Abstract
The oligomeric bifunctional HPr kinase/P-Ser-HPr phosphatase (HprK/P) regulates many metabolic functions in Gram-positive bacteria by phosphorylating the phosphocarrier protein HPr at Ser46. We isolated Lactobacillus casei hprK alleles encoding mutant HprK/Ps exhibiting strongly reduced phosphatase, but almost normal kinase activity. Two mutations affected the Walker motif A of HprK/P and four a conserved C-terminal region in contact with the ATP-binding site of an adjacent subunit in the hexamer. Kinase and phosphatase activity appeared to be closely associated and linked to the Walker motif A, but dephosphorylation of seryl-phosphorylated HPr (P-Ser-HPr) is not simply a reversal of the kinase reaction. When the hprKV267F allele was expressed in Bacillus subtilis, the strongly reduced phosphatase activity of the mutant enzyme led to increased amounts of P-Ser-HPr. The hprKV267F mutant was unable to grow on carbohydrates transported by the phosphoenolpyruvate:glycose phosphotransferase system (PTS) and on most non-PTS carbohydrates. Disrupting ccpA relieved the growth defect only on non-PTS sugars, whereas replacing Ser46 in HPr with alanine also restored growth on PTS substrates.
Keywords: bifunctional enzymes/carbohydrate metabolism/HPr/HPr kinase:P-Ser-HPr phosphatase/PEP:glycose phosphotransferase system
Introduction
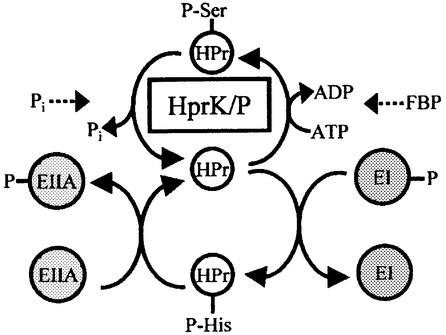
Protein phosphorylation is important for the regulation of most cellular functions in eukaryotes and prokaryotes. One of the best-studied bacterial protein kinases is the enzyme catalyzing the ATP-dependent, metabolite-activated phosphorylation of Ser46 in HPr (Deutscher and Saier, 1983; Deutscher et al., 1986), a phosphocarrier protein of the phosphoenolpyruvate (PEP):glycose phosphotransferase system (PTS) (Postma et al., 1993) (Figure 1). In Bacillus subtilis, seryl-phosphorylated HPr (P-Ser-HPr) controls the expression of ∼10% of the genome (Moreno et al., 2001). The P-Ser-HPr-regulated genes are implicated in many cellular functions such as nitrogen metabolism, carbon catabolite repression (CCR) or activation (Deutscher et al., 1994), stress response, cytochrome c synthesis and regulation of central metabolic pathways such as glycolysis or the tricarboxylic acid (TCA) cycle (Stülke and Hillen, 2000; Deutscher et al., 2001). To carry out these functions, P-Ser-HPr interacts with the catabolite control protein A (CcpA) (Deutscher et al., 1995; Jones et al., 1997), a member of the LacI/GalR repressor family (Henkin et al., 1991). P-Ser-HPr functions as catabolite co-repressor by allowing CcpA to bind to catabolite response elements (cre) (Fujita et al., 1995), operator sites located in front of P-Ser-HPr/CcpA-controlled genes and operons (Weickert and Chambliss, 1990; for reviews see Stülke and Hillen, 2000; Deutscher et al., 2001). P-Ser-HPr has been suggested also to regulate carbohydrate uptake via the PTS, since it is a poor substrate for the PEP-dependent protein kinase enzyme I (EI) (Deutscher et al., 1984, 1994; Ye and Saier, 1996). EI phosphorylates HPr at His15 (Figure 1), which represents the first step of the PTS phosphorylation cascade leading to the phosphorylation of carbohydrates during their transport (Postma et al., 1993). In addition, P-Ser-HPr has been reported to inhibit the activity of several non-PTS permeases (Ye and Saier, 1995; Gauthier et al., 1997; Dossonnet et al., 2000; Viana et al., 2000), thereby preventing the entry of the inducer for the corresponding catabolic operon. This phenomenon was therefore called inducer exclusion in analogy to a similar regulatory function carried out by unphosphorylated EIIAGlc in Gram-negative bacteria (Postma et al., 1993).
Fig. 1. Schematic presentation of ATP- and PEP-dependent HPr phosphorylation. The ATP-dependent phosphorylation of HPr by HprK/P at Ser46 is stimulated by FBP, whereas dephosphorylation of P-Ser-HPr is activated by Pi. Within the PTS phosphorylation cascade, HPr is phosphorylated intermediately by EI at His15 and transfers its phosphoryl group to various EIIAs.
Interestingly, HPr kinase and the first discovered bacterial protein kinase, isocitrate dehydrogenase kinase from Escherichia coli, are bifunctional enzymes also catalyzing the dephosphorylation of their substrates (LaPorte and Koshland, 1982; Kravanja et al., 1999; Dossonnet et al., 2000; Huynh et al., 2000). These two enzymes exhibit no similarity to eukaryotic protein kinases or P-protein phosphatases nor do they exhibit similarity to each other. HPr kinase/P-Ser-HPr phosphatase (HprK/P) contains a Walker motif A similar to nucleotide-binding proteins (Galinier et al., 1998). Experiments aimed at localizing domains containing the Idh kinase or P-Idh phosphatase activity provided no clearcut results, since mutations leading to normal kinase but reduced phosphatase activity affected amino acids in all parts of the protein (Ikeda and La Porte, 1991; Miller et al., 2000). We tried to address the question of how the opposing activities of bifunctional enzymes are organized and controlled by carrying out random mutagenesis with Lactobacillus casei hprK. We were able to isolate hprK alleles encoding enzymes exhibiting almost normal kinase, but strongly reduced phosphatase activity. The mutations affected either the nucleotide-binding site or a C-terminal conserved region, which is in close contact with the Walker motif A of a neighboring subunit in the HprK/P hexamer, and completely disturbed carbon metabolism when introduced into B.subtilis.
Results
hprK mutations leading to reduced expression of a CCR-sensitive reporter gene fusion
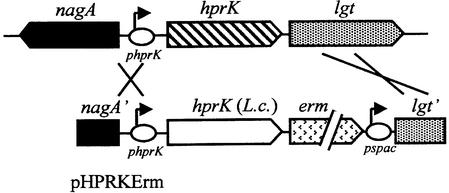
Random mutagenesis with the hprK gene of L.casei was carried out as described in Materials and methods. The resulting hprK alleles were inserted in the integrative plasmid pHPRKErm and subsequently used to replace the hprK gene in the B.subtilis strain QB7144, which carries a xylose-inducible catabolite repression-sensitive ynaJ′– lacZ fusion (Galinier et al., 1999). In the integrants, the L.casei hprK alleles are under control of the B.subtilis hprK promoter (Figure 2). L.casei hprk was used, since it differs sufficiently from B.subtilis hprK to prevent random recombination. Integration of the L.casei wild-type hprK led to the formation of blue colonies on solid CSK medium containing xylose and 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal), whereas addition of glucose repressed ynaJ′–lacZ expression (white colonies). Trans formants synthesizing HprK/P with unbalanced enzyme activities leading to increased amounts of P-Ser-HPr were expected to form white colonies even in the absence of glucose. Out of 4000 clones tested, seven formed pale blue (TG102 and TG103) or white colonies on xylose- and X-Gal-containing medium. Pale blue or white clones were also obtained when QB7144 was transformed with chromosomal DNA isolated from the seven strains. β-galactosidase assays confirmed that the xylose-induced expression of the ynaJ′–lacZ fusion in these strains was 2- to 100-fold lower compared with QB7144 (Table I).
Fig. 2. Replacement of B.subtilis hprK with the L.casei hprK alleles. The integrative plasmid pHPRKErm carries the 3′ part of nagA, the B.subtilis hprK promoter, the L.casei hprK gene, an erythromycin resistance cassette, the spac promoter and the 5′ part of lgt. Double crossover allows replacement of the chromosomal B.subtilis hprK with the L.casei hprK and expression of lgt and the downstream genes from the spac promoter.
