Abstract
Analysis of the Haloarcula marismortui large ribosomal subunit has revealed a common RNA structure that we call the kink-turn, or K-turn. The six K-turns in H.marismortui 23S rRNA superimpose with an r.m.s.d. of 1.7 Å. There are two K-turns in the structure of Thermus thermophilus 16S rRNA, and the structures of U4 snRNA and L30e mRNA fragments form K-turns. The structure has a kink in the phosphodiester backbone that causes a sharp turn in the RNA helix. Its asymmetric internal loop is flanked by C–G base pairs on one side and sheared G–A base pairs on the other, with an A-minor interaction between these two helical stems. A derived consensus secondary structure for the K-turn includes 10 consensus nucleotides out of 15, and predicts its presence in the 5′-UTR of L10 mRNA, helix 78 in Escherichia coli 23S rRNA and human RNase MRP. Five K-turns in 23S rRNA interact with nine proteins. While the observed K-turns interact with proteins of unrelated structures in different ways, they interact with L7Ae and two homologous proteins in the same way.
Keywords: K-turn/ribosome/RNA motif/RNA–protein interactions/RNA structure
Introduction
RNA molecules form complex structures containing A-form helices and non-helical regions that are often designated as loops or bulges in secondary structure diagrams (Shen et al., 1995). These regions, however, form definite three-dimensional structures, and several non-A-form RNA structural motifs have been identified, including U-turns, S-turns, A-platforms and tetraloops (Hermann and Patel, 1999; Moore, 1999). The prevalence of these motifs in RNA generally, and their association with specific sequences, suggest that their existence in RNAs of unknown conformation can be inferred on the basis of sequence alone. Therefore, to enable the prediction of RNA structures from their sequences, it is essential to compile a comprehensive library of such structural motifs identifiable in sequences. Furthermore, cellular RNA molecules almost always exist bound to proteins. A large number of RNA motifs that bind proteins are currently known (Burd and Dreyfuss, 1994), and a better understanding of how each interacts with RNA would facilitate attempts to predict the structures of ribonucleoprotein (RNP) complexes.
The recently determined atomic structures of the Haloarcula marismortui large ribosomal subunit (Ban et al., 2000) and the Thermus thermophilus small ribosomal subunit (Schluenzen et al., 2000; Wimberly et al., 2000) have increased the size of the database of known RNA structures by ∼8-fold, providing an unusual opportunity for discovery of new RNA motifs. Here we report an analysis of the H.marismortui 50S ribosomal subunit that has led to the identification of a new RNA motif, which we term the kink-turn, or K-turn. This helix–internal loop–helix motif has an unusual conformation, which includes a kink in its phosphodiester backbone that bends the RNA helix axis by ∼120°. These motifs are important sites for protein recognition in the H.marismortui large subunit. K-turns interact with nine of the large subunit’s 31 proteins: L4, L7Ae, L10, L15e, L19e, L24, L29, L32e and L37Ae. Furthermore, proteins S11 and S17 in the T.thermophilus 30S subunit interact with K-turns, and one of these is positioned to form an intersubunit bridge in the 70S ribosome. In addition, while this manuscript was in preparation, the structure of a U4 snRNA fragment that contains a K-turn bound to human 15.5 kDa spliceosomal protein was published (Vidovic et al., 2000). Although most K-turns bind proteins, they also mediate RNA tertiary structure interactions.
Results
Overview of the motif
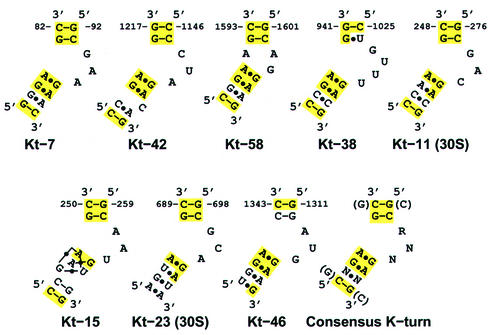
The K-turn is a two-stranded, helix–internal loop–helix motif comprising ∼15 nucleotides (Figures 1 and 2). The first helical stem, the ‘canonical stem’ or ‘C-stem’, ends at the internal loop with two Watson–Crick base pairs, typically C–Gs, while the second helical stem, the ‘non-canonical stem’ or ‘NC-stem’, which follows the internal loop, starts with two non-Watson–Crick base pairs, typically sheared G–A base pairs. The internal loop between the helical stems is always asymmetrical, and usually has three unpaired nucleotides on one strand and none on the other (Figure 1). The 5′-most nucleotide in the long strand of the loop stacks on the C-stem, the second extends to stack on the NC-stem, and the third protrudes into solution. Because of the kink in the phosphodiester backbone in this strand, the orientation of the axes of the C-stem and the NC-stem differ by 120°. The K-turn occurs six times in H.marismortui 23S rRNA, and twice in T.thermophilus 16S rRNA. Each one is designated ‘KT-#’, with KT standing for kink-turn and the number indicating the helix of rRNA in which it is found (Leffers et al., 1987). Although these eight K-turns vary somewhat in sequence, each has essentially the same distinctive three-dimensional form, and a consensus sequence can be derived (Figure 1).
