Abstract
Many eukaryotic cell surface proteins are anchored to the plasma membrane via glycosylphosphatidylinositol (GPI). The GPI transamidase mediates GPI anchoring in the endoplasmic reticulum, by replacing a protein’s C-terminal GPI attachment signal peptide with a pre-assembled GPI. During this transamidation reaction, the GPI transamidase forms a carbonyl intermediate with a substrate protein. It was known that the GPI transamidase is a complex containing GAA1 and GPI8. Here, we report two new components of this enzyme: PIG-S and PIG-T. To determine roles for PIG-S and PIG-T, we disrupted these genes in mouse F9 cells by homologous recombination. PIG-S and PIG-T knockout cells were defective in transfer of GPI to proteins, particularly in formation of the carbonyl intermediates. We also demonstrate that PIG-S and PIG-T form a protein complex with GAA1 and GPI8, and that PIG-T maintains the complex by stabilizing the expression of GAA1 and GPI8. Saccharomyces cerevisiae Gpi16p (YHR188C) and Gpi17p (YDR434W) are orthologues of PIG-T and PIG-S, respectively.
Keywords: endoplasmic reticulum/glycosylphosphatidyl inositol/post-translational modification/transamidase
Introduction
Many eukaryotic cell surface proteins are anchored to the plasma membrane via a glycosylphosphatidylinositol (GPI) moiety. GPI anchoring is essential for the expression of those proteins on the cell surface. GPI is widely utilized in unicellular and higher eukaryotes (McConville and Ferguson, 1993; Kinoshita et al., 1995).
GPI is synthesized by sequential additions of sugars and ethanolamine phosphates to phosphatidylinositol in the endoplasmic reticulum (ER) (Udenfriend and Kodukula, 1995; Kinoshita and Inoue, 2000). The backbone structure of GPI is common among different species. Pre-formed GPI is attached to proteins in the ER. Precursor proteins to be modified with a GPI have two signals. One, at the N-terminus, is a signal required for translocation across the ER membrane. The other, at the C-terminus, is a GPI attachment signal. The GPI attachment signal peptide is recognized by the GPI transamidase, which cleaves the signal peptide and replaces it with GPI. During this reaction, the GPI transamidase attacks the peptide bond at the N-terminus of the signal peptide, generating a carbonyl enzyme–substrate protein intermediate. An amino group of the terminal ethanolamine of GPI then attacks the carbonyl intermediate, completing attachment of GPI to the newly formed C-terminus (Udenfriend and Kodukula, 1995; Sharma et al., 1999).
The GPI attachment signal consists of three portions: a stretch of three amino acids including the amino acid to which GPI attaches (ω site), a terminal hydrophobic segment of 12–20 amino acids and a hydrophilic spacer segment of usually <10 amino acids between them (Udenfriend and Kodukula, 1995; Furukawa et al., 1997). The length of the spacer region (Furukawa et al., 1997; Bucht et al., 1999) and the extent of hydrophobicity in the hydrophobic region (Waneck et al., 1988) affect GPI anchoring efficiency but there is no consensus sequence in the signals. Well known characteristics of the signal are the restrictions at ω and ω+2 sites. The ω site is restricted to six amino acids with small side chains (Micanovic et al., 1990; Moran et al., 1991; Nuoffer et al., 1993). The ω+2 site, the second residue C-terminal to the ω site, is also limited to relatively small amino acids, whereas the ω+1 site tolerates any amino acid except proline and tryptophan (Gerber et al., 1992).
Requirements for GPI attachment signals are different between mammalian cells and parasitic protozoa. GPI attachment signals of trypanosome variant surface glycoproteins did not function in mammalian cells (Moran and Caras, 1994). This difference is proposed to be due mainly to the size of amino acids at the ω to ω+2 sites. These amino acids of parasitic protozoa are larger than those used in human proteins. Therefore, parasite GPI transamidase is assumed to accommodate and tolerate larger amino acids in its catalytic site (Moran and Caras, 1994). It would, therefore, be possible to generate inhibitors that inhibit parasite GPI transamidase but not mammalian transamidase. Such selective inhibitors of GPI synthesis would be potent therapeutic drugs for parasitic diseases (Ferguson, 2000).
However, the GPI transamidase has not been characterized fully. Two components, GAA1 and GPI8, are required in both mammalian and Saccharomyes cerevisiae GPI transamidases (Hamburger et al., 1995; Benghezal et al., 1996; Yu et al., 1997; Hiroi et al., 1998; Ohishi et al., 2000). We previously demonstrated that GAA1 and GPI8 form a protein complex (Ohishi et al., 2000). GPI8 is most probably a catalytic component because it is associated with substrate proteins (Spurway et al., 2001; Vidugiriene et al., 2001) and because it has homology to members of a family of cysteine proteases including a jack bean asparaginyl endopeptidase that has transamidase activity (Benghezal et al., 1996; Eisenhaber et al., 2001). Human and yeast GPI8s with a mutation at putative active cysteine and histidine residues were non-functional (Meyer et al., 2000; Ohishi et al., 2000). Treatments of trypanosome microsomal membranes and recombinant GPI8 with some alkylating reagents inactivated transamidase activity (Sharma et al., 1999, 2000). On the other hand, the function of GAA1 is still unclear due to a lack of homology to other proteins.
In the present study, we cloned PIG-S and PIG-T, two new components of the GPI transamidase, and demonstrate that these components are essential for transfer of GPI to proteins, particularly for formation of the carbonyl intermediates. PIG-S and PIG-T form a protein complex with GAA1 and GPI8, and PIG-T has a critical role in maintaining the complex.
Results
Identification of two new components of GPI transamidase
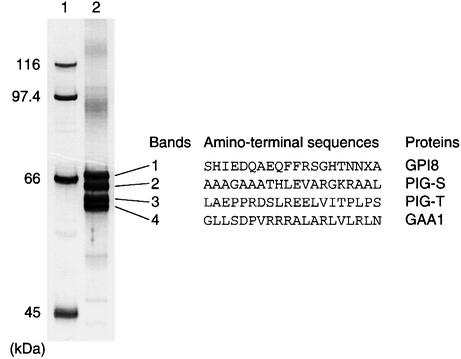
To characterize better the GPI transamidase complex, we stably expressed FLAG-glutathione S-transferase (GST)-tagged human GPI8 in class K cells, GPI8-deficient human K562 cells (Yu et al., 1997), and isolated the complex by two-step affinity purification (Figure 1). An analysis of the complex by SDS–PAGE demonstrated four bands with comparable intensities at positions 60–70 kDa (lane 2). We sequenced their N-termini. The sequence of band 1 coincided with amino acids 28–47 predicted from GPI8 cDNA (when X is assumed to be tryptophan). Amino acids 1–27 of GPI8 conform to a signal sequence for translocation across the ER membrane. The fact that residue 28 is the N-terminus of the expressed protein confirms that the N-terminal 27 residues encode a signal sequence. Consistent with our previous report that GPI8 and GAA1 form an NP-40-resistant complex, the sequence of band 4 coincided with amino acids 2–21 of GAA1. A lack of the first methionine in the expressed protein is consistent with the cytoplasmic orientation of the N-terminus of GAA1 because it is frequently observed in cytoplasmically located N-termini of proteins (Arfin and Bradshaw, 1988). The molecular size of GAA1 estimated from the mobility was ∼60 kDa, which was lower than the predicted molecular size of 70 kDa. The reason for this difference is not clear. Bands 2 and 3 were new proteins that we termed PIG-S and PIG-T, respectively.