Table I. Expression of the ynaJ′–lacZ fusion in QB7144 and its derivatives carrying various hprK alleles.
| Strain | hprK allele | β-galactosidase activitya |
|---|---|---|
| QB7144 | hprK+ B.subtilis | 568 |
| TG100 | hprK+ L.casei | 562 |
| M181A | G58S G270R | 7 |
| TG101 | V267F | 5 |
| TG102 | D34V L152M H223Y | 247 |
| TG103 | N272I | 253 |
| TG104 | G160S | 106 |
| TG200 | G270E | 6 |
| TG201 | E163K | 17 |
| TG203 | V267F and ccpA | 582 |
| TG113 | hprK+ B.subtilis | 563 |
| TG114 | V265F B.subtilis | 35 |
aβ-galactosidase activity was measured in cells grown in CSK medium containing 0.2% xylose. Presented are the mean values of three independent experiments, which exhibited standard deviations of less than ±10%.
The hprK mutations are located in two conserved regions
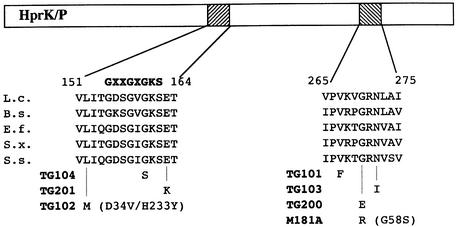
To identify the mutations responsible for the reduced expression from the ynaJ promoter, the L.casei hprK alleles of the integrants exhibiting reduced β-galactosidase activity were amplified by PCR and entirely sequenced. In all cases, at least one mutation could be detected in the hprK genes. These mutations are listed in Table II. The hprK alleles integrated in strains TG102 and M181A contained three and two mutations, respectively, and were not analyzed further. In M181A, the G270R mutation is probably responsible for the reduced ynaJ′–lacZ expression, since G270 was also affected in strain TG200 (G270E mutation). In summary, the hprK mutations leading to reduced expression from the ynaJ promoter are located in two defined regions. One is the presumed ATP-binding site (Walker motif A) extending from position 151 to 164 in the amino acid sequence of L.casei HprK/P, and the other is a C-terminal region (position 265–275), which, similarly to the ATP-binding site, is well conserved in all HprK/Ps (Figure 3).
Table II. Mutations in hprK of L.casei leading to the observed reduced ynaJ′–lacZ expression.
| Mutant | Codon changes | Amino acid replacements |
|---|---|---|
| M181A | GGT→AGT; GGG→AGG | G58S; G270R |
| TG101 | GTT→TTT | V267F |
| TG102 | GAT→GTT; CTG→ATG; CAT→TAT | D34V; L152M; H233Y |
| TG103 | AAC→ATC | N272I |
| TG104 | GGC→AGC | G160S |
| TG200 | GGG→GAG | G270E |
| TG201 | GAA→AAA | E163K |
Fig. 3. Location of the mutations leading to reduced P-Ser-HPr phosphatase activity within two conserved regions of HprK/P. Aligned are the corresponding conserved regions of HprK/P from L.casei (L.c.), B.subtilis (B.s.), Enterococcus faecalis (E.f.), Staphylococcus xylosus (S.x.) and Streptococcus salivarius (S.s.). The consensus sequence of the Walker motif A is presented above the aligned sequences. Numbering refers to L.casei HprK/P.
The V267FLc hprK allele prevents growth on PTS and non-PTS sugars
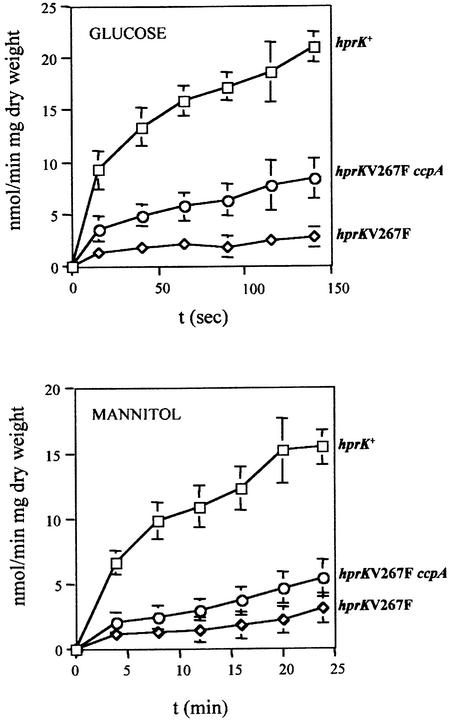
The V267F replacement in L.casei HprK/P was found to exert the strongest repressive effect on the expression of the ynaJ′–lacZ fusion (Table I). The V267F hprK-containing integrant TG101 was therefore chosen to test in a more general way the effect of this mutation on sugar metabolism (Table III). In contrast to a strain carrying wild-type L.casei hprK, the V267FLc hprK mutant was unable to grow on minimal medium supplemented with the PTS substrates glucose, fructose, mannitol or maltose (Reizer et al., 1999) or with the non-PTS substrates gluconate, ribose or glucitol. Among the carbon sources tested, glycerol was the only one that could be utilized by the V267FLc hprK mutant. We also measured the transport activity of the V267FLc hprK mutant with the PTS substrates glucose and mannitol. The strain carrying wild-type L.casei hprK (TG100) exhibited normal uptake of these two carbohydrates, whereas transport of these PTS substrates was almost completely abolished in the V267FLc hprK mutant TG101 (Figure 4). CCR was not responsible for the reduced PTS activity, since introducing a ccpA mutation into strain TG101 providing the double mutant TG203 had little effect on mannitol transport, and glucose uptake was 3- to 4-fold lower than in the hprK+ strain (Figure 4).
Table III. Growth of various B.subtilis strains in C mineral medium supplemented with different carbon sources.
| Strain and relevant genotypea | PTS sugars |
Non-PTS sugars |
||||||
|---|---|---|---|---|---|---|---|---|
| Glucose | Fructose | Mannitol | Maltose | Glycerol | Gluconate | Ribose | Glucitol | |
| TG100 | + | + | + | + | + | + | + | + |
| hprK+ | ||||||||
| TG101 | – | – | – | – | + | – | – | – |
| hprKV267F | ||||||||
| TG202 | + | + | + | + | + | + | + | + |
| ccpA | ||||||||
| TG203 | +/– | – | – | +/– | + | + | + | + |
| ccpA hprKV267F | ||||||||
| TG107 | + | + | + | + | + | + | + | + |
| ptsH1 | ||||||||
| TG108 | + | + | + | + | + | + | – | + |
| ptsH1 hprKV267F | ||||||||
| TG122 | + | + | + | + | + | – | + | + |
| ptsH1 crh hprKV267F | ||||||||
aStrains were grown on solid C minimal medium supplemented with the indicated carbon sources (0.2% w/v) at 37°C and growth was checked after 2 days. (+), growth with a colony size >3 mm; (–), no growth at all or formation of tiny translucent colonies <0.5 mm in diameter; (+/–), intermediate growth with a colony size between 1 and 2 mm. Glutamate (0.25 mM) was added to the minimal medium to allow growth of ccpA mutants (Faires et al., 1999).
Fig. 4. Glucose and mannitol transport studies were carried out with B.subtilis strains containing the L.casei wild-type (squares) or V267F hprK allele (diamonds), or with the V267FLc hprK ccpA double mutant (circles).
To test whether expression of ptsG, which codes for the glucose-specific EIICBA, was lowered in strain TG101, the L.casei wild-type and the V267F hprK alleles were introduced into strain QB7035, which carries a ptsG′–′lacZ fusion (Stülke et al., 1997), providing strains TG109 and TG110, respectively. β-galactosidase activity measured in both strains after growth in the presence and absence of glucose was stimulated similarly by the presence of glucose: from 97 to 1171 U for strain TG109 and from 119 to 1018 U for the V267FLc hprK mutant TG110. These results established that the V267F hprK allele does not affect the synthesis of the glucose-specific EII but rather its transport activity.