Fig. 1. Secondary structure diagrams of the eight K-turns found in the H.marismortui 50S and T.thermophilus 30S subunit structures and a derived consensus sequence. The names indicate which helix each example of the motif is found in, according to the helix numbering scheme of Leffers et al. (1987). Solid lines represent Watson–Crick pairings between bases, and black dots represent mismatched base pairings. Yellow shading indicates nucleotides that conform to the derived consensus sequence.
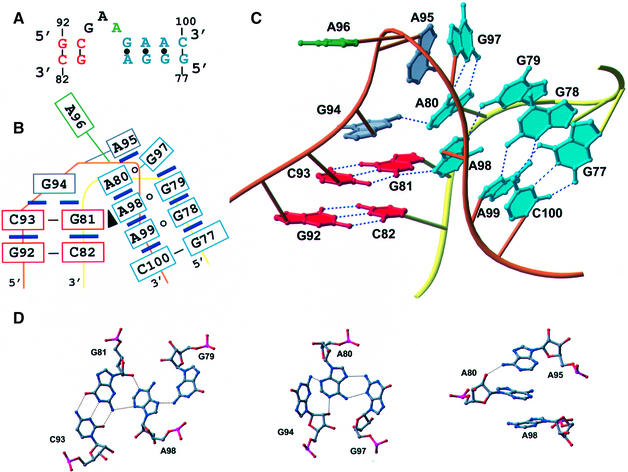
Fig. 2. Structure of KT-7. (A) Secondary structure with residues in the C-stem labeled red and the NC-stem blue. The bulged nucleotide is green. (B) Schematic representation of the relative base-stacking and pairing interactions. A black triangle represents an A-minor interaction. (C) Three-dimensional representation of KT-7 with the phosphate backbone of the kinked strand in orange and the unkinked strand in yellow. Hydrogen bonds are indicated by dashed lines. (D) Atomic details of individual base–base and base–backbone hydrogen bonds.
Structure of KT-7
The K-turn found in helix 7 (KT-7) of H.marismortui 23S rRNA contains the largest number of features seen repeatedly among the ribosomal K-turns, and is therefore the archetypical example of the motif (Figure 2). The C-stem of KT-7 consists of Watson–Crick base pairs C82–G92 and C93–G81. The C-stem is capped by an unpaired guanosine, G94, while the C2′-endo sugar pucker of A95 allows it to stack on the NC-stem. A96 protrudes markedly and makes no interactions with the rest of the motif. Stabilized by a hydrogen bond between the pro(S) phosphate oxygen of A95 and the 2′-OH of A96, the phosphate backbone kinks by ∼120°. G97 in the NC-stem forms a non-planar sheared base pair with A80, with G97 approaching A80 at an angle of ∼50°. A98 stacks beneath A80 and pairs with G79, giving rise to a cross-strand adenosine stack and a type I A-minor interaction (Nissen et al., 2001). An additional G–A pair (G78–A99) continues the coaxial stack before Watson–Crick base pairing is restored with G77–C100.
The close helical packing between the C- and NC-stems, which is seen in all K-turns, appears to be very important for stabilizing the structure of KT-7. As a consequence of the conserved G–A base pair in the NC-stem, A98 is able to make a type I A-minor interaction (Nissen et al., 2001) with C93–G81 in the minor groove of the C-stem (Figure 2C and D). The requirement for this type I A-minor interaction may account for the conservation of C–G in the C-stem and A–G in the NC-stem. Additional interactions between the two helical stems are provided by A80, which close packs with G94, and the sugars of C82 and A99, which also pack together. These hydrophobic interactions are reinforced by a series of sugar–base and sugar–sugar hydrogen bonds. The overall importance of base stacking in K-turns is demonstrated by the observation that all bases except A96 are involved in stacking interactions.
