Abstract
Early embryonic development involves complex events such as the regulation of cell division and the establishment of embryonic polarity. To identify genes involved in these events, we collected four-dimensional time-lapse video recordings of the first three cell divisions and analysed terminal phenotypes after RNA interference of 147 embryonic lethal genes previously identified in a systematic screen of Caenorhabditis elegans chromosome I. Over half gave defects in early processes such as meiosis, the assembly or position of the first mitotic spindle, cytokinesis, and proper nuclear positioning. For some phenotypic classes, the majority of genes are involved in a shared biochemical process. In addition, we identified loss-of-function phenotypes for genes of unknown function, but for which homologues exist in other organisms, shedding light on the function of these uncharacterized genes. When applied to the whole genome, this approach should identify the vast majority of genes required for early cell processes, paving the way for a greatly improved understanding of these processes and their regulation at the molecular level.
Keywords: Caenorhabditis elegans/embryogenesis/RNAi/video recordings
Introduction
In Caenorhabditis elegans, early embryonic development follows a stereotypical series of events that is essentially invariant in wild-type animals (reviewed in Kemphues and Strome, 1997; illustrated in Figure 1A–I). These include the completion of meiosis following fertilization, pronuclear migration, spindle assembly, asymmetric positioning of the spindle in the zygote, cytokinesis, and blastomere-specific cell cycle timing and establishment of correct spindle orientation in the subsequent cell divisions. Although some of the genes required for these early events are known, the majority of those involved have yet to be discovered. These early events can be observed using time-lapse video microscopy, and the reproducibility of wild-type development allows the identification of relatively subtle defects in mutants. These features make C.elegans an excellent system for studying basic processes such as cell division and the establishment of cell polarity.
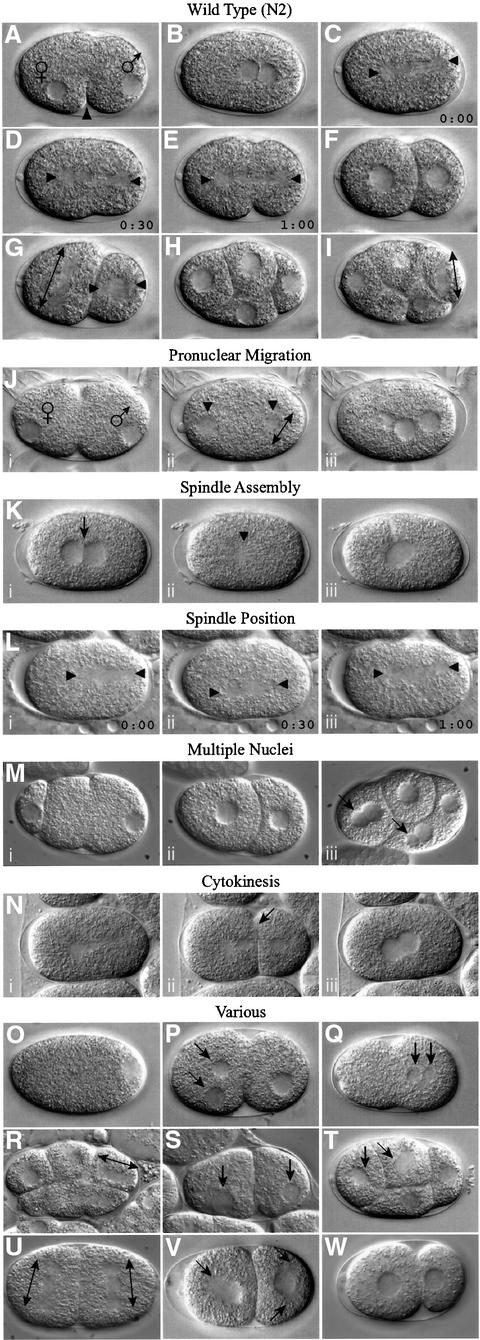
Fig. 1. Wild-type and RNAi mutant embryos. (A–I) Development of a wild-type embryo from the one- to the eight-cell stage. After fertilization, the oocyte nucleus completes meiosis and two polar bodies are extruded (not shown). (A) Subsequently the female and male pronuclei become visible. (A and B) The single oocyte pronucleus migrates to meet the sperm pronucleus in the posterior of the one cell embryo; this is accompanied by a pseudocleavage furrow [arrowhead in (A)]. (C–E) The first mitotic spindle is set up, elongating in the posterior direction through a rocking movement (arrowheads show ends of the spindle). (F) Two-cell embryo; first cleavage results in a larger anterior cell (AB) and a smaller posterior cell (P1). (G) The anterior AB cell divides before P1, along the short axis of the egg (AB spindle is shown by a double-headed arrow); P1 will divide along the anterior–posterior axis due to nucleocentrosomal rotation, which places centrosomes along this axis (arrowheads). (H) Four-cell embryo. (I) Division of P2 cell; the double-headed arrow shows orientation of the spindle. (J–N) Three time points from video recordings of each of five different RNAi mutants. (J) Pronuclear Migration; T06G6.9. Pronuclei fail to migrate. The embryo in (Jii) is similar stage to (C); pronuclear envelopes have broken down and sperm pronucleus