To exclude the possibility that the effects caused by the L.casei V267F hprK allele on B.subtilis carbohydrate utilization were due to the heterologous system used in the above experiments, a corresponding B.subtilis hprK mutant was constructed by replacing the wild-type gene in strain QB7144 with the B.subtilis V265F hprK allele. Xylose-induced expression of the ynaJ′–lacZ fusion in the B.subtilis V265F hprK mutant TG114 was strongly repressed compared with the isogenic hprK+ strain TG113 (Table I). In addition, the V265F hprK mutant exhibited growth characteristics on PTS and non-PTS sugars identical to those observed with the V267FLc hprK mutant (data not shown).
The hprK mutations primarily lower the phosphatase activity of HprK/P
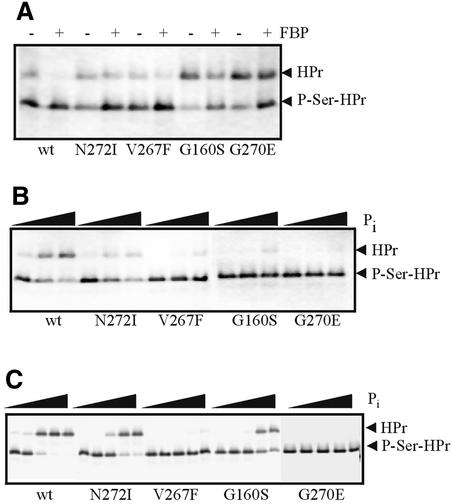
To test whether the hprK mutations would unbalance the two opposing activities of the bifunctional HprK/P, which might be responsible for the observed changes in ynaJ′– lacZ expression and carbohydrate metabolism, four L.casei hprK alleles were inserted into His tag expression vectors and the encoded HprK/Ps were purified and tested for HPr kinase and phosphatase activity. In the V267F, G270E and N272I mutant HprK/Ps, the HPr kinase activity was only slightly reduced, whereas G160S mutant HprK/P exhibited low HPr kinase activity (Figure 5A). However, in all cases, the HPr kinase activity was still stimulated by the presence of 10 mM fructose-1,6-bisphosphate (FBP), as previously observed for wild-type HprK/P (Dossonnet et al., 2000). The phosphatase activity, which is stimulated by inorganic phosphate (Pi) (Dossonnet et al., 2000), was much more strongly affected in the four mutant enzymes. Even in the presence of 2 mM Pi, no phosphatase activity could be detected with V267F and G270E mutant HprK/Ps, and only low activity was observed with the other two mutant enzymes (Figure 5B). Pi has been shown to inhibit the FBP-stimulated formation of P-Ser-HPr by L.casei wild-type HprK/P, probably by stimulating the opposing P-Ser-HPr phosphatase activity (Dossonnet et al., 2000). However, even when the Pi concentration was increased up to 40 mM, no or only slight inhibition of P-Ser-HPr formation in the presence of 10 mM FBP was observed with V267F and G270E HprK/Ps (Figure 5C). The diminished inhibitory effect of Pi on HPr phosphorylation by the different mutant HprK/Ps correlated well with the reduced ynaJ′–lacZ expression observed in strains carrying the corresponding hprK alleles (Table I).
Fig. 5. HPr kinase and P-Ser-HPr phosphatase assays with L.casei wild-type and N272I, V267F, G160S and G270E mutant HprK/Ps. (A) HPr kinase assays with 20 ng of HprK/P in the absence (–) and presence (+) of 10 mM FBP. Samples were incubated for 150 s at 37°C. (B) Phosphatase assays with 40 ng of HprK/P and increasing concentrations of Pi (0, 0.2 and 2 mM). Samples were incubated for 5 min at 37°C. (C) Inhibition of P-Ser-HPr formation in the presence of 200 ng of HprK/P and 10 mM FBP by increasing concentrations of Pi (0, 1, 5, 20 and 40 mM). Samples were incubated for 10 min at 37°C.
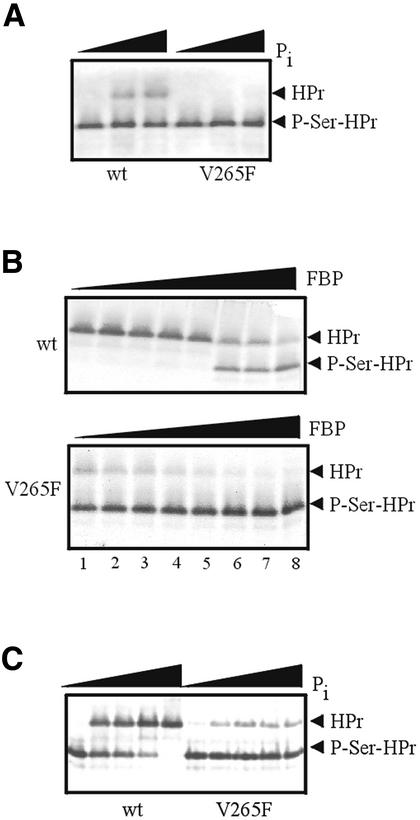
The effects of FBP and Pi on HPr phosphorylation were also determined for B.subtilis wild-type and V265F HprK/P. As observed with the corresponding L.casei mutant enzyme, V265F HprK/P exhibited no detectable phosphatase activity (Figure 6A). As a consequence, in the presence of 5 mM Pi and 2 mM FBP, HPr was almost completely phosphorylated by the mutant enzyme, whereas HPr was completely dephosphorylated when wild-type HprK/P was used under identical conditions (compare lanes 5 of Figure 6B). Even when the concentration of Pi was increased to 40 mM, phosphorylation of HPr by V265F HprK/P was barely diminished (Figure 6C). The chosen concentrations of FBP and Pi, which can vary over a wide range depending on whether the organisms grow on a rapidly metabolizable carbon source or not, lie within the range reported for Gram-positive bacteria (Thompson and Torchia, 1984).
Fig. 6. HPr kinase and P-Ser-HPr phosphatase assays with B.subtilis wild-type and V265F mutant HprK/P. (A) Phosphatase assays with 40 ng of HprK/P and increasing concentrations of Pi (0, 0.2 and 2 mM). Samples were incubated for 5 min at 37°C. (B) Stimulation of the HPr kinase activity (200 ng of HprK/P) by increasing amounts of FBP (0, 0.2, 0.5, 1, 2, 5, 10 and 20 mM) in the presence of 5 mM Pi. Samples were incubated for 10 min at 37°C. (C) Inhibition of P-Ser-HPr formation in the presence of 200 ng of HprK/P and 10 mM FBP by increasing concentrations of Pi (0, 5, 10, 20 and 40 mM). Samples were incubated for 10 min at 37°C.
Is P-Ser-HPr dephosphorylation a reversal of the kinase reaction?
The finding that the mutations leading to low phosphatase activity of HprK/P were located either directly in the Walker motif A or in a conserved C-terminal region in contact with the ATP-binding site of an adjacent subunit (Fieulaine et al., 2001) suggested that kinase and phosphatase activity are closely associated and that the Walker motif A might be the active site for both reactions. One possible explanation was that P-Ser-HPr dephosphorylation was a reversal of the kinase reaction. However, the presence of ADP at up to 5 mM had no stimulatory effect on the phosphatase activity of HprK/P (data not shown). Nevertheless, in the presence of 5 mM ADP, ∼15% of the radioactive products of the phosphohydrolase reaction with [32P]P-Ser-HPr and HprK/P were found to co-migrate with [γ-32P]ATP during thin-layer chromatography (TLC), whereas the major radioactive spot co-migrated with labeled Pi. When 5 mM Pi, which stimulates the phosphatase activity, was also present during [32P]P-Ser-HPr dephosphorylation, no product co-migrating with [γ-32P]ATP could be detected (data not shown).