Fig. 1. Identification of two new components of GPI transamidase. The transamidase complex was purified from the NP-40 extract of class K cells stably expressing FLAG-GST-tagged GPI8 by two-step affinity purification. A sample of protein complex (equivalent to 2 × 108 cells) was separated on a 10% SDS–polyacrylamide gel under reducing conditions and silver stained (lane 2). Bands were numbered 1–4 from top to bottom. The N-terminal amino acid sequences (X is an unidentified amino acid) and protein identities are indicated on the right. Lane 1, molecular size markers.
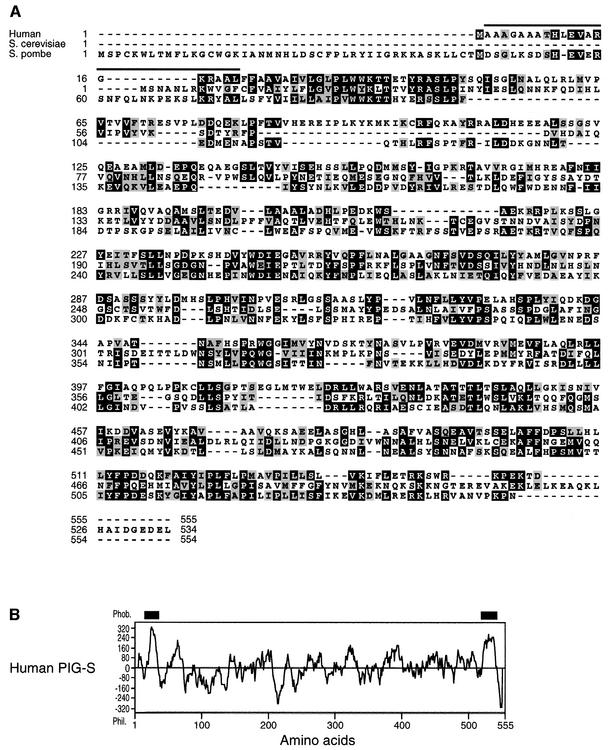
We found several mouse and human expressed sequence tags (ESTs). Using these EST sequences, we obtained mouse and human genomic sequences of the PIG-S gene. Based on these sequences, we designed PCR primers and amplified a human cDNA containing a full coding region. Human PIG-S consists of 555 amino acids and its residues 2–21 coincided with the N-terminal sequence of band 2 protein (Figure 2A). PIG-S has two transmembrane domains near the N- and C-termini (Figure 2B). If a lack of N-terminal methionine in the expressed PIG-S protein indicates a cytoplasmic orientation for the N-terminus, the large hydrophilic region in the middle of the molecule would be luminally oriented. PIG-S homologues are found in S.cerevisiae and Schizosaccharomyces pombe (Figure 2A), and in Droso phila melanogaster (AAF56365) and Caenorhabditis elegans (CAA93093) (not shown). The S.cerevisiae PIG-S consists of 534 amino acids and has 23% amino acid identity with human PIG-S. The S.pombe PIG-S consists of 554 amino acids and has 27 and 19% amino acid identity with human and S.cerevisiae PIG-S, respectively. Both S.cerevisiae and S.pombe PIG-S have similar hydrophobicity profiles to human PIG-S (not shown).
Fig. 2. (A) Alignment of protein sequences of PIG-S homologues. Human PIG-S (accession No. AB057723) and its S.cerevisiae (S69714) and S.pombe (T38067) homologues are aligned using the Multiple Alignment Program software (Huang, 1994). The human PIG-S sequence determined by N-terminal sequencing is overlined. Black and grey boxes are identical and similar amino acids, respectively. (B) Hydrophobicity profile of human PIG-S protein according to the Kyte and Doolittle method (Kyte and Doolittle, 1982). Putative transmembrane domains near the N- and C-termini are indicated by thick bars.
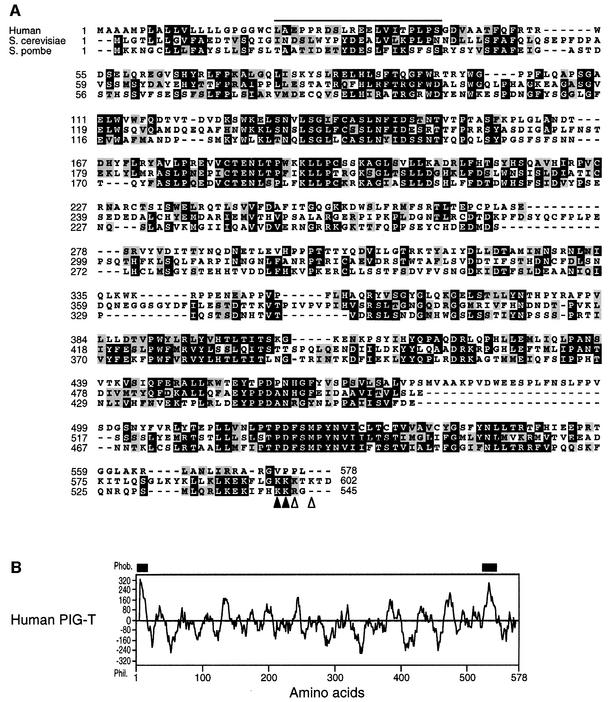
We found a hypothetical human protein of 578 amino acids in the database whose predicted amino acids 22–41 coincided with the N-terminal 20 residues of PIG-T (Figure 3A). The first 21 amino acids that form a hydro phobic region (Figure 3B), therefore, encode a signal peptide. PIG-T homologues are found in S.cerevisiae and S.pombe (Figure 3A), and in D.melanogaster (AAF46367) and C.elegans (CAA96629) (not shown). The S.cerevisiae PIG-T consists of 602 amino acids having 26% identity with human PIG-T. The S.pombe homologue has 545 amino acids with 27% identity with human PIG-T. A hydrophobicity plot of human PIG-T demonstrated, in addition to the N-terminal signal peptide, a putative transmembrane domain near the C-terminus (Figure 3B). Two yeast homologues have di-lysine ER retention signals at the C-terminus (Figure 3A). Therefore, PIG-T is a type-I transmembrane protein with a large luminal domain and a short cytoplasmic tail.