has set up a small spindle (double-headed arrow). (K) Spindle Assembly; F32H2.3. (Kii) The mitotic spindle fails to set up [the arrowhead marks a small clear area where pronuclei have broken down; compare to spindle in (D)]. Note that the pronuclei meet centrally [arrow in (Ki)]. (L) Spindle Position; F21C3.5. The position of the mitotic spindle (marked by arrowheads) is unstable. Compare (Li–Liii) to (C–E). Individual images have been taken at 30 s intervals. (M) Multiple Nuclei; F56C11.5. Two cells each have two nuclei after division [arrows in (Miii)]. (N) Cytokinesis; Y18D10A.17. A normal cytokinesis furrow [arrow in (Nii)] regresses (Niii). (O–W) Single pictures illustrating other phenotypes. (O) 1-Cell Arrest; R12E2.3. (P) Meiosis; M01E11.6. Multiple female pronuclei are visible (arrows). (Q) Pronuclear Envelope; F56A3.3. The pronuclei have an indistinct appearance in comparison to (B). (R) Spindle Orientation; Y18D10A.5. Embryos have an abnormal P2 spindle orientation [compare to (I)]. (S) Nuclear Position; Y65B4B_10.b. Nuclei (arrows) are not centrally located in comparison with (F). (T) Nuclear Morphology; F33H2.5. Nuclei (arrows) are irregularly shaped and somewhat indistinct in comparison to the clear round nuclei in (H). (U) Spindle Orientation/Position; F20G4.3. AB and P1 cells divide at the same time, and both spindles exhibit a transverse orientation [compare to (G)]. The identical size of the two cells is reflective of a centrally located P0 spindle. (V) Aster Morphology (from Multiple Nuclei class); F26E4.1. Asters are very large and can easily be seen as clear regions next to the nuclei (marked by arrows). (W) Cytoplasmic Appearance; F26H9.6. The cytoplasm appears clear and less granular than wild type (F). Table I gives further information about the examples shown here. In all images, posterior is to the right.
The C.elegans genomic sequence is essentially complete, with 19 099 genes predicted (The C. elegans Sequencing Consortium, 1998). This information, coupled with the technique of RNA interference (RNAi), make it possible to ask which genes have a role in early embryonic development. RNAi causes the mRNA of the gene of interest to be specifically degraded following introduction of the corresponding double-stranded RNA into the animal, making it an ideal method for investigating gene function (Fire et al., 1998; Montgomery et al., 1998). We previously made a bacterial library for RNAi of ∼90% of C.elegans chromosome I genes and identified those required for embryonic development (Fraser et al., 2000). Here we present a phenotypic analysis of 147 of these genes. These results provide a starting point for future studies of processes such as meiosis, mitosis, cytokinesis and cell polarity.
Results
Identifying chromosome I genes required for early development
To identify genes involved in early embryonic processes, we analysed 147 genes that we previously showed to have a strong embryonic lethal phenotype using RNAi (Fraser et al., 2000). After RNAi, we video recorded embryonic development up to the eight-cell stage at least twice for each gene, and analysed these recordings for any deviation from wild-type development. After video recording, embryos were left to develop until the terminal stage, and the resulting phenotype was analysed visually for completion of embryonic cell divisions and for the extent of differentiation. Complete results are given in Table I, and video recordings are available at http://www.wormbase.org
Table I. Phenotypic classification of 147 C.elegans chromosome I emb genes.