The V267FLc hprK mutation increases the intracellular amount of P-Ser-HPr
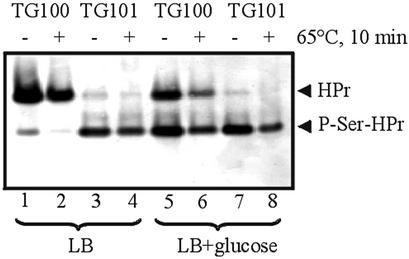
The enhanced formation of P-Ser-HPr observed in vitro with the mutant HprK/Ps suggested that expression of the mutant hprK alleles in B.subtilis might also lead to increased amounts of P-Ser-HPr in vivo. To test this assumption, we carried out western blots with crude extracts prepared from the hprK+ strain TG100 or the V267FLc hprK mutant TG101 grown in Luria Bertani (LB) medium or in glucose-containing LB medium as described in Materials and methods. LB-grown TG100 cells contained ∼85% unphosphorylated HPr (Figure 7, lane 1) and 15% P-His-HPr, which was converted to unphosphorylated HPr when kept for 10 min at 65°C (Figure 7, lane 2) (Waygood et al., 1985). There was almost no P-Ser-HPr present in LB-grown TG100 cells. Growth on LB plus glucose increased the amount of P-Ser-HPr in TG100 to ∼50%. The residual HPr was present in unphosphorylated form and there was little or no P-His-HPr (Figure 7, lanes 5 and 6). In contrast, in LB- and LB plus glucose-grown V267FLc hprK mutant cells, almost all HPr was present as P-Ser-HPr (Figure 7, lanes 3, 4, 7 and 8). No doubly phosphorylated HPr could be detected under the experimental conditions employed, either in TG100 or in the V267FLc hprK mutant strain.
Fig. 7. Western blot with crude extracts prepared from LB- and LB plus glucose-grown TG100 and TG101 cells. Aliquots of the crude extracts were either loaded directly on a non-denaturing poly acrylamide gel or kept at 65°C for 10 min, which causes complete hydrolysis of the P-His bond, but leaves P-Ser-HPr intact. After electrophoresis, which allows separation of the various HPr forms (HPr, P-HPr and doubly phosphorylated HPr), HPr bands were detected with rabbit polyclonal antibodies directed against B.subtilis HPr.
The ptsH1 mutation restores growth on PTS and non-PTS carbohydrates
If the increased amount of P-Ser-HPr in the QB7144 derivatives expressing the L.casei hprK alleles was responsible for the observed permanent CCR and for their failure to grow on PTS and non-PTS carbohydrates, introduction of the ptsH1 mutation (Eisermann et al., 1988), which prevents phosphorylation of HPr at Ser46, was expected to relieve the growth defects and the permanent CCR. Chromosomal DNA of strains TG100 and TG101 was therefore used to transform the ptsH1 strain GM1222 and the isogenic ptsH+ strain GM1221 as described in Materials and methods. In contrast to the V267FLc hprK mutant TG101, the V267FLc hprK ptsH1 double mutant TG108 obtained was capable of growing on the PTS carbohydrates glucose, fructose, mannitol and maltose and on the non-PTS carbohydrates glycerol, gluconate and glucitol, similarly to ptsH+ (TG105) and ptsH1 (TG107) strains carrying L.casei wild-type hprK (Table III). However, growth on the non-PTS carbohydrate ribose could not be restored by the ptsH1 mutation. We therefore inactivated the crh gene, which codes for an HPr-like protein also phosphorylated by HprK/P, and obtained the ptsH1 crh double mutant TG122. Similarly to P-Ser-HPr, P-Ser-Crh can function as catabolite co-repressor/co-activator for certain genes and operons in B.subtilis (Galinier et al., 1997, 1999; Martin-Verstraete et al., 1999; Presecan-Siedel et al., 1999; Zalieckas et al., 1999). The ribose operon seems to be one of these P-Ser-Crh-controlled transcription units, since, in contrast to the ptsH1 mutant TG108, the ptsH1 crh double mutant TG122 was capable of growing on ribose. Strain TG122 had lost the capacity to grow on gluconate as the sole carbon source. Poor growth on gluconate has been observed previously with ptsH1 crh double mutants and is not related to the V267F hprK mutation.
Inactivation of ccpA restores growth only on non-PTS carbohydrates
The finding that compared with an hprK+ strain expression of the ptsG′–′lacZ fusion was not altered in the V267FLc hprK mutant TG110 suggested that permanent CCR might not be the only cause of the impaired growth of the hprK mutants on PTS substrates such as glucose. Additional effects of P-Ser-HPr on PTS transport activity were thought to become detectable in strain TG203 carrying the V267F hprK mutation and a ccpA disruption, which was expected to cause a relief from CCR without altering the elevated amount of P-Ser-HPr. In TG203, β-galactosidase activity was indeed inducible with xylose, similarly to the hprK+ strain TG100 (Table I), confirming that the ccpA mutation caused a relief from permanent CCR. In addition, TG203 had regained the capacity to grow on the non-PTS substrates glucitol, gluconate and ribose. However, it was not able to grow on the PTS substrates fructose and mannitol, and barely grew on the PTS sugars glucose and maltose, suggesting that PTS transporters are also regulated by P-Ser-HPr at the activity level in a CCR-independent manner.
Discussion
The phosphocarrier protein HPr, which participates in PTS-catalyzed sugar transport, is the target of two phosphorylation reactions (Figure 2) and represents the major regulator of carbon metabolism in Gram-positive bacteria. PEP-dependent phosphorylation via EI and P-His-HPr controls the activity of catabolic enzymes, antiterminators and transcriptional activators, whereas P-Ser-HPr plays a role in inducer exclusion and CCR (Stülke and Hillen, 2000; Deutscher et al., 2001). Formation of P-Ser-HPr is regulated by the bifunctional enzyme HprK/P, which phosphorylates HPr during high throughput through glycolysis, whereas it dephosphorylates P-Ser-HPr when the concentration of glycolytic intermediates drops.
Bifunctional enzymes carrying opposing enzyme activities have been detected in eukaryotic cells, including human (Heine-Suner et al., 1998), animal (Hasemann et al., 1996) and plant cells (Draborg et al., 1999), and prokaryotic cells (Johansson and Gest, 1977; LaPorte and Koshland, 1982; Garcia and Rhee, 1983; Zhu et al., 2000). Many of these enzymes catalyze the modification/demodification (phosphorylation, adenylylation, uridylylation) of proteins implicated in the regulation of essential cellular functions. In most cases, effector molecules bind to these bifunctional enzymes and specifically stimulate or inhibit one of the two opposing activities. Although the structure of some bifunctional enzymes has been determined (Hasemann et al., 1996; Thompson et al., 1998) and in a few cases domains could be identified harboring one of the two opposing activities (Jaggi et al., 1997; Lee et al., 1997), knowledge about the regulation of the enzymatic activities of bifunctional enzymes and why there is an advantage in having two opposing activities organized on a single polypeptide chain remains very limited.
We report here the isolation and characterization of mutant HprK/Ps, in which the balance between kinase and phosphatase activities was changed in favor of the kinase activity. Expression of the corresponding hprK alleles led to enhanced conversion of HPr to P-Ser-HPr and to CCR even in the absence of a repressing sugar. Surprisingly, the hprK mutants were unable to grow on most PTS and non-PTS carbohydrates. This inability was clearly related to the increased amounts of P-Ser-HPr or P-Ser-Crh, since introducing ptsH1 or ptsH1 crh mutations restored growth on all carbohydrates. Only glycerol was metabolized normally by the hprK mutants. Although the glpFK operon coding for the glycerol facilitator and glycerol kinase is preceded by a potential cre (Miwa et al., 2000), CcpA-dependent CCR probably plays no or only a minor role for the glpFK operon, and other CcpA-independent CCR mechanisms seem to exist (Deutscher et al., 1994).
Disruption of the ccpA gene, which prevents CCR probably without diminishing the elevated amounts of P-Ser-HPr in the hprK mutants, restored full growth only on non-PTS, but not on PTS carbohydrates. P-Ser-HPr is a poor substrate for the PEP-dependent phosphorylation by EI. If almost all HPr is converted to P-Ser-HPr, as was observed with the V267FLc hprK mutant, PTS substrates are transported so slowly that they no longer support growth. In wild-type cells, formation of P-Ser-HPr in response to increasing amounts of FBP and other glycolytic intermediates is probably used as a feedback mechanism to slow the PEP-dependent phosphorylation of HPr and consequently PTS transport. The slow growth of the V267FLc hprK ccpA double mutant on glucose and maltose (Table III) and the elevated glucose transport activity observed with this strain compared with the V267FLc hprK mutant (Figure 4) are probably due to the synthesis of glucose- or maltose-specific non-PTS transporters such as GlcP, GlcU or YvdHI (Deutscher et al., 2001), which might be subject to CcpA-dependent CCR.