Fig. 3. (A) Alignment of protein sequences of PIG-T homologues. Human PIG-T (AB057724) and its S.cerevisiae (S46687) and S.pombe (T39499) homologues are aligned using the Clustal W software. The human PIG-T sequence determined by N-terminal sequencing is overlined. Di-lysine ER retention signals in S.cerevisiae (KXKXX) and S.pombe (KKXX) are marked by open and closed triangles, respectively. (B) Kyte and Doolittle hydrophobicity plot of human PIG-T protein. The N-terminal signal peptide and a putative transmembrane domain near the C-terminus are indicated by thick bars.
Since PIG-S and PIG-T do not have significant homology to proteins with known functions, it is not possible to predict their functions; however, their structure and membrane topology are consistent with the idea that GPI is transferred to proteins on the luminal side of the ER (Udenfriend and Kodukula, 1995).
PIG-S and PIG-T are essential components of GPI transamidase
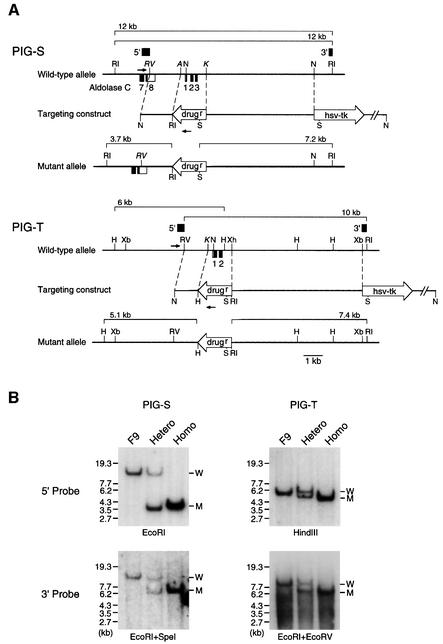
To study functions of PIG-S and PIG-T, we disrupted these genes in F9 embryonal carcinoma cells by homologous recombination (Figure 4A). In both cases, we replaced a region that includes an exon containing the initiation codon with a drug resistance gene. After the second round of recombinations, wild-type alleles disappeared and mutant alleles of predicted sizes appeared, as shown by Southern blot analysis (Figure 4B), indicating successful disruptions.
Fig. 4. Generation of PIG-S and PIG-T knockout mouse F9 cells. (A) Disruption of mouse PIG-S (upper) and PIG-T (lower) genes. Structures of the mouse PIG-S and PIG-T genes (top), targeting constructs (middle) and expected mutant alleles (bottom) are shown. Exons are indicated as boxes (closed boxes, coding regions; open boxes, untranslated regions). Exon 1 of the PIG-S gene is located 1.6 kb downstream to exon 8 of the aldolase C gene. First and second round disruptions of both PIG-S and PIG-T were performed with targeting constructs bearing puromycin and neomycin resistance genes, respectively. Thin arrows represent PCR primers for screening the recombinants. Probes for Southern analysis are indicated by thick bars. The predicted sizes of fragments hybridized with 5′ and 3′ probes are indicated above each genomic structure. A, ApaI; H, HindIII; N, NotI; K, KpnI; RI, EcoRI; RV, EcoRV; S, SpeI; Xb, XbaI; Xh, XhoI (italicized sites in the figure are not unique). (B) Southern analysis of PIG-S and PIG-T knockout F9 cells. Digested genomic DNA from wild-type cells (F9), heterozygous (Hetero) and homozygous (Homo) knockout cells for PIG-S (left panels) and PIG-T (right panels) genes were hybridized with the 5′ probe (top) and 3′ probe (bottom) shown in (A). Hybridized fragments of wild-type (W) and mutant (M) alleles, and size markers are indicated on the right and left, respectively. Enzymes used for digestion are shown below each panel.
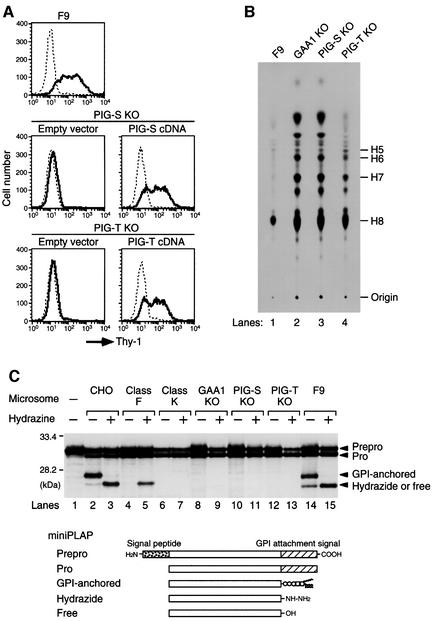
We first examined the surface expression of GPI-anchored proteins. PIG-S and PIG-T knockout cells did not expreess Thy-1, whereas parental F9 cells did. The surface expression of Thy-1 was restored by transfection of the corresponding cDNA (Figure 5A). We next tested whether GPI synthesis was affected. We metabolically labelled GPIs with [3H]mannose and analysed them by thin-layer chromatography (TLC) (Figure 5B). PIG-S and PIG-T knockout cells (lanes 3 and 4), like GAA1 knockout cells (lane 2), synthesized and accumulated large amounts of mature forms of GPI, H7 and H8, and their precursors H5 and H6. In contrast, only H8 was accumulated to some extent in wild-type F9 cells, as reported (lane 1) (Ohishi et al., 2000). Therefore, PIG-S and PIG-T are not required for GPI synthesis. Thus, they are essential for attachment of GPI to proteins.
Fig. 5. PIG-S and PIG-T are essential components of GPI transamidase. (A) Parent F9 cells, and PIG-S and PIG-T knockout cells transfected with an empty vector or corresponding human cDNA were analysed for the surface expression of Thy-1 by flow cytometry 2 days after transfection. Solid and dotted lines indicate staining with anti-Thy-1 antibody and with secondary reagent alone, respectively. (B) Biosynthesis of GPI anchor intermediates. Cells were metabolically labelled with [3H]mannose for 60 min in the presence of tunicamycin. Mannolipids were extracted and analysed by TLC. Identities of spots according to Hirose et al. (1992) and origin are indicated on the right. H5 and H6, GPI bearing two and three mannoses, respectively; H7 and H8, mature forms of GPI. (C) In vitro assay for GPI transamidase. MiniPLAP mRNA was translated in vitro using rabbit reticulocyte lysates and the microsomal membranes prepared from the indicated cells in the absence (–) or presence (+) of hydrazine. The class F cell is defective in biosythesis of GPI. The class K cell is a mutant of GPI8. The translation product in the absence of the microsomal membranes is prepro-miniPLAP (lane 1). Identities of other bands according to Kodukula et al. (1991) are shown on the right. Schematic representations of the products are shown below.