Variations in K-turns
Although no two of the eight K-turns in the ribosome have the same sequence, each has between 7 and 10 matches to the 10 nucleotide consensus of the 15 nucleotide motif, and all possess the same basic three-dimensional structure as KT-7 (Figure 3). The average backbone r.m.s.d. of these eight K-turns is 1.7 Å. Substitutions, insertions and deletions are tolerated by the K-turn motif. For example, the C–G base pair in the C-stem that borders the internal loop can be replaced by either a U–G or G–C with minor consequences. Whereas the U–G in KT-38 allows the ideal type I A-minor interaction, the G–C in KT-46 shifts the relative position of the two stems, creating a non-optimal type I A-minor interaction, and the hydrogen bonding between sugars on either side of the kink is not maintained. The proximity of the two stems in KT-46 is supported instead by hydrophobic sugar packing between C1343 and A1318. Sequence variation is also tolerated in the NC-stem, as seen in KT-23 in 16S rRNA, where a reverse-Hoogsteen A–U pair replaces the second A–G base pair usually seen in the NC-stem. The A of this non-Watson–Crick A–U pair still makes a type I A-minor interaction with the C–G of the C-stem. The NC-stem of KT-23 also contains an A–A mismatch. Nevertheless, its backbone r.m.s.d. from KT-7 is 0.77 Å.
Fig. 3. Backbone representations of nine K-turn structures after superposition on KT-7. The average backbone r.m.s.d. of these structures to KT-7 is 1.7 Å.
The identity of the protruded nucleotide also varies, which is not surprising because it makes no base-specific contacts with the rest of the motif. Although the protruded nucleotide is always present, there is some variability in the length of the internal loop. Predominantly, there are three nucleotides on one strand and none on the other (3 + 0), but examples with four nucleotides in one strand and none on the other (4 + 0), as well as two on one strand and one on the other (2 + 1), are also observed.
Insertions of nucleotides into the helices of K-turns result in some alterations in backbone position. KT-42 contains an unpaired C after the second sheared G–A in the NC-stem. This insertion is accommodated by a slight bulging of the kinked strand in the NC-stem, which results in a further widening of this stem and flattening of its minor groove. KT-38, which has an additional nucleotide in its internal loop, maintains the distinctive K-turn structure by having two protruded nucleotides.
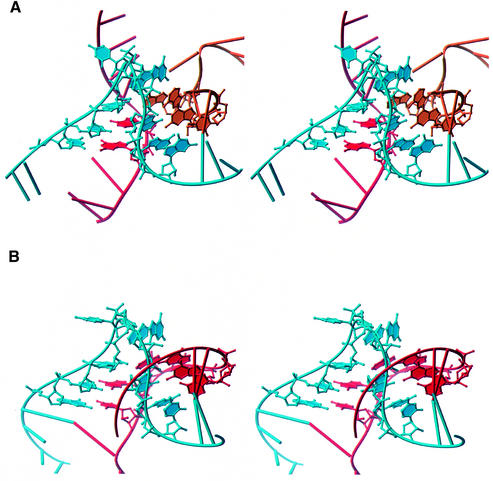
There is one place in the H.marismortui large ribosomal subunit where three RNA strands come together and adopt a structure similar to a K-turn, around nucleotide 46. This three-stranded variant has a similar kinked strand, while the complementary unkinked strand of the C- and NC-stems is donated by different pieces of RNA. The K-turn-like structure at nucleotide 46 has an r.m.s.d. of 1.3 Å relative to KT-7, and is illustrated in Figure 4. The occurrence of three-stranded K-turns suggests that the K-turn may be energetically stable, as a large entropic cost has to be overcome to bring distant strands together.
Fig. 4. (A) Stereo view of the three-stranded K-turn located around nucleotide 46 in H.marismortui 23S rRNA. (B) Stereo image of KT-7 shown in the same relative orientation.
Protein recognition of the K-turn motif
The K-turn is an important RNA recognition motif for the ribosomal proteins in the 50S subunit: five of the six K-turns in H.marismortui 23S rRNA make significant interactions with at least one ribosomal protein, and nine of the 28 observed proteins interact with K-turns (Figure 5). One of these, KT-46, also interacts extensively with two distant regions of the rRNA, demonstrating that K-turns also function to stabilize RNA tertiary structure. There is considerable variation in the way that K-turns interact with proteins in the ribosome. Four principal surface features are recognized: (i) the widened major groove of the C-stem; (ii) the flattened minor groove of the NC-stem; (iii) the sharply kinked sugar–phosphate backbone and the protruded nucleotide; and (iv) the exposed base planes. Recognition of these features involves complementary surfaces on proteins that allow the burial of significant hydrophobic surface area. Interaction with the protruded nucleotide can be accomplished in several different ways (Figure 6). In the following sections, we describe examples of individual protein–K-turn interactions observed in the H.marismortui 50S subunit.
