Abstract
Cystathionine β-synthase (CBS) is a unique heme- containing enzyme that catalyzes a pyridoxal 5′-phosphate (PLP)-dependent condensation of serine and homocysteine to give cystathionine. Deficiency of CBS leads to homocystinuria, an inherited disease of sulfur metabolism characterized by increased levels of the toxic metabolite homocysteine. Here we present the X-ray crystal structure of a truncated form of the enzyme. CBS shares the same fold with O-acetylserine sulfhydrylase but it contains an additional N-terminal heme binding site. This heme binding motif together with a spatially adjacent oxidoreductase active site motif could explain the regulation of its enzyme activity by redox changes.
Keywords: cystathionine β-synthase/cysteine biosynthesis/heme protein/pyridoxal 5′-phosphate/X-ray crystal structure
Introduction
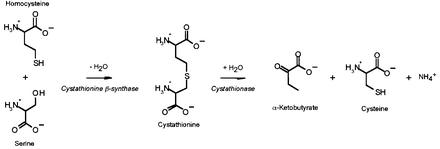
Cystathionine β-synthase (CBS, l-serine hydrolyase, EC 4.2.1.22) is the first enzyme of the transsulfuration pathway in which the potentially toxic homocysteine is converted to cysteine (Figure 1). Deficiency of CBS activity is the most common cause of homocystinuria, an inherited metabolic disease characterized by dislocated eye lenses, skeletal problems, vascular disease and mental retardation (Mudd et al., 2001). There have now been >100 mutations described in this gene (Kraus et al., 1999). Hyperhomocysteinemia, a condition characterized by small increases in plasma concentrations of homocysteine, represents an independent risk for vascular disease (Yap et al., 2000).
Fig. 1. Transsulfuration pathway.
The human CBS is a homotetramer consisting of 63 kDa subunits, which binds two cofactors, pyridoxal 5′-phosphate (PLP) and heme (Skovby et al., 1984; Kery et al., 1994). Each CBS subunit of 551 amino acid residues binds two substrates (homocysteine and serine) and is further regulated by S-adenosyl-l-methionine (AdoMet) (Kery et al., 1994). While the role of heme in CBS is unknown, catalysis by CBS can be explained solely by participation of PLP in the reaction mechanism (Kery et al., 1999). In fact, yeast CBS catalyzes the same reaction but does not contain heme (Jhee et al., 2000; Maclean et al., 2000). Limited proteolysis of the full-length enzyme yields the ‘active core’ of CBS (amino acid residues 40–413). The reduction in size is accompanied by a significant increase in the specific activity of the enzyme and a change in its oligomerization state. The active core enzyme is about twice as active as the full-length enzyme and forms dimers instead of tetramers. It binds both PLP and heme cofactors, but is no longer activated by AdoMet (Kery et al., 1998). This 45 kDa active core is the portion of CBS most homologous with the related enzymes in plants and bacteria, O-acetylserine sulfhydrylase (OASS) and O-acetyl-l-serine(thiol)lyase (OASTL) (Swaroop et al., 1992; Kraus, 1994). The C-terminal regulatory domain that is missing in the active core enzyme contains a recently identified protein folding motif called ‘CBS domain’, which is also found in inosine 5′-monophosphate dehydrogenase, chloride channels and several other proteins in various organisms (Bateman, 1997).
Since the CBS tetramer of the full-length enzyme has a strong tendency to aggregate, physical studies are very difficult. We have recently crystallized recombinant human CBS comprising the amino acid residues 1–413 (Janosik et al., 2001). This truncated enzyme is similar to the above-mentioned active core in that the ∼140 C-terminal residues including the ‘CBS domain’ are missing. It is about twice as active as the wild-type CBS, forms dimers, and does not exhibit the aggregating properties of the full-length enzyme. We have now solved the X-ray structure of the truncated form of CBS by combining phase information obtained from molecular replacement (MR) and multiple anomalous dispersion (MAD). The crystals belong to the trigonal space group P31 with cell parameters a = b = 144.46 Å, c = 108.21 Å, and contain three dimers per asymmetric unit (Table I).