During the GPI anchoring process, GPI transamidase forms a carbonyl intermediate with the precursor protein. The carbonyl intermediate is then attacked by GPI. This process was reconstituted in an in vitro transcription/translation system developed by Udenfriend’s group (Kodukula et al., 1991). In this system, a radiolabelled model precursor protein, mini-placental alkaline phosphatase (miniPLAP), is translated in vitro and processed further by the microsomal membranes containing GPI transamidase (Figure 5C). We tested the microsomal membranes of PIG-S and PIG-T knockout cells in this system. In the absence of the microsomal membranes, prepro-miniPLAP, which still has the N-terminal signal peptide, was generated (lane 1). The microsomal membranes of wild-type CHO and F9 cells converted prepro-miniPLAP to pro-miniPLAP by removing the N-terminal signal peptide, and further to the GPI-anchored form (lanes 2 and 14). Small amounts of free forms resulting from hydrolysis of the carbonyl intermediates were also generated (lanes 2 and 14). In the presence of a potent nucleophile, hydrazine, the wild-type microsomes generated the hydrazide form but not the GPI-anchored form due to competition between GPI and excess hydrazine (lanes 3 and 15). Generation of hydrazide is an index of hydrazine-sensitive carbonyl intermediates of GPI transamidase and precursor proteins. Formation of the hydrazide form did not require the mature form of GPI because microsomes of class F cells that are defective in synthesis of mature GPI generated the hydrazide form but not the GPI-anchored form (lanes 4 and 5). As previously reported (Yu et al., 1997; Ohishi et al., 2000), microsomes of GAA1 knockout and GPI8 mutant class K cells did not generate hydrazide or GPI-anchored forms (lanes 6–9). Like these mutant cells, microsomes of PIG-S and PIG-T knockout cells did not generate hydrazide or GPI-anchored forms (lanes 10–13). Therefore, PIG-S and PIG-T are essential components of GPI transamidase and, in the absence of PIG-S or PIG-T, the carbonyl intermediate was not generated.
Saccharomyces cerevisiae GPI16 and GPI17 are orthologues of human PIG-T and PIG-S, respectively
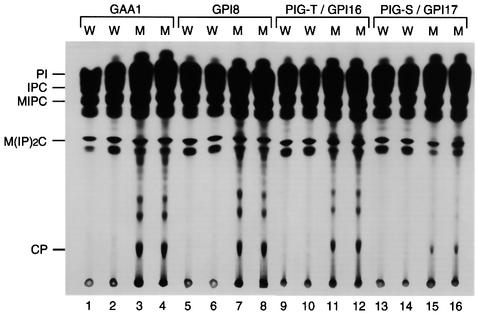
We termed S.cerevisiae homologues of PIG-T and PIG-S genes GPI16 and GPI17, respectively, according to a GPI# root of S.cerevisiae genes involved in GPI synthesis (Leidich et al., 1995). Consistent with a report that GPI synthesis is essential for growth of S.cerevisiae (Leidich et al., 1995), deletants of these genes are inviable (Winzeler et al., 1999). To test whether these genes are indeed involved in transfer of GPI to proteins, we made haploid strains of gpi16, gpi17, gaa1 and gpi8 deletants bearing galactose-regulatable expression plasmids of the corresponding deleted genes. The deletants grew at rates similar to the wild type in a galactose-containing medium. When they were transferred to a glucose-containing medium in order to deplete these proteins, growth of gaa1, gpi8 and gpi16 deletants began to decline at 12, 6 and 6 h, respectively, after transfer and ceased at 24, 12 and 12 h. In contrast, the gpi17 deletant continued to grow for >30 h at the same rate as the wild-type cells (data not shown). Since this seemed to be due to a long half-life of Gpi17p, we pre-cultured the gpi17 deletant in a raffinose-containing medium to decrease the level of Gpi17p. Growth of the gpi17 deletant began to decline at 6 h after transfer to a glucose-containing medium, but growth continued slowly for 33 h. We labelled gaa1, gpi8 and gpi16 deletants in the stationary state and the slowly growing gpi17 deletant with [3H]inositol, and analysed mannolipids by TLC (Figure 6). None of the wild-type cells bearing the same plasmids accumulated the complete precursor (CP) of GPI (lanes 1, 2, 5, 6, 9, 10, 13 and 14). As previously reported, gaa1 and gpi8 deletants accumulated CP and its precursors (lanes 3, 4, 7 and 8) (Hamburger et al., 1995; Benghezal et al., 1996). The gpi16 deletant accumulated comparable amounts of CP (lanes 11 and 12). The gpi17 deletant also accumulated CP, although at lower levels, perhaps due to continuous growth (lanes 15 and 16). These results indicate that yeast Gpi16p and Gpi17p are involved in transfer of GPI to proteins.
Fig. 6. Saccharomyces cerevisiae GPI16 and GPI17 are orthologues of PIG-T and PIG-S, respectively. Wild-type, and gaa1, gpi8, gpi16 and gpi17 deletant haploid yeast cells bearing a GAL1-regulatable expression plasmid for the respective ORF were pre-cultured in 2% galactose (for gaa1, gpi8 and gpi16 deletants) or 2% raffinose (for the gpi17 deletant) and transferred to 5% glucose-containing medium. gaa1, gpi8, gpi16 and gpi17 deletants were metabolically labelled with [3H]inositol at 24, 12, 12 and 24 h, respectively, after transfer. Mannolipids were extracted and analysed by TLC. The identities of spots (Hamburger et al., 1995; Benghezal et al., 1996) are indicated on the left. Four lines analysed in groups were derived from the same ascus. W, wild type; M, mutant.
PIG-T is critical for formation of GPI transamidase complex
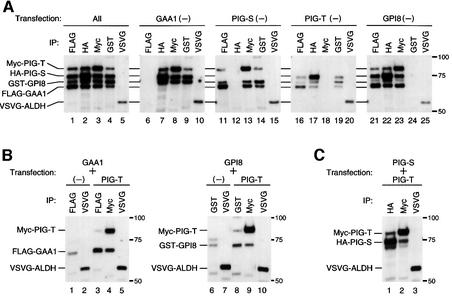
In the absence of any one of the four components, no transamidase activity was detected. Moreover, generation of the carbonyl intermediate was not seen (Figure 5). This could be due either to a lack of one functional component or to a lack of complex formation. We examined the formation of GPI transamidase complex under these conditions. We used CHO cells because expression levels of the components were very low in F9 cells. The four differentially tagged components and aldehyde dehydrogenase (ALDH), an ER resident protein used as a control, were transfected into CHO cells in various combinations. We used a very strong promoter, SRα, to overexpress transfected components relative to endogenous hamster proteins. Components co-precipitated with an immunoprecipitated component were analysed by western blotting. In cells transfected with all components, FLAG-GAA1 co-precipitated Myc-PIG-T, haemagglutinin (HA)-PIG-S and GST–GPI8, but not vesicular stomatitis virus glycoprotein (VSVG)–ALDH (Figure 7A, lane 1). HA-PIG-S (lane 2), Myc-PIG-T (lane 3) and GST–GPI8 (lane 4) also co-precipitated the other three components at levels comparable with FLAG-GAA1. None of the GPI transamidase components was precipitated with VSVG–ALDH (lane 5), showing the specificity of co-precipitation. In the absence of transfection of GAA1, three other components were expressed well and one component co-precipitated with the other two components (lanes 6–10). The levels of co-precipitation were comparable with those from transfectants with all four components (compare lanes 2, 3 and 4 with 7, 8 and 9). Similarly, in the absence of PIG-S (lanes 11–15) or GPI8 (lanes 21–25) transfection, the other three components formed complexes. In contrast, in the absence of PIG-T transfection, expression of FLAG- GAA1 and GST–GPI8 was markedly decreased, and only trace amounts of these two components were co-precipitated (lanes 16 and 19). HA-PIG-S was expressed well (lane 17). Therefore, expression of GAA1 and GPI8 was dependent upon PIG-T. PIG-S is stable by itself and is associated with a complex of three other components.