Although the high amounts of P-Ser-HPr almost completely prevented PTS carbohydrate transport in the V267FLc hprK mutant, the remaining low PTS phosphocarrier activity seemed to be sufficient to allow phosphorylation of PTS-controlled transcriptional regulators. Synthesis of EIICBAGlc, which is encoded by ptsG, is regulated by the antiterminator GlcT (Stülke et al., 1997). GlcT was proposed to be inactive when it becomes phosphorylated by P-EIICBAGlc. The finding that in a V267FLc hprK mutant expression of a ptsG′–′lacZ fusion was similarly low as in the isogenic hprK+ strain suggested that GlcT was inactivated normally by P-EIICBAGlc. Although glucose was barely transported by the V267FLc hprK mutant due to the interruption of the PTS phosphorylation cascade at the P-His-HPr level, the presence of glucose supposedly led to rapid dephosphorylation of P-EIICBAGlc and therefore to GlcT activation and the observed ptsG′–′lacZ induction.
The mutations that allowed dissection of the kinase and phosphatase activities of HprK/P affected two conserved regions: the Walker motif A and a C-terminal region extending from position 265 to 275 (Figure 3), which, according to the structure of L.casei HprK/P, is in contact with the Walker motif A of a neighboring subunit in the hexamer (Fieulaine et al., 2001). All mutations strongly diminished the P-Ser-HPr phosphatase activity. The HPr kinase activity was much less affected, even when the mutations were located in the ATP-binding site. These results suggest that the opposing activities of HprK/P are tightly associated and that the Walker motif A is implicated in both reactions. This assumption was supported further by the finding that Pi, the activator of the phosphatase reaction, binds to the Walker motif A (Fieulaine et al., 2001). However, P-Ser-HPr dephosphorylation is not simply a reversion of the kinase reaction, since the presence of 5 mM ADP had no stimulatory effect on the phosphatase activity. In addition, the major radioactive product obtained when [32P]P-Ser-HPr dephosphorylation was carried out in the presence of 5 mM ADP co-migrated with Pi during TLC and only a small amount of ATP was formed. If 5 mM Pi was also present, it completely prevented the formation of ATP. Since the intracellular concentration of Pi is in the range of 3–50 mM depending on the metabolic state of the bacteria (Thompson and Torchia, 1984), formation of ATP from ADP and P-Ser-HPr is probably not of physiological significance. Pi binds to the position in the Walker motif A of HprK/P normally occupied by the β-phosphate of the nucleotide (Fieulaine et al., 2001). Competition between Pi and ATP for the common binding site is probably responsible for the inhibition of the kinase activity of HprK/P by Pi. The most striking characteristic of the mutant HprK/Ps was the decrease or even loss of the inhibitory effect of Pi (Figures 5C and 6C). These results suggested that some of the hprK mutations described in this study cause structural changes, reducing the affinity for Pi without lowering the affinity for ATP too much. Interestingly, amino acids G160 and E163, which are affected in the hprK mutants TG104 and TG201, are part of the Pi-binding site (Fieulaine et al., 2001). Binding of FBP might lead to structural changes similar to those caused by the hprK mutations, since, in the presence of FBP, higher amounts of Pi are necessary to inhibit P-Ser-HPr formation. The question therefore arises of whether the conserved C-terminal region affected by four mutations might be part of the FBP-binding site (Jault et al., 2000).
With carbohydrates being the major source of carbon and energy, the almost complete inability of the hprK mutants to utilize carbohydrates drastically limits their growth capacities in their natural environment. Similarly to the V267F hprK mutation leading to the accumulation of P-Ser-HPr, the specific inactivation of the P-Idh phosphatase activity of the second well-characterized bacterial bifunctional protein kinase/P-protein phosphatase, IdhK/P of E.coli (LaPorte and Koshland, 1982), led to the accumulation of phosphorylated, barely active Idh and to growth arrest probably due to the loss of TCA cycle activity when >85% of Idh was phosphorylated (Ikeda and La Porte, 1991). The severe growth defects observed with mutants, in which the target proteins of these two bifunctional enzymes were present primarily in their modified form, might be one reason why organisms developed proteins in which modifying/demodifying enzyme activities are organized within a single polypeptide chain. If these enzyme activities were carried by two proteins, frameshift or any other mutations inactivating the demodifying enzyme would lead to the accumulation of the modified target protein and hence to severe growth defects. However, if the two opposing activities are tightly associated on one polypeptide chain, as observed with HprK/P, then most mutations will inactivate both enzyme activities. In agreement with this concept, mutants carrying a disrupted hprK gene had lost both activities and were able to grow on most carbohydrates (Galinier et al., 1998; Reizer et al., 1998; Dossonnet et al., 2000). To knock out only one of the two enzyme activities will require very specific mutations, such as those isolated during this study, which will occur with low frequency. Although this concept needs further experimental support, lowering the probability of lethal or growth-inhibiting mutations might be one advantage of organizing opposing enzyme activities on a single polypeptide chain.
Materials and methods
Bacterial strains and growth conditions
The B.subtilis strains used in this study are listed in Table IV. Cells were grown at 37°C under agitation in LB medium, C mineral medium or CSK medium (Martin et al., 1989) supplemented with different carbon sources at 0.2% (w/v). To allow a better growth of the ccpA mutant strains, 0.25 mM glutamate was added to the minimal medium (Faires et al., 1999). When required, erythromycin and chloramphenicol were used at concentrations of 5 µg/ml. Spectinomycin was used at 100 µg/ml and neomycin at 10 µg/ml. X-Gal was added at a final concentration of 20 µg/ml. β-galactosidase activity was determined in exponentially growing cells as previously described (Miller, 1972).
Table IV. Bacillus subtilis strains used in this study.
| Strain | Genotype | Reference or construction |
|---|---|---|
| GM1090 | trpC2 pheA1 ccpA::spc | M.Steinmetz |
| GM1221 | trpC2 pheA1 ΔlacA amyE::(gntRK′-lacZ) ptsH+ cat | Deutscher et al. (1994) |
| GM1222 | trpC2 pheA1 ΔlacA amyE::(gntRK′-lacZ) ptsH1 cat | Deutscher et al. (1994) |
| M181A | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK L.c. G58S G270R-erm-pspac)a | pHPRErm(L.c. G58S G270R)b→QB7144 |
| QB7035 | trpC2 amyE::(ptsG′-′lacZ cat) | Stülke et al. (1997) |
| QB7096 | trpC2 crh::aphA3 | Presecan-Siedel et al. (1999) |
| QB7144 | trpC2 amyE::(ynaJ′-lacZ cat) | Galinier et al. (1999) |
| TG100 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK+ L.c.-erm-pspac) | pHPRKErm(L.c.wt)→QB7144 |
| TG101 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK L.c. V267F-erm-pspac) | pHPRErm(L.c.V267F)→QB7144 |
| TG102 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK L.c.D34V L152M H233Y-erm-psapc) | pHPRKErm(L.c. D34V L152M H233Y)→QB7144 |
| TG103 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK L.c. N272I-erm-pspac) | pHPRKErm(L.c.N272I)→QB7144 |
| TG104 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK L.c. G160S-erm-pspac) | pHPRKErm(L.c.G160S)→QB7144 |
| TG105 | trpC2 pheA1ΔlacA amyE::(gntRK′-lacZ) hprK::(hprK+ L.c.-erm-pspac) | TG100→GM1221 |
| TG106 | trpC2 pheA1ΔlacA amyE::(gntRK′-lacZ) hprK::(hprK L.c.V267F-erm-pspac) | TG101→GM1221 |
| TG107 | trpC2 pheA1ΔlacA amyE::(gntRK′-lacZ) ptsH1 hprK::(hprK+ L.c.-erm-pspac) | TG100→GM1222 |
| TG108 | trpC2 pheA1ΔlacA amyE::(gntRK′-lacZ) ptsH1 hprK::(hprK L.c.V267F-erm-pspac) | TG101→GM1222 |
| TG109 | trpC2 amyE::(ptsG′-′lacZ cat) hprK::(hprK+ L.c.-erm-pspac) | TG100→QB7035 |
| TG110 | trpC2 amyE::(ptsG′-′lacZ cat) hprK::(hprK L.c.V267F-erm-pspac) | TG101→QB7035 |
| TG113 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK+ B.s.-erm-pspac) | pHPRKErm(B.s. wt)→QB7144 |
| TG114 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK B.s.V265F-erm-pspac) | pHPRKErm(B.s.V265F)→QB7144 |
| TG121 | trpC2 pheA1ΔlacA amyE::(gntRK′-lacZ) ptsH1 crh::aphA3 | QB7096→GM1222 |
| TG122 | trpC2 pheA1ΔlacA amyE::(gntRK′-lacZ) ptsH1 crh::aphA3 hprK:: (hprK L.c.V267F-erm-pspac) | TG101→TG121 |
| TG200 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK L.c. G270E-erm-pspac) | PHPRKErm(L.c.G270E)→QB7144 |
| TG201 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK L.c. E163K-erm-pspac) | PHPRKErm(L.c.E163K)→QB7144 |
| TG202 | trpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK+ L.c.-erm-pspac) ccpA::spc | GM1090→TG100 |
| TG203 | TrpC2 amyE::(ynaJ′-lacZ cat) hprK::(hprK L.c. V267F-erm-pspac) ccpA::spc | GM1090→TG101 |
aL.c. refers to hprK from L.casei, and B.s. to hprK from B.subtilis.
bArrows indicate transformation by plasmid or chromosomal DNA.