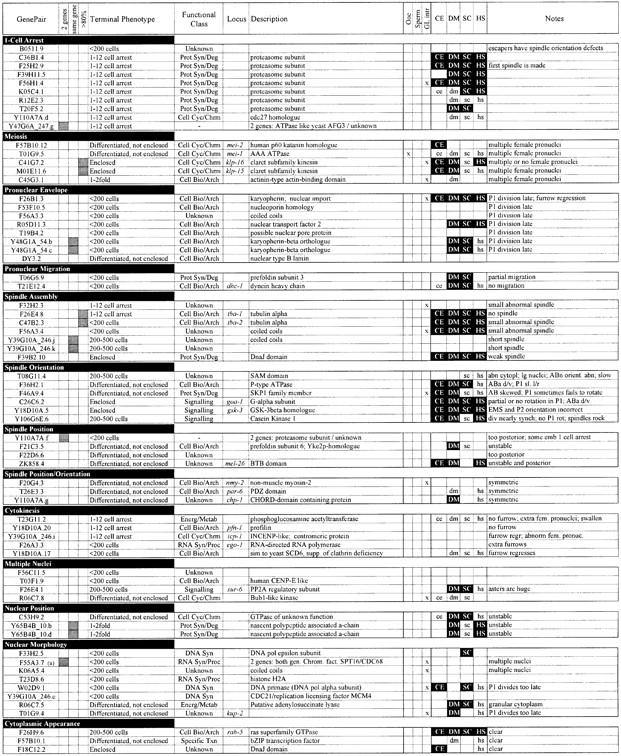
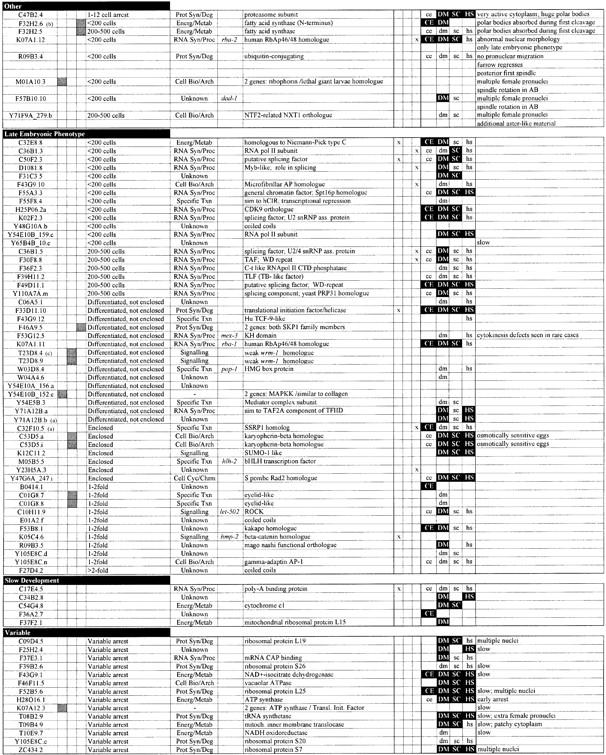
Phenotypes arising from RNAi of 147 embryonic lethal genes on chromosome I. Genes are grouped by phenotypic class. The following data are shown: the Research Genetics GenePair name; whether the sequence might target two or more unrelated genes [shaded box; determined in Fraser et al. (2000)]; whether the GenePair appears to target the same gene as another GenePair (shaded box); whether the sequence amplified by the GenePair is >80% identical to more than one predicted gene such that it might target two or more paralogous genes [shaded box; determined in Fraser et al. (2000)]; the terminal phenotype; the functional class [as defined in Fraser et al. (2000)]; the corresponding genetic locus name if it exists; a description of the gene sequence [as defined in Fraser et al. (2000)]; three columns showing whether the gene was found to have oocyte-specific (Ooc), sperm-specific (Sperm) or germline-intrinsic (GL intr) expression in Reinke et al. (2000); existence of matches (lower case) or homologues (filled box, white upper-case text) in C.elegans (CE), Drosophila melanogaster (DM), Saccharomyces cerevisiae (SC) or humans (HS) [as determined in Fraser et al. (2000)]; and additional phenotypic information. Phenotypic classes are defined as follows: ‘1-Cell Arrest’, embryos arrest as a single cell, usually without attempting mitosis; ‘Meiosis’, embryos have multiple or no female pronuclei; ‘Pronuclear Envelope’, the maternal and paternal pronuclear envelopes appear indistinct; ‘Pronuclear Migration’, pronuclei do not meet due either to complete or partial failure to migrate; ‘Spindle Assembly’, the mitotic spindle is absent or appears abnormal; ‘Spindle Orientation’, the orientation of division of at least one cell in the first three divisions is incorrect; ‘Spindle position’, correctly oriented spindles are mispositioned within cells, including spindles that are central, too posterior, skewed, or unstable; ‘Spindle Orientation/Position’, embryos display features of both classes; ‘Cytokinesis’, cytokinetic furrows fail to form, are incomplete, or occur ectopically; ‘Multiple Nuclei’, cells contain more than one nucleus, but meiosis is apparently normal as a single maternal pronucleus is present (probable chromosome segregation defect); ‘Nuclear Position’, nuclear position is unstable or abnormal; ‘Nuclear Morphology’, nuclei are either abnormally shaped or fail to reform after division, but pronuclei appear normal (the defect is usually first seen strongly in ABa and ABp); ‘Cytoplasmic Appearance’, cytoplasm appears clear and less granular than wild type; ‘Other’, genes where different recorded embryos showed different phenotypes or genes where a phenotype could not be assigned to a particular phenotypic class; ‘Late Embryonic Phenotype’, normal early embryonic development and a consistent terminal arrest phenotype; ‘Slow Development’, normal, but slow development; ‘Variable’, variable terminal arrest phenotype (early development is often slow).
aNote that genes C32F10.5, F55A3.7 and Y71A12B.b were found to be embryonic lethal subsequent to publication of Fraser et al. (2000).
bF32H2.6 shows 78% identity over 320 bp to F32H2.5.
cGenePair T23D8.4 does not target predicted gene T24D8.4, but the neighbouring predicted gene T24D8.9.