Fig. 7. PIG-T is critical for formation of the GPI transamidase complex. The four differentially tagged transamidase components and ALDH were transfected into CHO cells in various combinations (combination of transfection shown above panels). Cells were lysed in a buffer containing 1% NP-40 2 days after transfection. Aliquots of the lysates were immunoprecipitated (IP) individually with one of the five antibodies as indicated and western blotted against a mixture of five antibodies. Identities of bands are on the left. Size markers in kDa are on the right. (A) Combinations of all four (lanes 1–5) and three out of four (lanes 6–25) components. In lanes 21–23, bands between HA-PIG-S and FLAG-GAA1 are not GST–GPI8; they are derived from HA-PIG-S and migrate slightly more slowly than GST–GPI8. (B) Pairwise combinations of PIG-T and GAA1 (lanes 1–5) or GPI8 (lanes 6–10). (C) A combination of PIG-S and PIG-T.
To analyse further the association between components, we tested pairwise combinations. We transfected FLAG- GAA1 and VSVG–ALDH with or without Myc-PIG-T (Figure 7B, left panel). The expression of GAA1 was inefficient in the absence of PIG-T (lane 1) and was enhanced 8-fold by co-transfection of PIG-T (lane 3). About half of GAA1 was co-precipitated with Myc-PIG-T (lanes 3 versus 4), whereas none was co-precipitated with VSVG–ALDH (lane 5). FLAG-GAA1 that was not co-precipitated with Myc-PIG-T may, at least in part, have been associated with endogenous hamster PIG-T. Another possibility is that association of GAA1 with PIG-T is less stable in the absence of GPI8 and PIG-S than within the complete four-protein complexes, releasing a fraction of GAA1 during the immunoprecipitation and washes. Similar results were obtained with the combination of GPI8 and PIG-T. GPI8 was well expressed only in the presence of PIG-T (lane 6 versus 8) and was co-precipitated with PIG-T but not with VSVG–ALDH (lanes 9 and 10). Therefore, it appears that PIG-T stablizes GAA1 and GPI8 through binding to them. As shown in Figure 7C, HA-PIG-S co-precipitated Myc-PIG-T (lane 1) and vice versa (lane 2), suggesting that PIG-S and PIG-T were associated with each other. These results indicate that PIG-T plays a critical role in the maintenance of a complex of GPI transamidase by stabilizing the expression of GAA1 and GPI8 and linking them to PIG-S.
Discussion
GPI transamidase consists of at least four essential components
The affinity-purified protein complexes containing tandem-tagged GPI8 contained three other proteins, GAA1, PIG-S and PIG-T, at levels comparable with GPI8 (Figure 1). We demonstrated by disrupting PIG-S and PIG-T genes in mouse F9 cells that both PIG-S and PIG-T are essential for GPI transamidase (Figures 4 and 5). Taken together with the reports that GAA1 and GPI8 are essential GPI transamidase components (Yu et al., 1997; Ohishi et al., 2000), all four components are essential for the GPI transamidase.
We found through a database search that S.cerevisiae, S.pombe, C.elegans and D.melanogaster have structural homologues of PIG-S and PIG-T (Figures 2 and 3). We demonstrated that S.cerevisiae genes bearing the open reading frames (ORFs) YHR188C and YDR434W, termed GPI16 and GPI17, respectively, encode functional homologues of PIG-T and PIG-S, respectively (Figure 6). It seems, therefore, that GPI transamidases of these organisms also consist of at least four components.
The complex structure of the GPI transamidase is in common with another ER-resident enzyme involved in a similar post-translational modification, the oligosaccharyltransferase (OST). OST transfers oligosaccharide from lipid-linked oligosaccharide to asparagine residues within Asn-X-Ser/Thr sequences. OSTs of S.cerevisiae and very probably of humans as well consist of nine components. Five of them are essential components, whereas others, such as OST3, OST4, OST5 and OST6, are non-essential (Kelleher et al., 1992; te Heesen et al., 1993; Knauer and Lehle, 1999).
It is unclear whether GAA1, GPI8, PIG-S and PIG-T are the only components of the GPI transamidase. The isolated complex probably represents at least a core of the enzyme because it was stable in 1% NP-40 and did not contain any other protein at a significant level. It is possible that some components might have been lost during isolation if association is unstable. Establishment of a reconstituted enzyme assay for the GPI transamidase is required in order to determine whether the complex analysed in this study contains all the essential components.
A central role for PIG-T in formation of the GPI transamidase complex
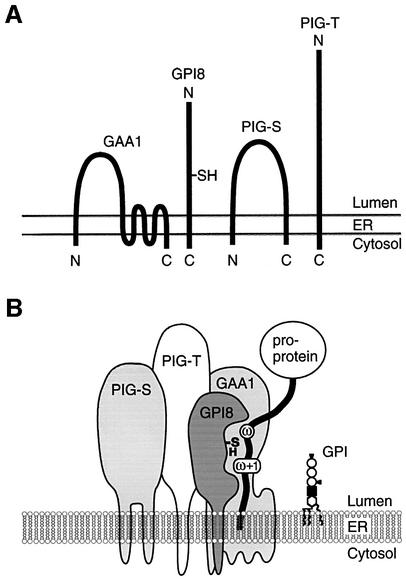
It is predicted that PIG-S has two transmembrane domains near the N- and C-termini with a large luminal domain (Figure 8A). PIG-T is a type-I membrane protein with a large luminal domain and a transmembrane domain near the C-terminus (Figure 8A). The luminal orientation of the major portions of PIG-S and PIG-T is consistent with the idea that the attachment of GPI to proteins occurs in the ER lumen (Udenfriend and Kodukula, 1995; Kinoshita and Inoue, 2000).