Table I. Data statistics.
| Data set | Native | λ1 | λ2 | λ3 |
|---|---|---|---|---|
| Space group | P31 | P31 | ||
| a, b (Å) | 144.46 | 144.52 | ||
| c (Å) | 108.21 | 108.16 | ||
| Wavelength | 0.9711 | 1.7411 | 1.7424 | 1.7300 |
| Resolution range (Å) | 50–2.6 | 50–3.1 | 50–3.15 | 50–3.15 |
| Unique reflections | 63 997 | 83 338 | 82 233 | 84 316 |
| Overall completeness (outermost shell) (%) | 82.4 (63.9) | 94.4 (67.1) | 93.8 (60.1) | 91.3 (68.6) |
| Rmergea (outermost shell) | 7.6 (30.9) | 12.2 (38.5) | 15.0 (44.3) | 16.9 (48.7) |
| Phasing powerb (acentric) | 0.585 | 1.045 | 0.716 | 0.633 |
| FOMc | 0.24658 | |||
| R factord (Rfree) | 25.7 (29.6) | |||
| R.m.s.d. from ideality | ||||
| bonds (Å) | 0.008 | |||
| angles (°) | 1.42 | |||
| Average B factors (Å2) | ||||
| protein | 37.6 | |||
| solvent | 24.1 | |||
| PLP | 17.4 | |||
| heme | 34.1 |
aRmerge = ∑hkl∑i|Ii – <I>|/∑hkl∑iIi
bPhasing power = (FH/lack of closure).
cFOM = figure of merit [(cosφ)2 + (sinφ)2]1/2
dR factor = ∑hkl|Fobs|–|Fcalc|/∑hkl|Fobs|
Results and discussion
Description of the structure
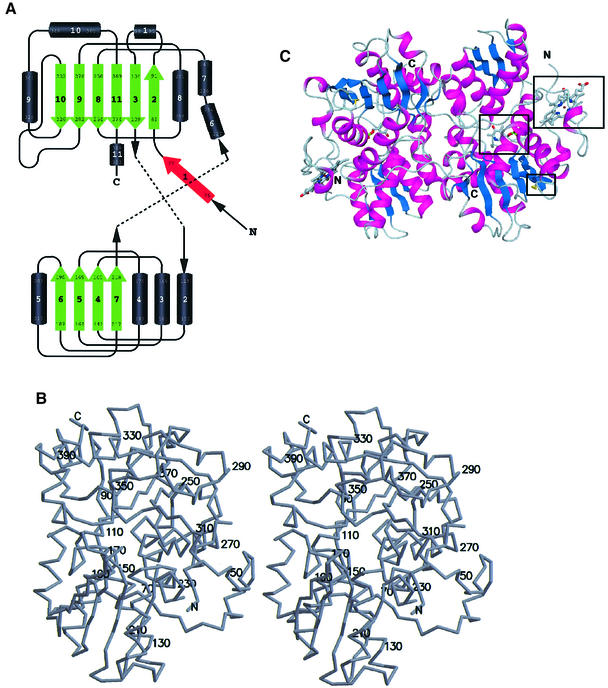
The fold of the truncated human CBS enzyme belongs to the β-family of vitamin B6 enzymes (Alexander et al., 1994) and resembles the fold of OASS from Salmonella typhimurium (Figure 2) (Burkhard et al., 1998). Three other PLP enzymes with known structure share the same fold type: tryptophan synthase (Hyde et al., 1988), threonine deaminase (TD) (Gallagher et al., 1998) and aminocyclopropane deaminase (Yao et al., 2000). A least square superposition of the Cα-positions of the structurally conserved parts between CBS and OASS yields a root mean square deviation (r.m.s.d.) of only 1.32 Å, while the differences between the structures are mainly located in the loop regions (residues 95–104, 282–298 and 359–369). The monomer is composed of 11 α-helices, seven short 310 helices and two β-sheets consisting of four (in the N-terminal domain) and six strands (in the C-terminal domain), respectively (Figure 2A). The additional β-strand 1 interacts with strand 2 of the C-terminal sheet of the other monomer in the dimer in a parallel manner. The α-helices 1 and 11, β-strand 1 and the N-terminal heme binding site of CBS are missing in OASS, while in turn OASS has an additional α-helix that is not present in CBS (helix 8 in Burkhard et al., 1998). The heme binding motif lacks any secondary structure with the exception of a short 310 helix. It has been shown that OASS, and to a lesser extent tryptophan synthase, undergoes a large conformational change from the open, uncomplexed to the closed, complexed conformation upon ligand binding. This conformational change involves a rigid body rotation of the so-called moveable domain (Schneider et al., 1998; Burkhard et al., 1999, 2000). The Cα-backbone of CBS in this region (residues 186–222) more closely resembles the open conformation of OASS.