We identified a wide range of defects, and assigned these to 17 distinct phenotypic classes (Figure 1; Table I). Over 50% of the genes analysed gave an RNAi defect detectable in the first three cell cycles, including defects in meiosis, the assembly or positioning of the mitotic spindle, cytokinesis, and the position or morphology of the nuclei (Figures 1 and 2).
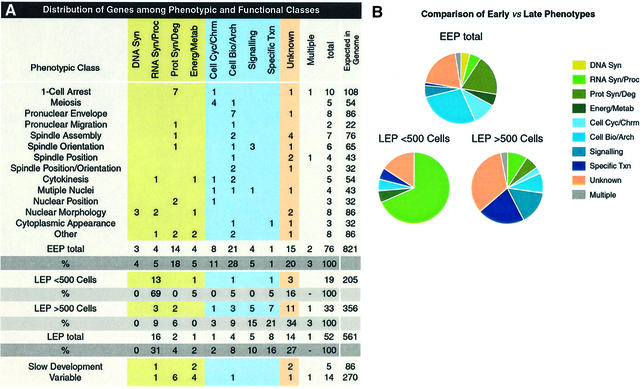
Fig. 2. Distribution of genes among phenotypic and functional classes. (A) The first column gives phenotypic classes. EEP, early embryonic phenotype; LEP, late embryonic phenotype. Each row gives the number of genes with the given RNAi phenotype in each functional class, the total number in that class identified in this screen, and the total number in that class expected to be found in the genome by RNAi (see Materials and methods for the latter calculation). (B) Pie charts showing the distribution of functional classes among genes within the EEP, LEP <500 and LEP >500 phenotypic groupings. Functional classes were used as defined in Fraser et al. (2000).
For some phenotypic classes, most of the genes identified are involved in a shared biochemical process. For example, 8 of 10 genes having a one-cell arrest RNAi phenotype encode components of either the proteasome or the anaphase promoting complex (APC) (Figure 1O; Table I). This cell cycle arrest is consistent with their known function in the regulation of cell cycle proteins; previous study of one component of the APC complex, EMB-30, yielded a similar phenotype (Zachariae and Nasmyth, 1999; Furuta et al., 2000). A second striking example is the Pronuclear Envelope phenotypic class, in which the pronuclear and subsequent nuclear envelopes have an indistinct appearance (Figure 1Q). The majority of genes in this class encode components of the nuclear envelope or are involved in nuclear transport (e.g. a nuclear lamin and karyopherin; Table I), suggesting that this phenotype is due to an improperly formed nuclear envelope. A third example is the Nuclear Morphology class. Many genes in this class encode proteins involved in DNA synthesis (e.g. DNA primase; Table I) or proteins involved in chromatin regulation or condensation (e.g. histone H2A; Table I). Thus, improper packaging or synthesis of DNA results in abnormal nuclear morphology. Applying this information to previously identified mutants with the above phenotypes (e.g. Gönczy et al., 1999) should facilitate their cloning by a candidate gene approach.
We identified many other genes with early RNAi phenotypes that provide novel functional information about previously studied genes or pathways. For example, we found that RNAi of the C.elegans homologue of glycogen synthase kinase 3 (gsk-3), a component of the Wnt pathway, produces embryos with a spindle orientation defect in the P2 cell (Figure 1R). Previous studies of gsk-3 did not identify this phenotype, but work on gsk-3 and other components of the Wnt pathway has revealed a requirement for the orientation of a different cell division (Schlesinger et al., 1999). It may be that the Wnt pathway has a more general role in orienting mitotic spindles than previously thought, or it might be important for the fate of the P2 cell. One gene we identified in the Cytokinesis class is similar to the yeast gene SCD6, a suppressor of clathrin deficiency. The yeast function is consistent with a role in the regulation of vesicle trafficking, and work in C.elegans has suggested that vesicle fusion is necessary for completion of the cytokinetic furrow (Nelson et al., 1996; Jantsch-Plunger and Glotzer, 1999); the RNAi phenotype of the C.elegans SCD6 homologue supports this view. In the Spindle Position class, we identified a homologue of prefoldin 6 (human KE2/yeast GIM1/YEK2). Previous work in yeast suggested that this protein might be involved in promoting the formation of functional α- and γ-tubulin (Geissler et al., 1998).