Fig. 8. (A) Membrane topology of GPI transamidase components. N- and C-termini are shown. GAA1 has a large luminal domain and multiple transmembrane regions. Both N- and C-termini of GAA1 are predicted to be on the cytoplasmic side (Hamburger et al., 1995). PIG-S has two transmembrane regions near the N- and C-termini. GPI8 (Benghezal et al., 1996) and PIG-T are type-I transmembrane proteins. (B) A model of GPI transamidase. The GPI transamidase is a complex consisting of at least four components. PIG-T is a central component interacting with GAA1, GPI8 and PIG-S. The catalytic component, GPI8, recognizes substrate proproteins and attacks a peptide bond between ω and ω+1 sites with a sulfhydryl group of its active site cysteine to form a carbonyl intermediate. It is unclear how GPI is presented to the carbonyl intermediate to complete transamidation.
We showed that stable expression of GPI8 and GAA1 is dependent on their associations with PIG-T (Figure 7B). PIG-S is stable by itself but it also associates with PIG-T (Figure 7A and C). In the absence of PIG-S, a complex of GAA1, PIG-T and GPI8 is stably formed. These results suggest that PIG-T is located at the centre of the complex and critical in maintenance of the GPI transamidase. Figure 8B shows a model of the GPI transamidase drawn based on these results.
It was reported that GPI anchoring ability was lost when the microsomal membranes of Trypanosoma brucei were washed at high pH (Sharma et al., 2000), whereas similar treatment of the mammalian microsomal membranes had no effect (Vidugiriene and Menon, 1995). Moreover, the GPI anchoring was restored by adding the high pH extract of trypanosomal membranes or recombinant GPI8 protein of the related protozoa, Leishmania mexicana, to the washed membranes (Sharma et al., 2000). These results suggest that trypanosomal GPI8 is a soluble protein. Consistent with this, GPI8s of T.brucei (K.Nagamune, Y.Fujinaga and T.Kinoshita, unpublished data) and L.mexicana (Hilley et al., 2000) lack a transmembrane domain, whereas human and S.cerevisiae GPI8s are type-I membrane proteins. It is suggested, therefore, that GPI8 and PIG-T associate with each other via their luminal domains.
Functional roles of four components of the GPI transamidase
Several lines of evidence indicate that GPI8 is a catalytic component that generates a carbonyl intermediate with a substrate protein: (i) GPI8 has homology with cysteine proteases; (ii) GPI8 has cysteine and histidine that are conserved in cysteine proteases and are essential for the GPI transamidase activity; (iii) Spurway et al. (2001) demonstrated physical association of GPI8 with a substrate protein using chemical cross-linkers; and (iv) Vidugiriene et al. (2001) showed physical interaction of GPI8 with the GPI attachment signal peptide of the substrate protein, particularly a hydrophilic portion of the signal peptide.
The GPI attachment signal peptide must be recognized before generation of the carbonyl intermediate. Because the GPI attachment signal peptide consists of hydrophilic and hydrophobic segments, and because GPI8 from trypanosome and leishmania lack a transmembrane domain, it is unlikely that the GPI attachment signal is recognized only by GPI8. Three other components have at least one transmembrane domain. In the absence of any one of the other three components, the carbonyl intermediate was not generated (Figure 5C). As discussed above, the protein complex was not formed well without PIG-T, so it is not possible to speculate whether PIG-T is required for the recognition of the GPI attachment signal. Without GAA1 or PIG-S, the protein complexes were formed well. Therefore, it is possible that GAA1 and/or PIG-S are required for the signal recognition. It is also possible, however, that a lack of GAA1 or PIG-S indirectly affected the signal recognition by causing a conformational change of the protein complex. Vidugiriene et al. (2001) reported that the GPI attachment signal peptide was photo-cross-linked to a 60 kDa protein. This can be GAA1 or PIG-S. They also demonstrated photo-cross-linking between the substrate protein and a 120 kDa protein. Since none of the four components is of that molecular size, there may be yet another component of the GPI transamidase. We have no information at the moment about the recognition of the other substrate, GPI.
Materials and methods
Cells
Mouse class F cells defective in PIG-F (Inoue et al., 1993) and human class K cells defective in GPI8 (Yu et al., 1997) were gifts from Dr R.Hyman (Salk Institute, CA) and Dr M.E.Medof (Case Western Reserve University, OH), respectively. They were cultured in high glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS). CHO-K1 cells were cultured in Ham’s F-12 medium containing 10% FCS.
Isolation and amino acid sequencing of GPI transamidase components
To allow two-step affinity purification of the GPI transamidase complex, we tagged the C-terminus of human GPI8 with FLAG and GST (Maeda et al., 2001) and subcloned it into a pME vector bearing a PGK (phosphoglycerokinase)-driven puromycin resistance gene cassette (Ohishi et al., 2000). We transfected it into class K cells and selected stable transfectants with puromycin at 2 µg/ml.
For purification of the GPI transamidase, 1010 cells were lysed in 500 ml of an IP buffer [50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP-40 and complete protease inhibitor cocktail (Roche Diagnostics)]. Cell debris was removed by centrifugation at 10 000 g for 20 min and the supernatants were clarified by centrifugation at 100 000 g for 1 h. We incubated the resulting lysate with 2 ml of a slurry of anti-FLAG M2 antibody resin (Sigma) for 6 h at 4°C, washed it five times in IP buffer and eluted bound proteins five times with 5 ml of FLAG peptide in IP buffer at 500 µg/ml. Eluates were combined and mixed with 100 µl of glutathione–Sepharose slurry (Amersham Pharmacia Biotech). After five washes in IP buffer, the transamidase complex was eluted with 100 µl of an SDS–PAGE sample buffer containing 2-mercaptoethanol (2-ME). A sample equivalent to 2 × 109 cells was electrophoresed on a 10% gel and transferred to a PVDF membarne. Coomassie Blue-stained bands were excised and their N-terminal sequences were determined with a G1005A Hewlett-Packard Protein Sequencing System.
Cloning human PIG-S and PIG-T cDNAs
Searching the EST database in the National Center for Biotechnology Information (NCBI) with the N-terminal sequence of band 2 protein and the TBLASTN program, we found several mouse and human ESTs. Using these and a human genomic sequence (AC005726) containing the PIG-S gene, we determined the ORF of human PIG-S in silico. For PIG-T, we searched the non-redundant protein database in the NCBI with the N-terminal sequence of band 3 protein using the BLASTP program, and found a hypothetical protein and its cDNA sequence (AL121742) and a human genomic sequence (AL021578). A sequence for the first four amino acids (Met-Ala-Ala-Ala) was lacking. We identified the most likely initiation codon by comparing the sequences of 5′-RACE products from mouse brain Marathon-Ready cDNA (Clontech) and the human genomic sequence. Based on these sequences, we designed PCR primers and amplified the entire coding regions of PIG-S and PIG-T cDNA from human placental Marathon-Ready cDNA (Clontech).