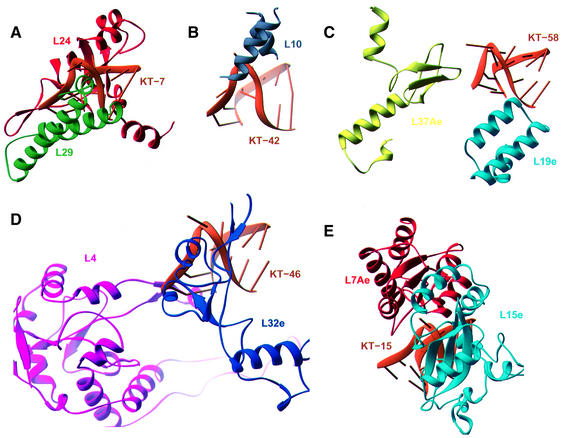
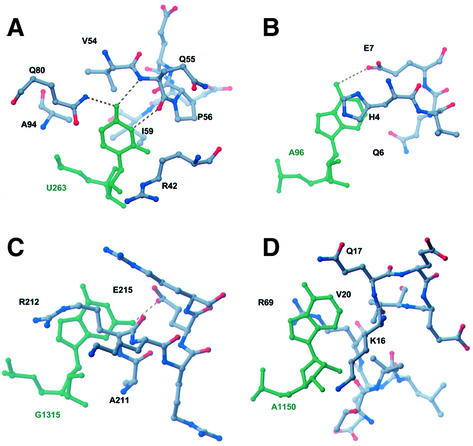
Fig. 5. Ribbon representations indicating the structural diversity of protein–K-turn complexes found in the structure of the H.marismortui 50S subunit. (A) L24 and L29 interact with opposite sides of KT-7. (B) Helical fragments of L10 bind to the bulged nucleotide in KT-42. (C) L19e and L37Ae make limited interactions with KT-58. (D) The β-extension of L4 interacts through the widened major groove of KT-46’s C-stem, while L32e recognizes its kinked backbone and protruded base. (E) L7Ae and L15e surround KT-15 with the majority of interactions involving hydrophobic packing made possible by the severe bend in the RNA helix.
Fig. 6. Recognition of the protruded nucleotide in ribosomal K-turns by specific amino acid contacts. Amino acids from (A) L7Ae and L15e, (B) L29, (C) L32e and (D) L10 interact with their respective K-turns by recognition of the protruded nucleotide (green).
KT-7 interacts extensively with ribosomal proteins L24 and L29 (Figure 5A). L24 has been shown to be an assembly initiator protein (Spillmann and Nierhaus, 1978) and it appears to stabilize domain I of 23S rRNA by bridging three RNA elements, one of which is KT-7. It binds the flattened minor groove and the exposed base planes of KT-7, burying ∼750 Å2 of total surface area (Lee and Richards, 1971). A hydrophobic patch in the protein, consisting of Leu16, His17, His20 and Leu67, packs against the backbone and the smooth N1-C2-N3 edge of A99 in the minor groove of the NC-stem. In addition, the conserved Asp105 recognizes the Watson–Crick face of G97, while Lys107 inserts into a small hydrophobic cavity between the base planes of G97 and G79. The L29 protein contacts the opposite face of KT-7, with His4 stacking on the protruded nucleotide A96 (Figure 6B).
Ribosomal proteins L7Ae and L15e bind to KT-15, which is also located in domain I (Figure 5E). KT-15 is the only RNA element to which L7Ae binds in the 50S subunit. This interaction buries ∼650 Å2 of surface area and it appears to be base specific, with the conserved amino acids Asn34 and Glu35 making hydrogen bonds to G264 and G246 in the NC-stem. The binding site for the protruded nucleotide U263 is located at the interface between L7Ae and L15e, with hydrophobic moieties from both proteins forming the binding pocket (Figures 5E and 6A). These residues include Val54, Gln55, Pro56, Ile59, Gln80 and Ala94 in L7Ae, and Arg42 and Leu46 in L15e. A purine would not be accommodated in this binding pocket without significant structural rearrangements. Specificity for uracil over cytosine is derived from the close approach of the peptide backbone, which allows hydrogen bonding between the O4 of U263 and the backbone amide of Gln55, as well as the side-chain amide of Gln80.