Fig. 2. Overall structure of truncated CBS. (A) Topology of the fold in CBS. Above, the C-terminal domain (with the first two strands of the β-sheet formed by the N-terminal residues); below, the N-terminal domain. Both domains are of the type α/β and contain a central β-sheet surrounded by several α-helices. Strand 1 (red) is part of the C-terminal β-sheet of the other monomer in the dimer. (B) Stereo drawing showing the overall fold of CBS with every twentieth residue labeled. (C) Schematic representation of the tertiary fold of a dimer of CBS. The central β-sheets are colored in blue and the surrounding α-helices are colored in magenta. In ball-and-stick representation and marked by a black box are the active site PLP, the heme and the oxidoreductase motif. The view is down the non-crystallographic 2-fold axis, which relates the two subunits of the dimer.
The dimer interface is mainly hydrophobic in character and is composed of the side chains of the residues Ile76, Leu77, Ile80, Thr87, Val90, Ile92, Ile152, Leu156, Val160, Val180, Ala183, Leu184, Ile339, Ala340, Leu344, Leu345, Val378, Met382 and Leu386. The central part of the dimer interface is formed by the residues Phe111 and Phe112 close to the 2-fold dimer axis, thus Phe112 of monomer A interacts with Phe112 of monomer B and vice versa. But polar interactions also contribute to the dimer interactions. On the other hand, the guanidium group of Arg379 is completely buried within the core of the dimer interface but is not involved in any polar interactions between monomers, but rather forms hydrogen bonds to Gly115 and Asn380 of the same monomer.
The active site
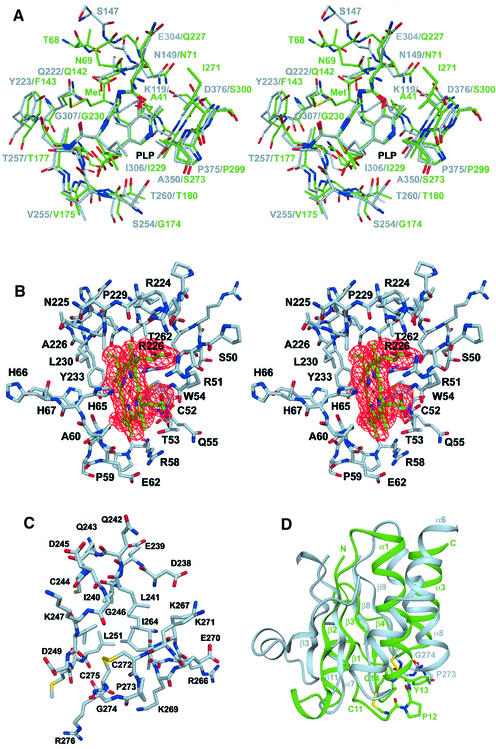
The coenzyme PLP is deeply buried in a cleft between the N- and C-terminal domains, and the active site is accessible only via a narrow channel. The cofactor is linked to the ε-amino group of Lys119 via a Schiff base linkage forming the so-called ‘internal aldimine’ (Figure 3A) (Christen and Metzler, 1985; Kery et al., 1999). The nitrogen of the pyridine ring forms a hydrogen bond to the Oγ of Ser349 similar to the other enzymes of the β-family of PLP enzymes, OASS, tryptophan synthase and TD (Gallagher et al., 1998). Another hydrogen bond is formed between the 3′ hydroxyl group of PLP and the Nδ2 of Asn149. This residue is coplanar with the pyridine ring and thus allows the expected ring tilt upon transaldimination. The phosphate binding loop is located between β-strand 8 and α-helix 8 and is composed of the residues Gly256, Thr257, Gly258, Gly259 and Thr260. These residues form an extended hydrogen bonding network with the phosphate moiety of PLP, thus anchoring the cofactor to the protein matrix. In addition, the positive pole of the helix dipole from α-helix 8 compensates for the negative charge of the phosphate group.
Fig. 3. Structural details of CBS. (A) The active site region of CBS (gray) in a superposition with the active site of OASS (green). The sequences as well as the structure of the two enzymes are very similar. A superposition of the 25 structurally most conserved residues yields an r.m.s.d. of 1.6 Å of their Cα positions. The substrate analog of OASS methionine indicates the probable binding mode of the first substrate serine and also the region where the second substrate homocysteine is expected to bind. (B) The heme binding site of CBS with heme in green and the surrounding residues in gray. The two residues His65 and Cys52 are the ligands to the heme iron (dark red). The difference density for the cofactor is shown in red contoured at 3.5σ. (C) The oxidoreductase motif in ball-and-stick representation and (D) in a superposition with the structure of glutaredoxin. The structure of CBS is in gray, the one of glutaredoxin in green. The overall topology is very similar, but the active site motif in CBS is switched to the other helix compared with glutaredoxin.