We also identified a number of genes for which no function had previously been described (Table I). In many cases, homologues exist in other animals and our data provide an entry point for studying these novel conserved genes. Furthermore, some phenotypic classes predict the biochemical process involved (e.g. Nuclear Morphology and packaging or synthesis of DNA); thus, a possible biochemical function can be suggested for unknown proteins in these classes.
Terminal arrest phenotypes
Genes that did not give an RNAi phenotype detectable in the first three divisions were placed into one of three phenotypic classes based on their terminal phenotype (Table I; Figure 3). Genes in the Late Embryonic Phenotype (LEP) class had a consistent, reproducible terminal RNAi phenotype, those in the Variable class had a range of different arrest phenotypes, and those in the Slow Development class developed much slower than wild-type embryos, but otherwise appeared normal. Genes in this latter class are involved in a range of basal cell processes, but half of the genes in the Variable class encode components of the translation machinery, including five ribosomal proteins and a tRNA synthetase (Table I). After RNAi of these genes, general production of both maternal and zygotic proteins is likely to be reduced, and this may result in embryos with a variable amount of a range of different proteins, resulting in a variable arrest. In addition, RNAi of genes involved in translation occasionally resulted in multiple female pronuclei or multiple nuclei during cleavages, suggesting a possible defect in meiosis and/or mitosis (Table I, Notes column).
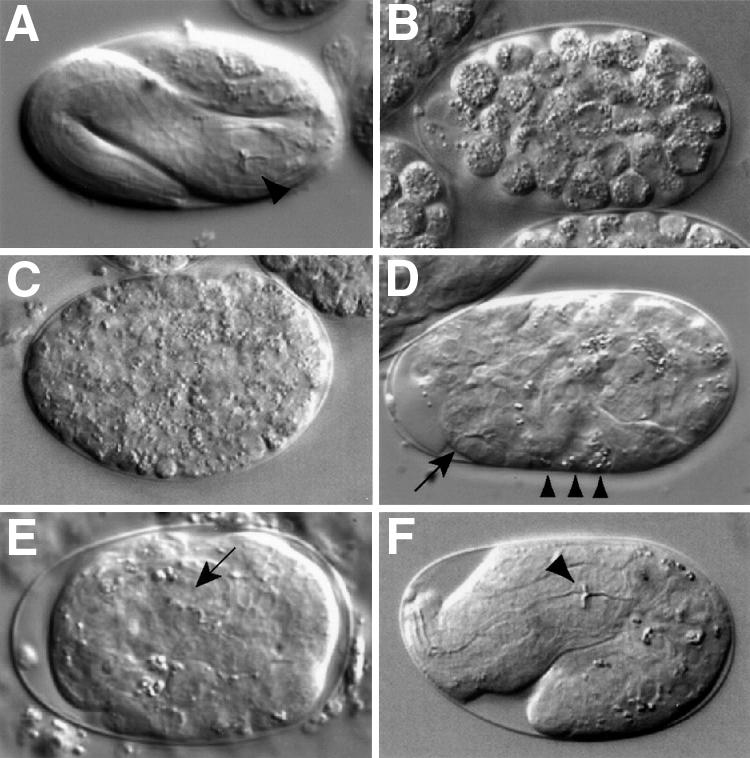
Fig. 3. Terminal phenotype classes. (A) Wild-type embryo at 3-fold stage of elongation. The pharyngeal grinder is marked with an arrowhead. (B) <200 cell arrest; K02F2.3. (C) 200–500 cell arrest; F30F8.8. (D) Differentiated, not enclosed; T23D8.4. The embryo is not enclosed within epidermal tissue [compare with (E)] such that pharyngeal tissue (arrow) and some gut cells (arrowheads) are found at the surface of the embryo. (E) Enclosed; C32F10.5. Pharynx (arrow) is surrounded by epidermal tissue. (F) 1- to 2-fold arrest; F53B8.1. Pharynx is properly formed with a normal grinder (arrowhead), but the embryo is not properly elongated [compare with (A)].
The LEP class is large and diverse, comprising genes that are involved in a range of different processes. The LEP class can be subdivided according to terminal phenotype, which in some cases can provide a possible indication of the function of a gene. For example, previous work has shown that mutants that fail to elongate past the 2-fold stage have defects either in the epidermis or in the underlying muscles (Williams and Waterston, 1994; Wissmann et al., 1997; Costa et al., 1998). In the 1- to 2-fold arrest class, we identified two genes involved in epidermal morphogenesis and for which mutants were previously shown to have this phenotype: hmp-2, which encodes a β-catenin, and let-502, related to Rho-binding kinases (Williams and Waterston, 1994; Wissmann et al., 1997; Costa et al., 1998). Based on the terminal arrest phenotype, some of the other genes in this class might be involved in muscle or epidermal structure or function (e.g. F53B8.1; Figure 3F). Other terminal arrest phenotypes (e.g. Enclosed) are less informative and comprise genes that clearly have a wide range of different functions.