Plasmid construction
The hprK gene from L.casei was amplified by PCR using genomic DNA and oligo1 and 2 (Table V). The 5′ end of the B.subtilis nagA gene, which is located upstream of hprK, was amplified by PCR using oligo3 and 4 as primers and B.subtilis 168 chromosomal DNA as template. The products were digested with KpnI–NcoI and SacI–NcoI, respectively, and cloned into pUC19 digested with KpnI and SacI to provide pHPRK. The erm gene and the spac promoter from plasmid pMUTIN (Vagner et al., 1998) were amplified by PCR using oligo5 and 6 (Table V). The resulting 1.6 kb fragment was digested with KpnI–EcoRI and ligated in one step to pUC19 digested with KpnI–HindIII and to the HindIII–EcoRI-digested 400 bp PCR fragment (amplified with oligo7 and 8) containing the 5′ part of the B.subtilis lgt gene providing plasmid pErm. The 1.4 kb SacI–KpnI fragment of pHPRK was ligated into SacI–KpnI-digested pErm. The resulting pHPRKErm contained the hprK gene of L.casei, which is under the control of the B.subtilis hprK transcription and translation initiation signals, an erythromycin resistance marker and the 5′ end of the lgt gene under control of the spac promoter (Figure 2). Pfu DNA polymerase (Promega) was used for all PCRs, and the correct sequence of all PCR products was confirmed by DNA sequencing. Escherichia coli NM522 (Stratagene) was used as a host for all plasmid constructions.
Table V. Oligonucleotides used in this study.
| oligo1 ACTCCATGGCAGACAGCGTG | Construction of |
| oligo2 TACGGTACCAATGAACTTCCAG | pHPRKErm |
| oligo3 GTCGAGCTCGGAAAAGCTAGC | |
| oligo4 TTGCCATGGTATGTTCCTCC | |
| oligo5 GAATGGTACCTCTAGCACAAA | |
| oligo6 CGGGAATTCAAGCTTAATTG | |
| oligo7 GCGGAATTCATTGAAGACGG | |
| oligo8 CGCAAGCTTCCAGAACGAAA | |
| HPrKBam GTGGGATCCATGGCAGACAGC | Cloning of L.casei hprK in pQE30 |
| HPrKBs1 CATACCATGGCAAAGGTTCG | Cloning of B.subtilis hprK in pHPRKErm |
| HPrKBs2 GAAGGATCCATGGCAAAGGTTCG | Cloning of B.subtilis |
| HPrKBs3 AACGGTACCTCCTATTCTTCTTG | hprK in pQE30 |
| V265F2 GACGATTCCCTTCCGCCCAGGCC | Mutagenesis of B.subtilis hprK |
Random mutagenesis of L.casei hprK
PCR-based random mutagenesis was used to introduce mutations into L.casei hprK. The hprK gene was amplified by PCR in two separate reactions using oligo1 and 2. The first reaction was performed with 200 µM dGTP, dTTP, dCTP and 20 µM dATP, and the second with 200 µM dATP, dTTP, dCTP and 20 µM dGTP. For both reactions, 30 cycles comprised of the following steps were carried out: 30 s at 94°C, 30 s at 50°C and 1.5 min of extension at 72°C. The reactions were performed in a total volume of 50 µl containing 2.5 U of Taq DNA polymerase and 20 pmol of each oligonucleotide. The 1 kb PCR products from both reactions were isolated, mixed, digested with NcoI and KpnI and used to replace the wild-type hprK gene with the different mutant alleles in pHPRKErm cut with the same enzymes. Strain NM522 was transformed with the pHPRKErm derivatives carrying the L.casei hprK alleles, and ∼10 000 E.coli clones were scraped off from the Petri dishes, pooled and grown for 2 h at 37°C in 0.5 l of LB medium containing 100 µg/ml ampicillin. Plasmid DNA was isolated from these cells and used to transform B.subtilis strain QB7144, which carries a xylose-inducible ynaJ′–lacZ fusion and therefore forms blue colonies when grown on solid LB medium containing xylose and X-Gal. The ynaJ gene is part of the xyn operon and supposedly encodes a β-xyloside-specific permease (Galinier et al., 1999). The pHPRKErm derivatives were integrated in the B.subtilis chromosome, thereby replacing B.subtilis hprK with the L.casei hprK alleles. In the integrants, L.casei hprK is expressed from the B.subtilis hprK promoter and lgt from the spac promoter (Figure 2). Integrants carrying wild-type L.casei hprK formed blue colonies when grown on solid LB medium containing xylose and X-Gal.
Construction of ccpA, ptsH1, crh and hprK mutants
Strains carrying the L.casei wild-type or V267FLc hprK allele were used to construct ccpA and ptsH1 mutants and a ptsH1 crh double mutant. To construct ccpA mutants, chromosomal DNA was isolated from strain GM1090 carrying a spectinomycin-disrupted ccpA gene. The isolated DNA was used to transform strains TG100 and TG101, and spectinomycin-resistant clones were selected, providing strains TG202 and TG203.
For the construction of the ptsH1 mutants carrying the L.casei wild-type or V267F hprK allele, chromosomal DNA was isolated from strains TG100 and TG101 and used to transform the ptsH1 strain GM1222 and its isogenic ptsH+ strain GM1221 (Deutscher et al., 1994). Erythromycin-resistant transformants were isolated, providing strains TG105–108. The ptsH1 crh double mutant TG121 was obtained by transforming GM1222 with chromosomal DNA of QB7096, in which the crh gene is disrupted with a kanamycin resistance cassette (Presecan-Siedel et al., 1999). DNA of strain TG101 was used to introduce the V267FLc hprK allele into TG121, providing strain TG122.
To obtain the B.subtilis hprK allele expressing V265F mutant HprK/P, the 3′ part of hprK was amplified by using oligonucleotides V265F2 (mutagenic) and HPrKBs3 (Table V). The resulting 160 bp DNA fragment was used as primer for PCR amplification in combination with the oligonucleotide HPrKBs1. The obtained 1 kb PCR product was digested with NcoI–KpnI and cloned in NcoI–KpnI-digested pHPRKErm, allowing the replacement of L.casei hprK with hprKV265FBs. Bacillus subtilis wild-type hprK was amplified by using oligonucleotides HPrKBs1 and HPrKBs3, and the PCR product was used to replace the L.casei hprK gene in pHPRKErm with B.subtilis hprK by following the procedure described for the V265FBs hprK allele. The B.subtilis hprK alleles were subsequently integrated in the chromosome of QB7144 by double recombination, providing strains TG113 (hprK+) and TG114 (V265F hprK).