K-turn 42 interacts with the ribosomal protein L10. The location of L10 at the base of the L7/L12 stalk was known from previous electron microscopy localization results (Oakes et al., 1986; Stoeffler and Stoeffler-Meilicke, 1986). In the model of the H.marismortui 50S reported by Ban et al. (2000), L10 was not included because of difficulty in map interpretation in this region. As refinement progressed, map quality improved and small segments of L10 were identified and modeled. Two α-helices corresponding to residues 12–29 and 63–73 have been included in this model. These residues contact KT-42, a conserved RNA structure that had been predicted to be the primary attachment site of L10 (Egebjerg et al., 1990). Arg63 is a conserved basic residue that lies across the major groove of the NC-stem, bridging phosphate oxygens of G1151 and G1210. The universally conserved Thr65 also makes a hydrogen bond to a phosphate oxygen of G1151. The α-helices create a hydrophobic binding pocket that is lined with Leu66, Val20, Arg63 and Lys16, into which A1150, the protruded nucleotide of KT-42, docks (Figures 4B and 5D).
KT-46 most fully illustrates the K-turn’s potential to interact with multiple proteins and RNA simultaneously. The β-extension in L4 contacts Watson–Crick base pairs in the widened major groove of the C-stem (Figure 5D). Residues 208–215 in L32e interact with the protruded nucleotide G1315 and the kinked sugar–phosphate backbone of KT-46 (Figure 6C). A337 and C338 in helix 20 of 23S rRNA pack against the exposed base planes in KT-46. Also, A1318 in the NC-stem binds the minor groove face of an A–U base pair in helix 2 of 23S rRNA, creating a type II A-minor interaction.
Discussion
Of the 28 proteins included in the crystal structure of the H.marismortui large ribosomal subunit, nine interact with K-turn motifs. They do so in various ways that complement the motif’s unique surface features, which include a widened major groove, a flattened minor groove, a severely kinked phosphodiester backbone and a series of exposed bases. These features enable a single K-turn motif to participate in many intermolecular interactions simultaneously, making it well suited to serve as a nucleation site around which large ribonucleoprotein assemblies can be built. Although these nine ribosomal proteins do not share a common structural domain that recognizes K-turns, there is at least one homologous family of RNA-binding domains that is specific for it. Holoarcuta marismortui L7Ae, yeast L30e and the human 15.5 kDa spliceosomal protein all contain an RNA-binding motif whose homologous relationship was identified initially from sequence comparisons by Koonin et al. (1994). The structures of all three of these proteins bound to their respective RNA ligands have recently been obtained (Mao et al., 1999; Ban et al., 2000; Vidovic et al., 2000). These proteins contain identical domain structures that bind K-turn RNA elements in the same fashion. It seems likely, therefore, that other proteins containing this RNA-binding motif will be found to bind to K-turns the same way. A sequence alignment searcher identified many other proteins that should have the same domain structure (Figure 7). It will be interesting to determine whether these homologous proteins, such as hoi-polloi, a Drosophila protein essential for nervous system development (Prokopenko et al., 2000), and Gle1P, an mRNA export factor (Murphy and Wente, 1996), also bind K-turns in the same way.
Fig. 7. Multiple sequence alignment of proteins displaying homology to H.marismortui L7Ae. Red boxes indicate consensus residues. The structures of H.marismortui L7Ae, S.cerevisiae L30e and the human homolog of Snu13p (15.5 kDa spliceosomal protein) have been solved and show striking three-dimensional similarity.