Establishment and characterization of mouse PIG-S and PIG-T knockout cells
Bacterial artificial chromosome (BAC) clones 176-d23 and 206-o17 containing mouse PIG-S and PIG-T genes, respectively, were obtained by screening Down-to-the-Well Mouse ES (Release I) BAC DNA Pools (Incyto Genomics). Construction of targeting vectors assembled from PGK-driven drug resistance gene cassettes, short (1.3–1.7 kb) and long (5.9–7.2 kb) genomic fragments (see Figure 4A) cloned in pBluescript II (pBS) (Stratagene) and a vector backbone of pPNT vector containing a PGK-driven human herpes simplex thymidine kinase gene, and disruption of genes in F9 cells were according to a previous report (Ohishi et al., 2000). Southern blot analysis and in vitro translation assay for GPI transamidase activity (Kodukula et al., 1991) were performed as described (Ohishi et al., 2000).
Yeast mutants and analysis of their GPI intermediates
The S.cerevisiae deletants for ORFs YHR188C, YDR434W, YLR088W and YDR331W, which encode Gpi16p, Gpi17p, Gaa1p and Gpi8p, respectively, were generated by the Saccharomyces Genome Deletion Project (Winzeler et al., 1999) and purchased from Research Genetics. GAL1-regulatable expession plasmids were constructed by insertion of ORFs amplified from genomic DNA of wild-type strain W303-1A to the centromeric plasmid p415 GAL1. We introduced each plasmid into deletion mutants defective in the corresponding genes and selected transfectants in SC-Leu (2% glucose) medium. To establish haploid wild-type and deletion mutants bearing the expression plasmid, spores were dissected on SC-Leu (2% galactose) medium. Cells from four spores derived from one ascus were pre-cultured in SC-Leu (2% galactose) (for gpi16, gaa1 and gpi8 deletants) or SC-Leu (2% raffinose) (for the gpi17 deletant) medium and then inoculated into SC-Leu (5% glucose) medium.
Cells (8 × 107) were washed, resuspended in 0.5 ml of SC-Leu (2% glucose) without inositol and pre-incubated at 30°C for 30 min. A sample of 15 µCi of [3H]inositol (Amersham Pharmacia Biotech) was added, followed by incubation at 30°C for 90 min. The labelling reaction was stopped by addition of 10 µl of 0.5 M NaN3 and 0.5 M NaF on ice and washing twice in 10 mM NaN3 and 10 mM NaF. The cells were extracted by vortexing in 500 µl of CMW (chloroform:methanol:water 10:10:3) with 0.5 g of glass beads followed by shaking for 30 min. The extract was taken and the cells were re-extracted in 400 µl of CMW in a similar way. Pooled extracts were dried. Partitioning of GPI intermediates with n-butanol and TLC analysis were as previously reported (Horvath et al., 1994; Hamburger et al., 1995).
Mammalian expression plasmids
All expression plasmids were constructed on a pME18Sf vector, a gift from Dr K.Maruyama (Tokyo Medical and Dental University, Japan). Fragments encoding 3× HA, 3× VSVG and 6× Myc tags made by PCR with two partially complementary primers as previously reported (Nakajima and Yaoita, 1997) were inserted after the initiation codon or before the stop codon to construct expression plasmids for Myc-PIG-S, HA-PIG-S and VSVG–ALDH similarly to previous reports (Maeda et al., 1998; Watanabe et al., 1998). FLAG-GAA1 and GST–GPI8 were used in our previous report (Ohishi et al., 2000). Sequences of all primers used in this study are available upon request.
Flow cytometric analysis
Cells were stained with biotinylated anti-Thy-1 G7 followed by phycoerythrin-conjugated streptavidin (Biomeda) and analysed in a FACScan (Becton Dickinson).
Analysis of protein complexes
CHO cells (4 × 106) suspended in 0.4 ml of culture medium were electroporated with 4 µg each of five plasmids at 960 µF and 250 V. When a plasmid was omitted, an empty vector was added instead. Cells were cultured for 2 days and solubilized in 5 ml of IP buffer. The lysates were immunoprecipitated with 1 µg of anti-FLAG M2, anti-GST (Clontech), anti-Myc (Oncogene), anti-HA or anti-VSVG (Roche) plus protein G beads (Amersham Pharmacia Biotech). The precipitates were washed in IP buffer and treated in an SDS–PAGE sample buffer with 2-ME. Western blotting was performed with a mixture of five antibodies used for immunoprecipitation and horseradish peroxidase (HRP)-conjugated protein G (Bio-Rad Laboratories).
Acknowledgments
Acknowledgements
We thank Keiko Kinoshita for technical assistance. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Arfin S.M. and Bradshaw,R.A. (1988) Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry, 27, 7979–7984. [DOI] [PubMed] [Google Scholar]
- Benghezal M., Benachour,A., Rusconi,S., Aebi,M. and Conzelmann,A. (1996) Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J., 15, 6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Bucht G., Wikström,P. and Hjalmarsson,L. (1999) Optimising the signal peptide for glycosyl phosphatidylinositol modification of human acetylcholinesterase using mutational analysis and peptide-quantitative structure–activity relationship. Biochim. Biophys. Acta, 1431, 471–482. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Bork,P. and Eisenhaber,F. (2001) Post-translational GPI lipid anchor modification of proteins in kingdoms of life: analysis of protein sequence data from complete genomes. Protein Eng., 14, 17–25. [DOI] [PubMed] [Google Scholar]
- Ferguson M.A.J. (2000) Glycosylphosphatidylinositol biosynthesis validated as a drug target for African sleeping sickness. Proc. Natl Acad. Sci. USA, 97, 10673–10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y., Tsukamoto,K. and Ikezawa,H. (1997) Mutational analysis of the C-terminal signal peptide of bovine liver 5′-nucleotidase for GPI anchoring: a study on the significance of the hydrophilic spacer region. Biochim. Biophys. Acta, 1328, 185–196. [DOI] [PubMed] [Google Scholar]
- Gerber L.D., Kodukula,K. and Udenfriend,S. (1992) Phosphatidyl inositol glycan (PI-G) anchored membrane proteins. J. Biol. Chem., 267, 12168–12173. [PubMed] [Google Scholar]
- Hamburger D., Egerton,M. and Riezman,H. (1995) Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J. Cell Biol., 129, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilley J.D., Zawadzki,J.L., McConville,M.J., Coombs,G.H. and Mottram,J.C. (2000) Leishmania mexicana mutants lacking glycosylphosphatidylinositol (GPI):protein transamidase provide insights into the biosynthesis and functions of GPI-anchored proteins. Mol. Biol. Cell, 11, 1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi Y., Komuro,I., Chen,R., Hosoda,T., Mizuno,T., Kudoh,S., Georgescu,S.P., Medof,M.E. and Yazaki,Y. (1998) Molecular cloning of human homolog of yeast GAA1 which is required for attachment of glycosylphosphatidylinositols to proteins. FEBS Lett., 421, 252–258. [DOI] [PubMed] [Google Scholar]