Comparison of phenotype to predicted biochemical function
Further information on the type of gene involved in different cell processes can be gained from examination of the predicted cellular roles of the genes within each phenotypic class [the biochemical functional classes are defined in Fraser et al. (2000); Figure 2]. Of the genes having an early RNAi phenotype, the largest biochemical functional class contains proteins involved in cell biological and cell architecture processes—this includes components of the cytoskeleton and genes involved in vesicle fusion (Figure 2). In contrast, the largest class of genes with a LEP is RNA synthesis or processing (Figure 2A).
In wild-type embryos, tissue differentiation normally occurs when cell divisions are essentially complete (∼500 cells); we therefore subdivided the LEP class into those where embryos arrest prior to the 500-cell stage with no differentiation (LEP <500 cells) and those with >500 cells and obvious tissue differentiation (LEP >500 cells). The LEP <500 cell genes account for the majority of those involved in basal transcription or mRNA splicing (Figure 2). Normal early development of these is consistent with previous work showing that inhibitors of transcription allow embryonic development up to the beginning of gastrulation (Edgar et al., 1994; Powell-Coffman et al., 1996). Among the LEP >500 cell genes, the largest functional class contains proteins involved in specific transcription, such as transcription factors. This class includes previously studied genes such as pop-1, a TCF homologue that acts in the Wnt pathway (Lin et al., 1995), as well as genes not previously investigated, such as a homologue of Drosophila eyelid. eyelid encodes a DNA-binding domain protein of the BRIGHT family and appears to be antagonistic to wingless signalling (Treisman et al., 1997).
Another trend is that LEP >500 cell genes have a higher proportion of genes of unknown function than those with Early Embryonic Phenotypes (EEP; 34% versus 20%). This may reflect the fact that the basic processes of eukaryotic cell biology have been well studied in comparison to later developmental events. A high proportion (50%) of these genes of unknown function have homologues in other organisms (Table I) and are therefore likely to have conserved functions. Overall, the embryonic lethal genes analysed here are highly conserved: 84% have a match in humans, Drosophila or yeast (Table I), in comparison with 74% for all genes with an RNAi phenotype (Fraser et al., 2000).
Comparison with germline expression data
Recently, microarray experiments identified C.elegans oocyte-enriched, sperm-enriched and germline-intrinsic (germline expression not specific to sperm or oocytes) genes (Reinke et al., 2000). We examined the distribution of chromosome I phenotypes among these genes. The frequency of embryonic lethal RNAi phenotypes among the germline-intrinsic and oocyte-enriched genes is much higher than for genes on chromosome I in general (27 and 18%, respectively, versus 9%; Fraser et al., 2000; p <0.05), consistent with their germline expression. Interestingly, only one of 128 sperm-enriched genes on chromosome I had an RNAi phenotype (Fraser et al., 2000). The reason for this may be that genes involved in spermatogenesis appear resistant to RNAi or that our screening procedure was not suitable for identifying these genes. The lack of any other RNAi phenotype in almost all genes in this class is consistent with the proposal that these genes have sperm-specific functions; the one sperm-enriched gene that did give an RNAi phenotype produced sterility in the progeny, although we do not yet know whether this is the result of defective sperm (Fraser et al., 2000).
Genes with germline-intrinsic expression are found in most phenotypic classes, but show two clusterings. First, half of the genes in the Nuclear Morphology phenotypic class are germline intrinsic. Genes in this class are frequently involved in chromatin regulation or DNA replication. These genes would be expected to be germline expressed since this is the only tissue actively undergoing nuclear division in the adult. Secondly, in the LEP class, germline-intrinsic genes are clustered in the LEP <500 cell class, which contains many proteins involved in basal transcription. That so many of such genes are expressed at high levels in the germline can perhaps be explained by the fact that germline cytoplasm is rapidly turning over as oocytes are made. Phenotypic defects in the embryo indicate that these proteins may act both in the germline and the embryo.
Discussion
We have taken a systematic approach to identify genes required for early embryonic processes such as mitosis, the regulation of spindle position, cytokinesis, and the establishment of polarity. In this study, we present phenotypic information on 147 embryonic lethal genes that were initially identified in a genome-wide RNAi screen of C.elegans chromosome I (Fraser et al., 2000). Over half of the genes analysed have a detectable defect in the first three cell divisions. For these and for the genes without an early defect, the terminal arrest phenotypes provide further information on gene function. This work provides a starting point for further studies of cell division and other processes.