The conformation of the residues surrounding the cofactor is highly conserved between CBS and OASS (Figure 3A). In the two monomers of all three dimers the asparagine loop (residues 146–149; cf. Burkhard et al., 1999) adopts two slightly different conformations, indicating its flexibility and ability to bind the carboxylate group of the substrate by a local conformational change. In OASS this conformational change includes atom movements of >7 Å upon substrate binding. Preliminary X-ray data suggest that in CBS the substrate serine binds to the active site in a similar way to the substrate analog methionine in OASS (data not shown). Residues Tyr223 and Gly307 are probably the key residues for substrate specificity, as they are spatially adjacent to the substrate binding site.
The heme binding site
All dimers of the asymmetric unit contain two heme molecules that are located at distal ends of the dimers with the orientation of their ring planes normal to the protein surface (Figure 2C). The heme is bound in a hydrophobic pocket formed by residues 50–67, α-helices 6 and 8 and a loop preceding β-strand 10 (Figure 3B). The sulfhydryl group of Cys52 and the Nε2 atom of His65 axially coordinate the iron in the heme. This result confirms an earlier finding that the 5th and 6th coordination positions of the heme are a thiolate and a nitrogenous group, respectively (Omura et al., 1984). Unlike the globins or cytochrome c peroxidase, the Nδ1 of His65 is solvent accessible and lacking any hydrogen bonding partner from protein residues. The sulfur atom of Cys52 is deprotonized (Ojha et al., 2000) and forms additional polar interactions with the side chain of Arg266 and the main chain nitrogen of Trp54. The heme carboxylate groups are involved in ionic interactions with Arg51 and Arg224 and are partially solvent accessible. This is in contrast to heme enzymes like cytochrome c peroxidase and cytochrome P450cam, where the heme is completely buried in an internal cavity of the protein (Poulos, 1987). Since the iron ion is ligated from both sides by protein residues this makes an enzymatic role of the heme unlikely. The situation is more similar to the c-type cytochromes, which are involved in electron transfer.
Since the heme is not covalently attached to the protein it can be reversibly released under reducing conditions from CBS crystals in the presence of carbon monoxide (CO) (Bruno et al., 2001). Similarly, the heme can be fully dissociated from the reduced enzyme in CO-saturated solution. Under these conditions the enzyme retains ∼20% of original activity. In contrast to the reversible removal of heme from crystals, the heme cannot be introduced back in solution (J.P.Kraus, unpublished results). Under oxidizing conditions, the heme cannot be released, probably because CO does not bind to heme in its ferric state. It is likely that CO displaces one of the axial heme ligands, followed by a local unfolding of the N-terminal residues leading to a release of the prosthetic group. The displaced ligand is probably the cysteine, because the absorption spectrum of CBS treated with CO is similar to the spectra of other CO–heme–imidazole protein complexes (Taoka et al., 1999). It has been suggested that the redox state of the heme iron influences the catalytic rate in the full-length enzyme (Taoka et al., 1998). We find that the redox state of the heme has no impact on the CBS activity (J.P.Kraus, unpublished results) and that the heme itself is not required for catalysis (Bruno et al., 2001). Further evidence that the heme moiety is not involved in the catalytic steps comes from a CBS enzyme in which the first 70 residues were deleted, including the heme binding residues Cys52 and His65. This enzyme retains ∼25% of wild-type CBS activity (J.P.Kraus, unpublished results).
Oxidoreductase active site motif
The loop between α-helix 8 and β-strand 9 harbors a motif similar to the active site of disulfide oxidoreductases. The consensus sequence of this motif contains two cysteines that are linked by two residues, one of which is a proline (Table II). The two cysteines of oxidoreductases are involved in various redox reactions in the cell, because they can be reversibly oxidized and/or reduced by switching between a disulfide and dithiol form during the catalytic process. In CBS this motif consists of the sequence CPGC (residues 272–275, Figure 3C) and forms a β-turn. The two cysteines are oxidized and form a disulfide bridge. The disulfide bridge is in the same right-handed hook conformation as those in disulfide oxidoreductases and is located on the surface of the protein and hence is solvent accessible. The same two cysteines, however, are not solvent accessible in the full-length enzyme (J.P.Kraus, unpublished results).