Protein purification, and HPr kinase and P-Ser-HPr phosphatase assays
The different L.casei hprK alleles were amplified by PCR using oligo2 and HPrKBam as primers (Table V) (Dossonnet et al., 2000), cut with BamHI and KpnI and cloned into the expression vector pQE30 cut with the same enzymes. The B.subtilis V265F mutant hprK was amplified using oligo3 and HPrKBS2 as primers and also inserted into pQE30 cut with BamHI and KpnI. The E.coli M15[pREP4] strains transformed with the expression vector carrying the different hprK alleles were cultivated in 1 l of LB containing 100 mg of ampicillin and 25 mg of kanamycin. Expression of the different L.casei hprK alleles was induced with isopropyl-β-d-galactopyranoside (IPTG), and purification of the various mutant HprK/Ps on Ni-NTA columns (Qiagen) was carried out as previously described for the wild-type protein (Dossonnet et al., 2000). HprK/P-containing fractions were pooled, dialyzed at 4°C against 50 mM Tris–HCl pH 7, containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and stored in aliquots at –80°C. Bacillus subtilis HPr(His6) was purified as previously described (Galinier et al., 1997). P-Ser-HPr(His6) was prepared by ATP-dependent phosphorylation with purified B.subtilis HprK/P (Galinier et al., 1998) and was separated from unphosphorylated HPr (Deutscher et al., 1986).
The HPr(Ser) kinase and P-Ser-HPr phosphatase assays were performed in 20 µl of 50 mM Tris–HCl pH 7.4, containing 5 mM MgCl2, 1 mM ATP (only for the kinase reaction), 20–200 ng of HprK/P(His6) and varying amounts of FBP or potassium phosphate. The reactions were started by adding 2.5 µg of HPr(His6) (kinase assay) or P-Ser-HPr(His6) (phosphatase assay) (Dossonnet et al., 2000). After incubation at 37°C, the reactions were stopped by heating the assay mixtures for 5 min at 75°C. The different forms of HPr were separated by electrophoresis on non-denaturing 12.5% polyacrylamide gels that were stained with Coomassie Blue.
[32P]P-Ser-HPr dephosphorylation
Bacillus subtilis [32P]P-Ser-HPr(His6) was synthesized with [γ-32P]ATP and HprK/P (Galinier et al., 1998). The reaction products were loaded onto an Ni-NTA column and [32P]P-Ser-HPr(His6) was eluted with 300 mM imidazole and subsequently desalted on a PD10 column (Pharmacia) run with 20 mM ammonium bicarbonate. After lyophilizing, aliquots of the [32P]P-Ser-HPr(His6) preparation were used for dephosphorylation assays carried out for 30 min at 37°C in 20 µl of 50 mM Tris–HCl pH 7.4, containing 0.2 µg of HprK/P and either 5 mM ADP, 5 mM Pi or both compounds. The reaction was stopped by heating the samples for 5 min at 70°C. TLC was performed with 4 µl aliquots on Polygram CEL 300 PEI sheets (Macherey-Nagel) using 0.3 M KH2PO4 or K2HPO4 as solvent. [γ-32P]ATP and [33P]Pi were used as standards. Electrophoresis of 5 µl aliquots of the phosphatase assay mixtures on 15% polyacrylamide–1% SDS gels was used to determine to what extent [32P]P-Ser-HPr(His6) was dephosphorylated, which was always >95%, except in the control experiment where no HprK/P was added.
Western blots
Western blots were carried out with crude extracts prepared from strains TG100 and the V267FLc hprK mutant TG101 by following the method described by Gauthier et al. (1997). Cells were grown in LB medium or LB medium containing 0.5% glucose to an OD600 of between 0.25 and 0.35 before preparing crude extracts, which were separated on non-denaturing 10% polyacrylamide gels. On non-denaturing gels, HPr and phosphorylated HPr migrated to different positions and could be detected subsequently with polyclonal antibodies directed against His-tagged B.subtilis HPr.
Sugar transport studies
Bacillus subtilis strains were grown to an OD600 of between 0.6 and 0.7 in 25 ml of LB medium containing 0.2% of either glucose or mannitol to induce the corresponding carbohydrate transport system. Cells were collected by centrifugation, washed twice with 50 mM sodium phosphate buffer pH 7, containing 10 mM MgCl2 and resuspended in 500 µl of 50 mM Tris-maleate buffer pH 7.4, containing 5 mM MgCl2. Transport assays were performed at 37°C with 1 ml of cell suspensions (OD600 = 4) prepared with the latter buffer. At time 0, 14C-labeled sugars (0.5 mCi/mmol) were added to a final concentration of 0.5 mM. Aliquots of 100 µl were withdrawn at different time intervals, rapidly filtered through nitrocellulose filters (0.45 µm pore size) and washed twice with 5 ml of ice-cold transport buffer. Radioactivity retained in the filters was determined by liquid scintillation counting.
Acknowledgments
Acknowledgements
This research was supported by the CNRS, the INRA, the INA-PG, the Université Paris 7 and the MENRT. V.M. was a recipient of an FPU fellowship from the Ministerio de Educación y Cultura of Spain.
References
- Deutscher J. and Saier,M.H.,Jr (1983) ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc. Natl Acad. Sci. USA, 80, 6790–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J., Kessler,U., Alpert,C.A. and Hengstenberg,W. (1984) The bacterial phosphoenolpyruvate-dependent phosphotransferase system: P-Ser-HPr and its possible regulatory function. Biochemistry, 23, 4455–4460. [DOI] [PubMed] [Google Scholar]
- Deutscher J., Pevec,B., Beyreuther,K., Kiltz,H.-H. and Hengstenberg,W. (1986) Streptococcal phosphoenolpyruvate–sugar phosphotransferase system: amino acid sequence and site of ATP-dependent phosphorylation of HPr. Biochemistry, 25, 6543–6551. [DOI] [PubMed] [Google Scholar]
- Deutscher J., Reizer,J., Fischer,C., Galinier,A., Saier,M.H.,Jr and Steinmetz,M. (1994) Loss of protein kinase-catalyzed phosphoryl ation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J. Bacteriol., 176, 3336–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J., Küster,E., Bergstedt,U., Charrier,V. and Hillen,W. (1995) Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol. Microbiol., 15, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Deutscher J., Galinier,A. and Martin-Verstraete,I. (2001) Carbohydrate uptake and metabolism. In Sonenschein,A.L., Hoch,J.A. and Losick,R. (eds), Bacillus subtilis and its Closest Relatives: From Genes to Cells. American Society for Microbiology, Washington, DC, pp. 137–158.