One striking finding from our studies is that some phenotypic classes primarily contain genes involved in a shared biochemical process. For example, many genes whose RNAi resulted in one-cell arrest encode proteasome or APC components, those that cause altered pronuclear envelope morphology are often involved in nuclear transport or are components of the nuclear envelope, and those resulting in abnormal nuclear morphology are often involved in the packaging or synthesis of DNA. Such correlation between phenotype and cellular function can be used to inform analyses of genes whose biochemical function is unknown. Genes of unknown biochemical function occur in the phenotypic classes above, suggesting that these genes might encode new factors involved in the identified processes.
A similar RNAi study of genes on C.elegans chromosome III identified early embryonic phenotypes for 133 genes after video recording the first two cell divisions (Gönczy et al., 2000). The range of early phenotypes seen in that study is comparable to those observed here. However, two major differences in approach have allowed us to provide phenotypic information for a higher fraction of embryonic lethals analysed, and to carry out the analyses more rapidly. First, in addition to looking for defects during early cleavages, we also characterized genes with no early RNAi phenotype (nearly half of embryonic lethal genes) by examining terminal arrest phenotypes. This analysis identified a common arrest phenotype after RNAi of genes involved in basal transcription, and further identified candidate genes for late embryonic processes such as embryonic elongation. Secondly, we first determined which genes had an embryonic lethal RNAi phenotype using a rapid RNAi by feeding approach (Fraser et al., 2000) and then analysed those by video microscopy. In contrast, Gönczy et al. (2000) collected video recordings after RNAi of nearly all genes on chromosome III (2174 genes); an early embryonic phenotype was seen for only 6% of them. As 94% of chromosome III genes with an early embryonic RNAi phenotype were found to be embryonic lethal (Gönczy et al., 2000), our pre-screening step should not have caused us to miss many relevant genes. The video recording step is the most labour-intensive part of the process; thus, the pre-screening step reduces the effort required significantly, as <10% of genes give an embryonic lethal RNAi phenotype (Fraser et al., 2000; Gönczy et al., 2000).
RNAi of C.elegans ovary cDNAs was also recently used to identify genes involved in early development (Piano et al., 2000). The cDNA approach has the benefit of enriching for genes with an early role in development (∼30% of the genes analysed had an embryonic lethal RNAi phenotype, compared with 9% for chromosome I). However, the genes identified will be skewed towards those with high expression. In addition, a comparison with our chromosome I-wide results suggests that some classes of phenotypes are overrepresented in the cDNA approach. For example, Piano et al. (2000) found that nearly half of the genes with a detectable early RNAi phenotype had a one-cell arrest phenotype and were involved in protein synthesis or turnover, whereas this class represented only 5% of the genes in our study. A genome-wide screen has the advantage that nearly every gene can be assayed, irrespective of expression level.
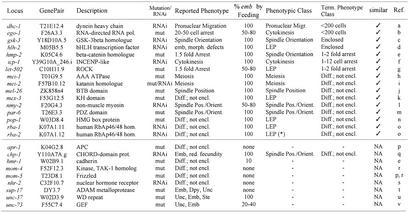
The efficiency of our screen can be estimated by com parison of the phenotypes we obtained to those previously reported for genes on chromosome I (Table II). Of the 25 sequenced genes on chromosome I previously shown to give an embryonic lethal phenotype and also analysed in Fraser et al. (2000), we found that 20 (80%) were embryonic lethal by RNAi (Fraser et al., 2000); this includes all of the seven previously shown to have defects in the first three cell divisions (dhc-1, icp-1, mei-1, mei-2, mel-26, nmy-2 and par-6). Of these 20, we recorded 17 in this study; in each case, we obtained the published phenotype (Table II). Therefore, our approach should be able to identify specific RNAi phenotypes for ∼80% of the non-redundant genes required for embryonic development.
Table II. Comparison of reported embryonic phenotypes with phenotypes presented in this study.
a, Gönczy et al., 1999; b, Smardon et al., 2000; c, Schlesinger et al., 1999; d, Krause et al., 1997; e, Costa et al., 1998; f, Kaitna et al., 2000; g, Wissmann et al., 1997; h, Clandinin and Mains, 1993; i, Srayko et al., 2000; j, Dow and Mains, 1998; k, Draper et al., 1996; l, Guo and Kemphues, 1996; m, Watts et al., 1996; n, Lin et al., 1995; o, Shi and Mello, 1998; p, Rocheleau et al., 1997, q, Shirasu et al., 1999; r, Thorpe et al., 1997; s, Sluder et al., 1997; t, Tax et al., 1997; u, Pflugrad et al., 1997; v, Steven et al., 1998.