Table II. Sequence comparison.
| Human | 248 LDMLVASVGTGGTITGIARKLKEKCPGCRIIGVDPEGSILAEP 290 |
| Rabbit | 248 LDMLVASAGTGGTITGIARKLKEKCPGCQIIGVDPEGSILAEP 290 |
| Rat | 245 VDMLVASAGTGGTITGIARKLKEKCPGCKIIGVDPEGSILAEP 287 |
| Dictyostelium | 202 IDMIVCTAGTGGTITGIARKIKERLPNCIVVGVDPHGSILAQP 244 |
| Yeast | 188 LRAVVAGAGTGGTISGISKYLKEQNDKIQIVGADPFGSILAQP 230 |
The structure of glutaredoxin from Escherichia coli can be roughly superimposed onto CBS (Figure 3D). In this superposition α-helix 1 of glutaredoxin fits onto α-helix 7 of CBS and α-helix 3 onto α-helix 8 of CBS, but in opposite directions. The four strands of the central β-sheet of glutaredoxin fit nicely onto strands 3, 8, 9 and 11 of the C-terminal domain of CBS. In this alignment the CXXC motifs are at similar positions but on the adjacent α-helices; in CBS the motif is found at a location corresponding to the N-terminal end of α-helix 3 of glutaredoxin, while in glutaredoxin it is located at the N-terminal end of helix 1. Thus, the motifs in the two proteins are in a similar environment with respect to their three-dimensional structure.
It is striking that CBS contains yet another motif that might be involved in redox reactions apart from the heme moiety. Moreover, as shown in Table II, this CPGC motif of CBS is also present in the sequences of the other mammalian CBS enzymes, which also contain the heme group, but is absent in the more distant species that are lacking the heme.
The regulatory domain
Full-length CBS contains a C-terminal regulatory domain of ∼140 residues, including the so-called ‘CBS domain’ (Bateman, 1997) of 53 residues. The C-terminal domain of CBS contains an autoinhibitory region that gets displaced from the active site upon binding of the allosteric activator AdoMet (J.P.Kraus, unpublished results).
The catalytic domain of TD has the same fold as CBS and also contains a C-terminal regulatory domain. A superposition of both structures suggests that the regulatory domains of TD and CBS are located at similar positions. The surface of CBS corresponding to the interface between the catalytic and regulatory domains of TD is largely hydrophobic, which is characteristic for most protein–protein interaction regions.
The fact that truncated CBS forms dimers rather than tetramers or higher order oligomers suggests that the regulatory domain is involved in tetramer formation. This idea is further supported by the fact that two CBS domains can associate e.g. as in the structure of inosine-5′-monophosphate dehydrogenase (Zhang et al., 1999). In TD, however, the regulatory domain is not involved in tetramerization; thus, the mechanism leading to higher order oligomers appears to be different in the two proteins.
Structural characterization of selected CBS mutations
Presently there are >100 CBS mutations known that lead to more or less severe phenotypes in the patient. Five mutations are located close to or at the heme binding site (R58W, H65R, R224H, A226T and R266G/K), eight mutations affect the active site region of CBS and binding of the cofactor PLP (G148R, N228K, T257M, G259S, E302K, G305R, G307S and T353M) and six mutations are located at the dimer interface (P88S, A114V, G116R, I152M, E176K and V180A). Twelve mutated residues are on the surface of the protein and exposed to solvent. Most of these mutations are expected to affect enzyme activity by a general destabilization of the protein structure, which is probably the case for one of the most frequent mutations in patients, I278T. Together with the other frequent mutation, G307S, they represent ∼40% of all mutant alleles. This second mutation (G307S), which confers a severe phenotype, probably influences binding of the second substrate homocysteine, as homocysteine is expected to bind to the protein in this region (Figure 3A). This is in agreement with the finding that patients with this mutation are not B6-responsive, because cofactor binding is not expected to be affected by this mutation.
Also the effects of some other mutations can be nicely explained by the crystal structure of CBS. Even though the mutation T257M is only found in a single patient, its effect on substrate and cofactor binding can easily be envisaged. Thr257 is hydrogen bonded to the phosphate group of the cofactor. The bulkier side chain of methionine would then occlude parts of the substrate binding pocket leading to a reduced enzyme activity. This mutation is not B6-responsive even though Thr257 is directly involved in cofactor binding. This is probably because an additional water molecule could easily replace the lost hydrogen bond to the phosphate group.