- Dossonnet V., Monedero,V., Zagorec,M., Galinier,A., Pérez-Martínez,G. and Deutscher,J. (2000) Phosphorylation of HPr by the bifunctional HPr kinase/P-Ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion, but not inducer expulsion. J. Bacteriol., 182, 2582–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draborg H., Villadsen,D. and Nielsen,T.H. (1999) Cloning, characteriz ation and expression of a bifunctional fructose-6-phosphate, 2-kinase/fructose-2,6-bisphosphatase from potato. Plant Mol. Biol., 39, 709–720. [DOI] [PubMed] [Google Scholar]
- Eisermann R., Deutscher,J., Gonzy-Tréboul,G. and Hengstenberg,W. (1988) Site-directed mutagenesis with the ptsH gene of Bacillus subtilis. Isolation and characterization of heat-stable proteins altered at the ATP-dependent regulatory phosphorylation site. J. Biol. Chem., 263, 17050–17054. [PubMed] [Google Scholar]
- Faires N., Tobisch,S., Bachem,S., Martin-Verstraete,I., Hecker,M. and Stülke,J. (1999) The catabolite control protein CcpA controls ammonium assimilation in Bacillus subtilis. J. Mol. Microbiol. Biotechnol., 1, 141–148. [PubMed] [Google Scholar]
- Fieulaine S., Morera,S., Poncet,S., Monedero,V., Guegen-Chaignon,V., Galinier,A., Janin,J., Deutscher,J. and Nessler,S. (2001) X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain. EMBO J., 20, 3917–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Miwa,Y., Galinier,A. and Deutscher,J. (1995) Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol. Microbiol., 17, 953–960. [DOI] [PubMed] [Google Scholar]
- Galinier A., Haiech,J., Kilhoffer,M.-C., Jaquinod,M., Stülke,J., Deutscher,J. and Martin-Verstraete,I. (1997) The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl Acad. Sci. USA, 94, 8439–8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinier A., Kravanja,M., Engelmann,R., Hengstenberg,W., Kilhoffer,M.-C., Deutscher,J. and Haiech,J. (1998) New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl Acad. Sci. USA, 95, 1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinier A., Deutscher,J. and Martin-Verstraete,I. (1999) Phosphoryl ation of either Crh or HPr mediates binding of CcpA to the Bacillus subtilis xyn cre and catabolite repression of the xyn operon. J. Mol. Biol., 286, 307–314. [DOI] [PubMed] [Google Scholar]
- Garcia E. and Rhee,S.G. (1983) Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII uridylyltrans ferase and uridylyl-removing enzyme. J. Biol. Chem., 258, 2246–2253. [PubMed] [Google Scholar]
- Gauthier M., Brochu,D., Eltis,L.D., Thomas,S. and Vadeboncoeur,C. (1997) Replacement of isoleucine-47 by threonine in the HPr protein of Streptococcus salivarius abrogates the preferential metabolism of glucose and fructose over lactose and melibiose but does not prevent the phosphorylation of HPr on serine-46. Mol. Microbiol., 25, 695–705. [DOI] [PubMed] [Google Scholar]
- Hasemann C.A., Istvan,E.S., Uyeda,K. and Deisenhofer,J. (1996) The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies. Structure, 4, 1017–1029. [DOI] [PubMed] [Google Scholar]
- Heine-Suner D., Diaz-Guillen,M.A., Lange,A.J. and Rodriguez de Cordoba,S. (1998) Sequence and structure of the human 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase heart isoform gene (PFKFB2). Eur. J. Biochem., 254, 103–110. [DOI] [PubMed] [Google Scholar]
- Henkin T.M., Grundy,F.J., Nicholson,W.L. and Chambliss,G.H. (1991) Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol. Microbiol., 5, 575–584. [DOI] [PubMed] [Google Scholar]
- Huynh P.L., Jankovic,I., Schnell,N.F. and Brückner,R. (2000) Characterization of an HPr kinase mutant of Staphylococcus xylosus. J. Bacteriol., 182, 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T. and La Porte,D. (1991) Isocitrate dehydrogenase kinase/phosphatase: aceK alleles that express kinase but not phosphatase activity. J. Bacteriol., 173, 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggi R., van Heeswijk,W.C., Westerhoff,H.V., Ollis,D.L. and Vasudevan,S.G. (1997) The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction. EMBO J., 16, 5562–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jault J.-M., Fieulaine,S., Nessler,S., Gonzalo,P., Di Pietro,A., Deutscher,J. and Galinier,A. (2000) The HPr kinase from Bacillus subtilis is a homo-oligomeric enzyme which exhibits strong positive cooperativity for nucleotide and fructose 1,6-bisphosphate binding. J. Biol. Chem., 275, 1773–1780. [DOI] [PubMed] [Google Scholar]
- Johansson B.C. and Gest,H. (1977) Adenylylation/deadenylylation control of the glutamine synthetase of Rhodopseudomonas capsulata. Eur. J. Biochem., 81, 365–371. [DOI] [PubMed] [Google Scholar]
- Jones B.E., Dossonnet,V., Küster,E., Hillen,W., Deutscher,J. and Klevit,R.E. (1997) Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J. Biol. Chem., 272, 26530–26535. [DOI] [PubMed] [Google Scholar]
- Kravanja M. et al. (1999) The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol. Microbiol., 31, 59–66. [DOI] [PubMed] [Google Scholar]
- LaPorte D.L. and Koshland,D.E. (1982) A protein with kinase and phosphatase activities involved in regulation of tricarboxylic acid cycle. Nature, 300, 458–460. [DOI] [PubMed] [Google Scholar]
- Lee Y.H., Olson,T.W., Ogata,C.M., Levitt,D.G., Banaszak,L.J. and Lange,A.J. (1997) Crystal structure of a trapped phosphoenzyme during a catalytic reaction. Nature Struct. Biol., 4, 615–618. [DOI] [PubMed] [Google Scholar]
- Martin I., Débarbouillé,M., Klier,A. and Rapoport,G. (1989) Induction and metabolite regulation of levanase synthesis in Bacillus subtilis. J. Bacteriol., 171, 1885–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Verstraete I., Deutscher,J. and Galinier,A. (1999) Phosphoryl ation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J. Bacteriol., 181, 2966–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Miller S.P., Chen,R., Karschnia,E.J., Romfo,C., Dean,A. and LaPorte,D.C. (2000) Locations of the regulatory sites for isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem., 275, 833–839. [DOI] [PubMed] [Google Scholar]
- Miwa Y., Nakata,A., Ogiwara,A., Yamamoto,M. and Fujita,Y. (2000) Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res., 28, 1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno M.S., Schneider,B.L., Maile,R.R., Weyler,W. and Saier,M.H.,Jr (2001) Catabolite repression mediated by CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol., 39, 1366–1381. [DOI] [PubMed] [Google Scholar]
- Postma P.W., Lengeler,J.W. and Jacobson,G.R. (1993) Phosphoenol pyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev., 57, 543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presecan-Siedel E., Galinier,A., Longin,R., Deutscher,J., Danchin,A., Glaser,P. and Martin-Verstraete,I. (1999) Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J. Bacteriol., 181, 6889–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J., Hoischen,C., Titgemeyer,F., Rivolta,C., Rabus,R., Stülke,J., Karamata,D., Saier,M.H. and Hillen,W. (1998) A novel protein kinase that controls carbon catabolite repression in bacteria. Mol. Microbiol., 27, 1157–1169. [DOI] [PubMed] [Google Scholar]
- Reizer J., Bachem,S., Reizer,A., Arnaud,M., Saier,M.H.,Jr and Stülke,J. (1999) Novel phosphotransferase system genes revealed by genome analysis—the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology, 145, 3419–3429. [DOI] [PubMed] [Google Scholar]
- Stülke J. and Hillen,W. (2000) Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol., 54, 849–880. [DOI] [PubMed] [Google Scholar]
- Stülke J., Martin-Verstraete,I., Zagorec,M., Rose,M., Klier,A. and Rapoport,G. (1997) Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol., 25, 65–78. [DOI] [PubMed] [Google Scholar]
- Thompson J. and Torchia,D.A. (1984) Use of 31P nuclear magnetic resonance spectroscopy and 14C fluorography in studies of glycolysis and regulation of pyruvate kinase in Streptococcus lactis. J. Bacteriol., 158, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson T.B., Thomas,M.G., Escalante-Semerena,J.C. and Rayment,I. (1998) Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium determined to 2.3 Å resolution. Biochemistry, 37, 7686–7695. [DOI] [PubMed] [Google Scholar]
- Vagner V., Dervyn,E. and Ehrlich,S.D. (1998) A vector for systematic gene inactivation in Bacillus subtilis. Microbiology, 144, 3097–3104. [DOI] [PubMed] [Google Scholar]
- Viana R., Monedero,V., Dossonnet,V., Vadeboncoeur,C., Pérez-Martínez,G. and Deutscher,J. (2000) Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol. Microbiol., 36, 570–584. [DOI] [PubMed] [Google Scholar]
- Waygood E.B., Erickson,E., El-Kabbani,O.A.L. and Delbaere,L.T.J. (1985) Characterization of phosphorylated histidine-containing protein (HPr) of the bacterial phosphoenolpyruvate:sugar phospho transferase system. Biochemistry, 24, 6938–6945. [DOI] [PubMed] [Google Scholar]
- Weickert M.J. and Chambliss,G.H. (1990) Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl Acad. Sci. USA, 87, 6238–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J.-J. and Saier,M.H.,Jr (1995) Cooperative binding of lactose and the phosphorylated phosphocarrier HPr(Ser-P) to the lactose/H+ symport permease of Lactobacillus brevis. Proc. Natl Acad. Sci. USA, 92, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J.-J. and Saier,M.H.,Jr (1996) Regulation of sugar uptake via the phosphoenolpyruvate-dependent phosphotransferase system in Bacillus subtilis and Lactococcus lactis is mediated by ATP-dependent phosphorylation of seryl residue 46 in HPr. J. Bacteriol., 178, 3557–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalieckas J.M., Wray,L.V.,Jr and Fisher,S.H. (1999) Trans-acting factors affecting carbon catabolite repression of the hut operon in Bacillus subtilis. J. Bacteriol., 181, 2883–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Qin,L., Yoshida,T. and Inouye,M. (2000) Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc. Natl Acad. Sci. USA, 97, 7808–7813. [DOI] [PMC free article] [PubMed] [Google Scholar]