla-Favera,R. (1998) Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev., 12, 1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer P., Zwilling,S., Hess,J. and Wirth,T. (1995) Functional characterization of the murine homolog of the B-cell-specific coactivator BOB.1/OBF.1. J. Biol. Chem., 270, 29870–29880. [DOI] [PubMed] [Google Scholar]
- Qin X.-F., Reichlin,A., Luo,Y., Roeder,R.G. and Nussenzweig,M.C. (1998) OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J., 17, 5066–5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter P. and Matthias,P. (1997) The B cell-specific coactivator OBF-1 (OCA-B, Bob-1) is inducible in T cells and its expression is dispensable for IL-2 gene induction. Immunobiology, 198, 207–216. [DOI] [PubMed] [Google Scholar]
- Sauter P. and Matthias,P. (1998) Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol. Cell. Biol., 18, 7397–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart D.B., Rolink,A., Kosco-Vilbois,M.H., Botteri,F. and Matthias,P. (1996a) B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature, 383, 538–542. [DOI] [PubMed] [Google Scholar]
- Schubart D.B., Sauter,P., Massa,S., Friedl,E.M., Schwarzenbach,H. and Matthias,P. (1996b) Gene structure and characterization of the murine homologue of the B cell-specific transcriptional coactivator OBF-1. Nucleic Acids Res., 24, 1913–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S., Wang,L. and Roeder,R.G. (2000) Functional analysis of the OCA-B promoter. J. Immunol., 164, 6372–6379. [DOI] [PubMed] [Google Scholar]
- Strubin M., Newell,J.W. and Matthias,P. (1995) OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer proteins. Cell, 80, 497–506. [DOI] [PubMed] [Google Scholar]
- Tang A.H., Neufeld,T.P., Kwan,E. and Rubin,G.M. (1997) PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell, 90, 459–467. [DOI] [PubMed] [Google Scholar]
- Tedder T.F., Tuscano,J., Sato,S. and Kehrl,J.H. (1997) CD22, a B lymphocyte-specific adhesion molecule that regulates antigen receptor signaling. Annu. Rev. Immunol., 15, 481–504. [DOI] [PubMed] [Google Scholar]
- Ye B.H. et al. (1997) The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nature Genet., 16, 161–170. [DOI] [PubMed] [Google Scholar]
- Yu X., Wang,L., Luo,Y. and Roeder,R.G. (2001) Identification and characterization of a novel OCA-B isoform. Implications for a role in B cell signaling pathways. Immunity, 14, 157–167. [PubMed] [Google Scholar]
- Zhang J., Guenther,M.G., Carthew,R.W. and Lazar,M.A. (1998) Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev., 12, 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling S., Dieckmann,A., Pfisterer,P., Angel,P. and Wirth,T. (1997) Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science, 277, 221–225. [DOI] [PubMed] [Google Scholar]