Because His65 is one of the two ligands of the heme iron, the mutation H65R would be expected to cause loss of heme binding. It has been shown that heme-free enzyme is still enzymatically active (Bruno et al., 2001); however, this mutation shows a severe phenotype (L.S.Chen, unpublished). This is difficult to explain by its PLP-dependent catalytic activity alone and points to the importance of heme during folding of CBS (Kery et al., 1994; Shan et al., 2001). Furthermore, recent work has shown that CBS subunits aggregate and are found in inclusion bodies when CBS is expressed in the absence of heme (J.P.Kraus, unpublished results).
The mutation G148R is located in the asparagine loop, which undergoes a large conformational change upon ligand binding in OASS (Burkhard et al., 1999). This loop forms an extended hydrogen bonding network to the carboxylate moiety of the ligand in OASS and is very likely to do the same in CBS. The homologous residue in OASS is also a glycine and seems to be important for the flexibility of this loop. It has, in both the open and the closed conformation of OASS, ϕ and ψ angles that are only allowed for a glycine residue. Even though a mutation to arginine would not interfere sterically with the substrate itself, the reduced flexibility of the asparagine loop would most probably reduce ligand binding, because the hydrogen bonding network to the carboxylate moiety of the ligand cannot be properly established. It is interesting to note that among the mutations discussed above, A226T, R224H, I278T and T353M, but not G307S, are functionally suppressed in a truncated CBS missing the last ∼140 residues. In addition, the common I278T mutation is suppressed by any of seven secondary missense mutations in the C-terminal domain of CBS (Shan et al., 2001).
Conclusions and outlook
Taken together, the crystal structure of the truncated form of CBS gives the first insights into structural details of this unique PLP-dependent heme enzyme. Since deficiency of CBS leads to homocystinuria, an inherited metabolic disease, structural insight into its enzymatic mechanism and its regulation are crucial for a better understanding of its physiological role. The identification of the residues involved in heme binding explains some of the previously described mutations leading to homocystinuria. This heme binding motif together with a spatially adjacent oxidoreductase active site motif could explain the regulation of its enzyme activity by redox changes. This intriguing hypothesis will now be investigated in more detail. Further more, CBS is also regulated by AdoMet through the inter action of this allosteric activator with the regulatory domain. To understand better the complicated regulatory mechan ism of its enzymatic activity, we are trying to achieve further structural information of the full-length enzyme.
Materials and methods
Crystallization and data collection
Expression of the truncated CBS protein and subsequent crystallization were carried out as described (Janosik et al., 2001). The X-ray diffraction data of the native crystal were collected at the beam-line BM1A of the SNBL at the ESRF in Grenoble at a temperature of 100 K. Furthermore, a MAD data set at three wavelengths around the absorption edge of the iron ion (λ = 1.74 Å) was collected at the BW7A beam-line (EMBL, DESY Hamburg) (Janosik et al., 2001). The crystals belong to the trigonal space group P31 with unit cell dimensions a = b = 144.46 Å, c = 108.21 Å; α = β = 90°, γ = 120° (Table I). The crystals contain three dimers per asymmetric unit corresponding to a solvent content of 46%.
X-ray structure determination
The structure was solved by combining phase information from MR and MAD of the heme iron. For molecular replacement a polyserine model of OASS was used as search model. Loops that were not conserved in a three-dimensional alignment between the structures of OASS (open and closed form), tryptophan synthase and TD were removed. The calculations were made with the program AMoRe (Navaza, 1994) and yielded a unique single solution in the rotation search, since all dimers have almost the same orientation with respect to each other. The direction of local 2-fold axes was found to be perpendicular to the crystallographic 3-fold axis in agreement with the self-rotation function (Janosik et al., 2001).
The phase information obtained by MR was sufficient to trace those parts of the protein that were homologous to the search model and to identify the positions of the heme irons. However, the N-terminal residues were completely missing in these electron density maps. For this reason the MAD data set was collected. The phases obtained with this method were combined with the MR phases. The quality of the maps resulting from the combined phases was sufficient to be interpreted so that the missing parts of the model could be built.
The electron density maps were averaged over all six monomers A–F and density modified (CCP4, 1994). During refinement, which was performed with the program CNS (Brünger et al., 1998), NCS symmetry restraints were applied on all monomers, using monomer A as reference. The model was built using the program O (Jones et al., 1991).