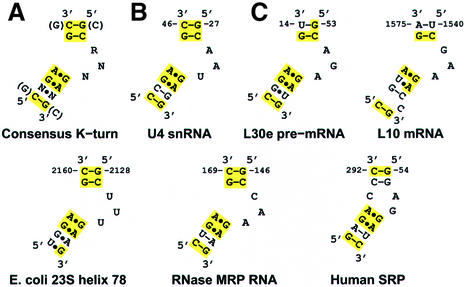
The U4 snRNA and L30e mRNA have secondary structures similar to those of the K-turns in the ribosome, with a 10- and a 9-nucleotide match to the consensus sequence, respectively (Figure 8). The three-dimensional structure reported for U4 snRNA has all the hallmarks of a K-turn: the signature protruded nucleotide, adjacent sheared G–A base pairs, and the hydrogen-bonding patterns typical of the ribosomal K-turns (Vidovic et al., 2000). Furthermore, it has the same structure since its backbone superimposes on the KT-7 backbone with an r.m.s.d. of 1.6 Å. Nevertheless, there are significant differences between the L30e mRNA model proposed from NMR studies (Mao et al., 1999) and the K-turns observed in the ribosome, despite the high degree of similarity at the sequence level. The structures of L30e and L7Ae are very similar, as is their interaction with the RNA. The backbone conformation of L30e mRNA resembles that of the ribosomal K-turns, but the positions of some of the nucleotide bases in L30e mRNA are different. Although the secondary structure of the mRNA can be drawn in the same way as that of the consensus K-turn, including tandem G–A base pairs adjacent to the internal loop (Figure 8), G–G and A–A mismatches are proposed instead. We are led to wonder, however, whether the L30e mRNA has the structural features present in the other nine known K-turn structures rather than the one proposed earlier (Mao et al., 1999).
Fig. 8. Secondary structure diagrams for the consensus and non-ribosomal K-turns. (A) Consensus derived from the eight K-turns found in the ribosome. (B) The secondary structure for the U4 snRNA K-turn adapted from Vidovic et al. (2000). (C) Predicted K-turn motifs in various non-ribosomal RNAs. The secondary structure we propose for L30e pre-mRNA is inconsistent with the NMR structure (Mao et al., 1999).
While the 10 known K-turns have strikingly similar structures, there are many ways in which these motifs interact with proteins. Thus, identification of a K-turn motif in an RNA secondary structure does not predict how or even whether it will be associated with proteins. However, the structurally homologous H.marismortui L7Ae, human 15.5 kDa spliceosome protein and Saccharomyces cerevisae L30e interact in nearly identical ways with K-turn motifs. This suggests that homologous RNA-binding proteins may interact with RNA motifs in similar ways.
Locations of K-turns in the ribosome structures
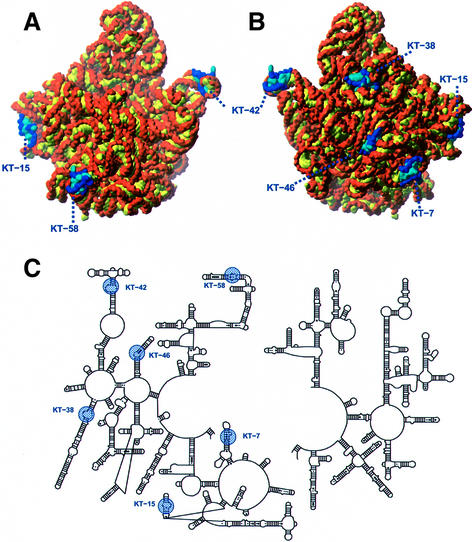
The K-turn motifs in 23S rRNA all appear at or near the surface of the particle in regions that are less well conserved among the kingdoms (Figure 9). KT-7, for example, is located at the attachment site of the ribo some to the membrane-embedded translocon proteins (Beckmann et al., 1997). Although L24 and L29 both make extensive contacts with KT-7, it has the potential for additional interactions via its exposed bases and widened major groove. Given its location, an interaction with some component of the translocon or signal recognition particle is conceivable.
Fig. 9. Location of the K-turns in the H.marismortui 50S structure. (A) The 50S particle shown in the crown view. (B) The back side of the subunit, rotated 180° around the vertical axis from the crown view. K-turns are in blue, with sugars and phosphates in dark blue, and bases in light blue. The remaining RNA has sugars and phosphates in orange, and bases in yellow. (C) The positions of K-turns in the schematic secondary structure diagram of 23S rRNA. K-turns are highlighted in blue.
KT-23 in the 30S subunit is situated in the location identified by Cate et al. (1999) as intersubunit bridge B7. It is oriented with its exposed bases placed so that they could pack against helix 68 in domain IV of 23S rRNA. KT-23 is a highly conserved feature of 16S rRNA and it is likely that it plays a structural role at the 50S/30S subunit interface.