- Hirose S., Prince,G.M., Sevlever,D., Ravi,L., Rosenberry,T.L., Ueda,E. and Medof,M.E. (1992) Characterization of putative glycoinositol phospholipid anchor precursors in mammalian cells. Localization of phosphoethanolamine. J. Biol. Chem., 267, 16968–16974. [PubMed] [Google Scholar]
- Horvath A., Sutterlin,C., Manning-Krieg,U., Movva,N.R. and Riezman,H. (1994) Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J., 13, 3687–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. (1994) On global sequence alignment. Comput. Appl. Biosci., 10, 227–235. [DOI] [PubMed] [Google Scholar]
- Inoue N., Kinoshita,T., Orii,T. and Takeda,J. (1993) Cloning of a human gene, PIG-F, a component of glycosylphosphatidylinositol anchor biosynthesis, by a novel expression cloning strategy. J. Biol. Chem., 268, 6882–6885. [PubMed] [Google Scholar]
- Kelleher D., Kreibich,G. and Gilmore,R. (1992) Oligosaccharyl transferase activity is associated with a protein complex composed of ribophorins I and II and a 48 kd protein. Cell, 69, 55–65. [DOI] [PubMed] [Google Scholar]
- Kinoshita T. and Inoue,N. (2000) Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol., 4, 632–638. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Inoue,N. and Takeda,J. (1995) Defective glycosyl phosphatidylinositol anchor synthesis and paroxysmal nocturnal hemoglobinuria. Adv. Immunol., 60, 57–103. [DOI] [PubMed] [Google Scholar]
- Knauer R. and Lehle,L. (1999) The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta, 1426, 259–273. [DOI] [PubMed] [Google Scholar]
- Kodukula K., Micanovic,R., Gerber,L., Tamburrini,M., Brink,L. and Udenfriend,S. (1991) Biosynthesis of phosphatidylinositol glycan-anchored membrane proteins. J. Biol. Chem., 266, 4464–4470. [PubMed] [Google Scholar]
- Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Leidich S.D., Kostova,Z., Latek,R.R., Costello,L.C., Drapp,D.A., Gray,W., Fassler,J.S. and Orlean,P. (1995) Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol: cloning of the GPI2 gene. J. Biol. Chem., 270, 13029–13035. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Tomita,S., Watanabe,R., Ohishi,K. and Kinoshita,T. (1998) DPM2 regulates biosynthesis of dolichol phosphate-mannose in mammalian cells: correct subcellular localization and stabilization of DPM1 and binding of dolichol phosphate. EMBO J., 17, 4920–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Watanabe,R., Harris,C.L., Hong,Y., Ohishi,K., Kinoshita,K. and Kinoshita,T. (2001) PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J., 20, 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville M.J. and Ferguson,M.A.J. (1993) The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J., 294, 305–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U., Benghezal,M., Imhof,I. and Conzelmann,A. (2000) Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry, 39, 3461–3471. [DOI] [PubMed] [Google Scholar]
- Micanovic R., Gerber,L.D., Berger,J., Kodukula,K. and Udenfriend,S. (1990) Selectivity of the cleavage/attachment site of phosphatidyl inositol-glycan-anchored membrane proteins determined by site-specific mutagenesis at Asp-484 of placental alkaline phosphatase. Proc. Natl Acad. Sci. USA, 87, 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P. and Caras,I.W. (1994) Requirements for glycosylphosphatidyl inositol attachment are similar but not identical in mammalian cells and parasitic protozoa. J. Cell Biol., 125, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P., Raab,H., Kohr,W.J. and Caras,I.W. (1991) Glycophospholipid membrane anchor attachment. Molecular analysis of the cleavage/attachment site. J. Biol. Chem., 266, 1250–1257. [PubMed] [Google Scholar]
- Nakajima K. and Yaoita,Y. (1997) Construction of multi-epitope tag sequence by PCR for sensitive Western blot analysis. Nucleic Acids Res., 25, 2231–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C., Horvath,A. and Riezman,H. (1993) Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J. Biol. Chem., 268, 10558–10563. [PubMed] [Google Scholar]
- Ohishi K., Inoue,N., Maeda,Y., Takeda,J., Riezman,H. and Kinoshita,T. (2000) Gaa1p and gpi8p are components of a glycosylphos phatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell, 11, 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D.K., Vidugiriene,J., Bangs,J.D. and Menon,A.K. (1999) A cell-free assay for glycosylphosphatidylinositol anchoring in African trypanosomes. J. Biol. Chem., 274, 16479–16486. [DOI] [PubMed] [Google Scholar]
- Sharma D.K., Hilley,J.D., Bangs,J.D., Coombs,G.H., Mottram,J.C. and Menon,A.K. (2000) Soluble GPI8 restores glycosylphosphatidyl inositol anchoring in a trypanosome cell-free system depleted of lumenal endoplasmic reticulum proteins. Biochem. J., 351, 717–722. [PMC free article] [PubMed] [Google Scholar]
- Spurway T.D., Dalley,J.A., High,S. and Bulleid,N.J. (2001) Early events in GPI-anchor addition: substrate proteins associate with the transamidase subunit Gpi8p. J. Biol. Chem., 276, 15975–15982. [DOI] [PubMed] [Google Scholar]
- te Heesen S., Knauer,R., Lehle,L. and Aebi,M. (1993) Yeast Wbp1p and Swp1p form a protein complex essential for oligosaccharyl transferase activity. EMBO J., 12, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udenfriend S. and Kodukula,K. (1995) How glycosylphosphatidyl inositol-anchored membrane proteins are made. Annu. Rev. Biochem., 64, 563–591. [DOI] [PubMed] [Google Scholar]
- Vidugiriene J. and Menon,A.K. (1995) Soluble constituents of the ER lumen are required for GPI anchoring of a model protein. EMBO J., 14, 4686–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidugiriene J., Vainauskas,S., Johnson,A.E. and Menon,A.K. (2001) Endoplasmic reticulum proteins involved in glycosylphos phatidylinositol-anchor attachment: photocrosslinking studies in a cell-free system. Eur. J. Biochem., 268, 2290–2300. [DOI] [PubMed] [Google Scholar]
- Waneck G.L., Stein,M.E. and Flavell,R.A. (1988) Conversion of a PI-anchored protein to an integral membrane protein by a single amino acid mutation. Science, 241, 697–699. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Inoue,N., Westfall,B., Taron,C.H., Orlean,P., Takeda,J. and Kinoshita,T. (1998) The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J., 17, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Yu J., Nagarajan,S., Knez,J.J., Udenfriend,S., Chen,R. and Medof,M.E. (1997) The affected gene underlying the class K glycosyl phosphatidylinositol (GPI) surface protein defect codes for the GPI transamidase. Proc. Natl Acad. Sci. USA, 94, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]