The final model comprises a total of 2090 amino acids, corresponding to ∼80% of all amino acids, six PLP, six heme and 154 water molecules. The final R factor and free R factors are rather high (25.7 and 29.6%, respectively). This can be explained by the rather poor quality of the crystals (high mosaicity) and by the fact that 20% of the protein residues were found to be disordered. These residues are probably disordered due to the truncation of the enzyme and are mainly located at the N- and C-terminus of the truncated protein. We are now addressing this problem by the design of more appropriate CBS truncation mutants lacking these disordered parts of the protein and hope to obtain better quality crystals using such improved deletion mutants.
Coordinates
The coordinates have been deposited in the Protein Data Bank (accession code 1JBQ).
Acknowledgments
Acknowledgements
We are grateful to J.N.Jansonius and U.Aebi for careful reading of the manuscript and for helpful discussions. This work was supported by grants of the Swiss National Science Foundation, by the M.E.Müller Foundation and the Canton Basel-Stadt, and by an NIH grant to J.P.K.
References
- Alexander F.W., Sandmeier,E., Mehta,P.K. and Christen,P. (1994) Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Regio-specific α, β and γ families. Eur. J. Biochem., 219, 953–960. [DOI] [PubMed] [Google Scholar]
- Bateman A. (1997) The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci., 22, 12–13. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bruno S., Schiaretti,F., Burkhard,P., Kraus,J.P., Janosik,M. and Mozzarelli,A. (2001) Functional properties of the active core of human cystathionine β-synthase crystals. J. Biol. Chem., 276, 16–19. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Rao,G.S., Hohenester,E., Schnackerz,K.D., Cook,P.F. and Jansonius,J.N. (1998) Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium. J. Mol. Biol., 283, 121–133. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Tai,C.H., Ristroph,C.M., Cook,P.F. and Jansonius,J.N. (1999) Ligand binding induces a large conformational change in O-acetylserine sulfhydrylase from Salmonella typhimurium. J. Mol. Biol., 291, 941–953. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Tai,C.H., Jansonius,J.N. and Cook,P.F. (2000) Identification of an allosteric anion-binding site on O-acetylserine sulfhydrylase: structure of the enzyme with chloride bound. J. Mol. Biol., 303, 279–286. [DOI] [PubMed] [Google Scholar]
- Christen P. and Metzler,D.E. (1985) Transaminases. John Wiley & Sons, New York, NY.
- Collaborative Computing Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Gallagher D.T., Gilliland,G.L., Xiao,G., Zondlo,J., Fisher,K.E., Chinchilla,D. and Eisenstein,E. (1998) Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure, 6, 465–475. [DOI] [PubMed] [Google Scholar]
- Hyde C.C., Ahmed,S.A., Padlan,E.A., Miles,E.W. and Davies,D.R. (1988) Three-dimensional structure of the tryptophan synthase α2β2 multienzyme complex from Salmonella typhimurium. J. Biol. Chem., 263, 17857–17871. [PubMed] [Google Scholar]
- Janosik M., Meier,M., Kery,V., Oliveriusova,J., Burkhard,P. and Kraus,J.P. (2001) Crystallization and preliminary X-ray diffraction analysis of the active core of human recombinant cystathionine β-synthase: an enzyme involved in vascular disease. Acta Crystallogr. D, 57, 289–291. [DOI] [PubMed] [Google Scholar]
- Jhee K.H., McPhie,P. and Miles,E.W. (2000) Domain architecture of the heme-independent yeast cystathionine β-synthase provides insights into mechanisms of catalysis and regulation. Biochemistry, 39, 10548–10556. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kery V., Bukovska,G. and Kraus,J.P. (1994) Transsulfuration depends on heme in addition to pyridoxal 5′-phosphate. Cystathionine β-synthase is a heme protein. J. Biol. Chem., 269, 25283–25288. [PubMed] [Google Scholar]
- Kery V., Poneleit,L. and Kraus,J.P. (1998) Trypsin cleavage of human cystathionine β-synthase into an evolutionarily conserved active core: structural and functional consequences. Arch. Biochem. Biophys., 355, 222–232. [DOI] [PubMed] [Google Scholar]
- Kery V., Poneleit,L., Meyer,J.D., Manning,M.C. and Kraus,J.P. (1999) Binding of pyridoxal 5′-phosphate to the heme protein human cystathionine β-synthase. Biochemistry, 38, 2716–2724. [DOI] [PubMed] [Google Scholar]
- Kraus J.P. (1994) Komrower Lecture. Molecular basis of phenotype expression in homocystinuria. J. Inherit. Metab. Dis., 17, 383–390. [DOI] [PubMed] [Google Scholar]
- Kraus J.P. et al. (1999) Cystathionine β-synthase mutations in homocystinuria. Hum. Mutat., 13, 362–375. [DOI] [PubMed] [Google Scholar]
- Maclean K.N., Janosik,M., Oliveriusova,J., Kery,V. and Kraus,J.P. (2000) Transsulfuration in Saccharomyces cerevisiae is not dependent on heme: purification and characterization of recombinant yeast cystathionine β-synthase. J. Inorg. Biochem., 81, 161–171. [DOI] [PubMed] [Google Scholar]
- Mudd S.H., Levy,H.L. and Kraus,J.P. (2001) Disorders of transsulfuration. In Scriver,C.R., Beaudet,A.L., Sly,W.S., Valle,D., Childs,B., Kinzler,K.W. and Vogelstein,B. (eds), The Metabolic and Molecular Bases of Inherited Disease. Vol. 1. McGraw-Hill, New York, NY, pp. 2007–2056.