Use of the K-turn consensus sequence to identify K-turns
Although variations are allowed in the K-turn motif, there are distinct preferences from which a consensus sequence can be derived that specifies 10 bases in a 15 nucleotide sequence (Figure 8). The consensus bases are key to maintaining the interactions resulting in the K-turn structures. With the exception of one U–G base pair, the C-stem is always composed of two G–C or C–G base pairs, with a strong preference for a C–G base pair adjacent to the internal loop. The latter is important for an optimal A-minor interaction. The C-stem is usually capped by a purine, while the nucleotide capping the NC-stem and the protruded nucleotide are variable. The NC-stem contains two sheared G–A base pairs, usually followed by another mismatch, before resuming canonical pairing. The A of the second G–A from the loop makes a stabilizing A-minor interaction in the minor groove of the C–G-containing C-stem. Furthermore, the G–A base pairs also play an important role in the formation of the cross-strand A-stack that is found in all K-turns. Thus, since there appears to be an important structural basis for the observed consensus sequence, discovery of this consensus sequence in RNA should be predictive of K-turn motif structures.
Using this consensus sequence we have examined the secondary structures of many non-translated RNAs, and predict that the three-dimensional structures of the 5′-UTR of L10 mRNA from Escherichia coli (Draper, 1989), helix 78 in domain V of E.coli 23S rRNA and human RNase MRP (Schmitt et al., 1993) should all contain K-turn motifs (Figure 8). L10 has been shown to bind its own mRNA (Yates et al., 1981) and we speculate that this binding is analogous to its interaction with KT-42 in 23S rRNA. Helix 78 is disordered in the H.marismortui 50S subunit crystals, and is not included in the model. It is near the putative L1 binding site (Egebjerg et al., 1991) and may interact with L1. RNase MRP is a ribonucleoprotein whose function is pre-rRNA processing (Lindahl and Zengel, 1995–96). A protein component of this complex, p38, has 32% sequence identity to the H.marismortui L7Ae. Therefore, we propose that p38 may bind the K-turn in the RNA component of RNase MRP in a manner that is similar to the binding of L7Ae to KT-15. The putative presence of this RNP structure in RNase MRP as well as box C/D snoRNAs (Watkins et al., 2000), which are also involved in rRNA modification, would be another important connection between the two.
The occurrence of K-turn motifs six times in H.marismortui 23S rRNA, twice in T.thermophilus 16S rRNA, in L30e mRNA and in U4 snRNA, as well as the inferred locations in box C/D snoRNAs, L10 mRNA, helix 78 of E.coli 23S rRNA and RNase MRP, underscore its importance and prevalence. It is very likely that the consensus secondary structure derived here for the K-turn motif will be useful for attempts to predict three-dimensional structures from RNA sequences.
Materials and methods
The full coordinates of the H.marismortui 50S subunit were refined against the 2.4 Å resolution diffraction amplitudes by successive rounds of gradient energy minimization and B-factor refinement using CNS (Brünger et al., 1998). The structures of the ribosomal proteins and rRNA were completely rebuilt into electron density maps calculated using 2Fo – Fc as coefficients and αcalc as phases prior to the modeling of solvent molecules and metal ions using the program O (Jones et al., 1991). In the process of rebuilding the structure, numerous adjustments were made in amino acid register in the proteins, as well as some amino acid sequence changes in L10e, L15e and L37Ae, which have not been sequenced in H.marismortui. The current structure includes 61 617 rRNA atoms, 28 800 protein atoms, 7898 water molecules, 88 monovalent cations, 117 magnesium ions, 22 chloride ions and 5 cadmium ions. The final Rfree is 22.3% using all data to 2.4 Å resolution. A manuscript describing the refinement process more fully and the refined structure in detail is currently in preparation.
Acknowledgments
Acknowledgements
The authors wish to thank Poul Nissen, Nenad Ban, Jeff Hansen and Lara Weinstein for many helpful discussions, and Joe Ippolito and Satwik Kamtekar for critical reading of the manuscript. D.J.K. is an HHMI predoctoral fellow. This work was supported by NIH grants GM22778 to T.A.S. and GM54216 to P.B.M., as well as by a grant from the Agouron Institute. The full coordinates of the refined structure of the H.marismortui 50S subunit have been deposited in the Protein Data Bank with accession number 1JJ2.