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Ojha S., Hwang,J., Kabil,O., Penner-Hahn,J.E. and Banerjee,R. (2000) Characterization of the heme in human cystathionine β-synthase by X-ray absorption and electron paramagnetic resonance spectroscopies. Biochemistry, 39, 10542–10547. [DOI] [PubMed] [Google Scholar]
- Omura T., Sadano,H., Hasegawa,T., Yoshida,Y. and Kominami,S. (1984) Hemoprotein H-450 identified as a form of cytochrome P-450 having an endogenous ligand at the 6th coordination position of the heme. J. Biochem. (Tokyo), 96, 1491–1500. [DOI] [PubMed] [Google Scholar]
- Poulos T.L. (1987) Heme Enzyme Crystal Structures. Elsevier Science Publishing Co., Inc., New York, NY.
- Schneider T.R., Gerhardt,E., Lee,M., Liang,P.H., Anderson,K.S. and Schlichting,I. (1998) Loop closure and intersubunit communication in tryptophan synthase. Biochemistry, 37, 5394–5406. [DOI] [PubMed] [Google Scholar]
- Shan X., Dunbrack,R.L.,Jr, Christopher,S.A. and Kruger,W.D. (2001) Mutations in the regulatory domain of cystathionine β-synthase can functionally suppress patient-derived mutations in cis. Hum. Mol. Genet., 10, 635–643. [DOI] [PubMed] [Google Scholar]
- Skovby F., Kraus,J.P. and Rosenberg,L.E. (1984) Biosynthesis of human cystathionine β-synthase in cultured fibroblasts. J. Biol. Chem., 259, 583–587. [PubMed] [Google Scholar]
- Swaroop M., Bradley,K., Ohura,T., Tahara,T., Roper,M.D., Rosenberg,L.E. and Kraus,J.P. (1992) Rat cystathionine β-synthase. Gene organiz ation and alternative splicing. J. Biol. Chem., 267, 11455–11461. [PubMed] [Google Scholar]
- Taoka S., Ohja,S., Shan,X., Kruger,W.D. and Banerjee,R. (1998) Evidence for heme-mediated redox regulation of human cystathionine β-synthase activity. J. Biol. Chem., 273, 25179–25184. [DOI] [PubMed] [Google Scholar]
- Taoka S., Widjaja,L. and Banerjee,R. (1999) Assignment of enzymatic functions to specific regions of the PLP-dependent heme protein cystathionine β-synthase. Biochemistry, 38, 13155–13161. [DOI] [PubMed] [Google Scholar]
- Yao M. et al. (2000) Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus. J. Biol. Chem., 275, 34557–34565. [DOI] [PubMed] [Google Scholar]
- Yap S., Naughten,E.R., Wilcken,B., Wilcken,D.E. and Boers,G.H. (2000) Vascular complications of severe hyperhomocysteinemia in patients with homocystinuria due to cystathionine β-synthase deficiency: effects of homocysteine-lowering therapy. Semin. Thromb. Hemost., 26, 335–340. [DOI] [PubMed] [Google Scholar]
- Zhang R., Evans,G., Rotella,F.J., Westbrook,E.M., Beno,D., Huberman,E., Joachimiak,A. and Collart,F.R. (1999) Characteristics and crystal structure of bacterial inosine-5′-monophosphate dehydrogenase. Biochemistry, 38, 4691–4700. [DOI] [PubMed] [Google Scholar]