References
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Beckmann R., Bubeck,D., Grassucci,R., Penczek,P., Verschoor,A., Blobel,G. and Frank,J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Yusupov,M.M., Yusupova,G.Z., Earnest,T.N. and Noller,H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Draper D.E. (1989) How do proteins recognize specific RNA sites? New clues from autogenously regulated ribosomal proteins. Trends Biochem. Sci., 14, 335–338. [DOI] [PubMed] [Google Scholar]
- Egebjerg J., Douthwaite,S.R., Liljas,A. and Garrett,R.A. (1990) Characterization of the binding sites of protein L11 and the L10.(L12)4 pentameric complex in the GTPase domain of 23 S ribosomal RNA from Escherichia coli. J. Mol. Biol., 213, 275–288. [DOI] [PubMed] [Google Scholar]
- Egebjerg J., Christiansen,J. and Garrett,R.A. (1991) Attachment sites of primary binding proteins L1, L2 and L23 on 23 S ribosomal RNA of Escherichia coli. J. Mol. Biol., 222, 251–264. [DOI] [PubMed] [Google Scholar]
- Hermann T. and Patel,D.J. (1999) Stitching together RNA tertiary architectures. J. Mol. Biol., 294, 829–849. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 46, 110–119. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Bork,P. and Sander,C. (1994) A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res., 22, 2166–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. and Richards,F.M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol., 55, 379–400. [DOI] [PubMed] [Google Scholar]
- Leffers H., Kjems,J., Ostergaard,L., Larsen,N. and Garrett,R.A. (1987) Evolutionary relationships amongst archaebacteria. A comparative study of 23 S ribosomal RNAs of a sulphur-dependent extreme thermophile, an extreme halophile and a thermophilic methanogen. J. Mol. Biol., 195, 43–61. [DOI] [PubMed] [Google Scholar]
- Lindahl L. and Zengel,J.M. (1995–96) RNase MRP and rRNA processing. Mol. Biol. Rep., 22, 69–73. [DOI] [PubMed] [Google Scholar]
- Mao H., White,S.A. and Williamson,J.R. (1999) A novel loop–loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nature Struct. Biol., 6, 1139–1147. [DOI] [PubMed] [Google Scholar]
- Moore P.B. (1999) Structural motifs in RNA. Annu. Rev. Biochem., 68, 287–300. [DOI] [PubMed] [Google Scholar]
- Murphy R. and Wente,S.R. (1996) An RNA-export mediator with an essential nuclear export signal. Nature, 383, 357–360. [DOI] [PubMed] [Google Scholar]
- Nissen P., Ippolito,J.A., Ban,N., Moore,P.B. and Steitz,T.A. (2001) RNA tertiary interactions in the large ribosomal subunit: The A-minor motif Proc. Natl Acad. Sci. USA, 98, 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Henderson,E., Scheinman,A., Clark,M. and Lake,J.A. (1986) Ribosome structure, function, and evolution: Mapping ribosomal RNA, proteins, and functional sites in three dimensions. In Hardesty,B. and Kramer,G. (eds), Structure, Function and Genetics of Ribosomes. Springer-Verlag, New York, NY, pp. 47–67.
- Prokopenko S.N., He,Y., Lu,Y. and Bellen,H.J. (2000) Mutations affecting the development of the peripheral nervous system in Drosophila: a molecular screen for novel proteins. Genetics, 156, 1691–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F. et al. (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- Schmitt M.E., Bennett,J.L., Dairaghi,D.J. and Clayton,D.A. (1993) Secondary structure of RNase MRP RNA as predicted by phylogenetic comparison. FASEB J., 7, 208–213. [DOI] [PubMed] [Google Scholar]
- Shen L.X., Cai,Z. and Tinoco,I.,Jr (1995) RNA structure at high resolution. FASEB J., 9, 1023–1033. [DOI] [PubMed] [Google Scholar]
- Spillmann S. and Nierhaus,K.H. (1978) The ribosomal protein L24 of Escherichia coli is an assembly protein. J. Biol. Chem., 253, 7047–7050. [PubMed] [Google Scholar]
- Stoeffler G. and Stoeffler-Meilicke,M. (1986) Immuno electron microscopy on Escherichia coli ribosomes. In Hardesty,B. and Kramer,G. (eds), Structure, Function and Genetics of Ribosomes. Springer-Verlag, New York, NY, pp. 28–46.
- Vidovic I., Nottrott,S., Hartmuth,K., Luhrmann,R. and Ficner,R. (2000) Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell, 6, 1331–1342. [DOI] [PubMed] [Google Scholar]
- Watkins N.J. et al. (2000) A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren, R.J., Carter,A.P., Vonrheinn,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Yates J.L., Dean,D., Strycharz,W.A. and Nomura,M. (1981) E.coli ribosomal protein L10 inhibits translation of L10 and L7/L12 mRNAs by acting at a single site. Nature, 294, 190–192. [DOI] [PubMed] [Google Scholar]