Comparison of phenotypes of previously known loci to those identified in this study. Shown are the locus, the GenePair covering that locus, the gene description, whether the gene was previously studied using a mutant (mut) or RNAi, the reported phenotype, whether it was identified as embryonic lethal (Emb) in Fraser et al. (2000), our phenotypic class, our terminal phenotype class, whether our phenotype matches the published phenotype, and reference for the published phenotype. The upper part of the table lists the genes in this study and hence those that could be compared with published phenotypes; the lower part lists the genes without comparison either because we obtained no or low embryonic lethality or maternal sterility (Ste), or because the published phenotype was not detailed enough. A tick indicates our phenotypic class or terminal class, or both are similar to published. We could not compare the chp-1 phenotypes as no specific phenotype (besides embryonic lethality and reduced fecundity) was reported. *For K07A1.12, our two analyses yielded two different phenotypic classes: Late Embryonic Phenotype/Differentiated, not enclosed in one case and Nuclear Morphology/<200 cell arrest in the other case. Note that air-2 is missing from this list because it was not analysed in Fraser et al. (2000). LEP, late embryonic phenotype; NA, not applicable.
Scaling this approach up to the entire genome, we estimate that 821 genes with an early RNAi phenotype and 561 with a late one could be identified by RNAi (see Materials and methods for calculation). Figure 2 gives individual numbers for each phenotypic class; for example, RNAi might identify 54 genes involved in meiosis. Moreover, of the 821 early genes that could be identified by RNAi, ∼162 of these are expected to be of unknown biochemical function, and half will have homologues in other organisms. This work will therefore provide insights into the functions of these novel genes. When complete, an analysis of early RNAi phenotypes should identify the majority of genes involved in a wide range of cell processes and thus allow rapid progress to be made in understanding their molecular mechanisms.
Materials and methods
dsRNA generation
RNAi was accomplished either by injection or by feeding as in Kamath et al. (2000). Supplementary table I (Supplementary data are available at The EMBO Journal Online) shows the number of recordings made for each gene, which method was used, and in which a mutant phenotype was observed. Overall, the classified phenotype was observed by injection and feeding in 76% of the cases (n = 93). Twenty-one per cent showed the classified phenotype by injection only, and in 3% a phenotype was seen by feeding only. For injections, dsRNA for each gene was generated as follows: PCR using 25 pmol of T7 oligo (5′-CGTAATACGACT CACTATAG-3′; 95°C for 30 s, 52°C for 30 s, 72°C for 90 s, 25 cycles) was performed on bacteria containing the gene-specific fragment of interest cloned into the L4440 vector (Timmons and Fire, 1998; Fraser et al., 2000). Inserts in this vector are flanked by inverted T7 RNA polymerase promoters; hence, in vitro transcription of these PCR fragments using T7 polymerase generates sense and antisense strands in the same transcription reaction. PCR products were used directly as templates in transcription reactions (4.5 h at 37°C, 10 min at 72°C, RiboMAX™ RNA Production kit from Promega). dsRNA was analysed on a gel for verification of its size and concentration. Transcription reactions containing dsRNAs were diluted with 10 mM Tris–HCl pH 7.2, 0.1 mM EDTA about five times to a concentration of 0.5–1.0 mg/ml for injection. dsRNA was injected into the distal arms of both gonads of young hermaphrodites. Injected worms were kept at 22°C for 24 h prior to dissection of embryos. When feeding was used, L4 hermaphrodites were placed on induced feeding bacteria and left for 3 days at 15°C before dissection of embryos.
Video recordings
After RNAi, hermaphrodites were placed on a coverslip in egg buffer (118 mM NaCl, 40 mM KCl, 3 mM CaCl2, 3 mM MgCl2, 5 mM HEPES pH 7.2), cut open to release embryos, and then the coverslip was mounted onto a 3% agar pad and sealed with petroleum jelly. In general, the development of at least two embryos (from two different mothers) was video recorded per GenePair using Improvision OpenLab™ software. In some cases in the 1-Cell Arrest and Variable classes, progeny of only one mother were analysed. Four-dimensional time-lapse video recordings consisting of 12 optical sections were taken every 30 s for 100 time points, usually beginning before pronuclear migration and ending after the third round of divisions. From each recording, a single focal plane movie consisting of a middle focal plane was made for analysis and for deposition into the C.elegans database WormBase (http://www.wormbase.org). After completion of recordings, slides were kept at 15°C for 24 h prior to collecting a single series of 24 focal planes for analysis of the terminal phenotype.
Data analyses
Names given in all figures and tables are GenePair names and not names of predicted genes. GenePair names do not always match current gene names as gene predictions can change as they are improved, but GenePair names will always match the phenotype identified. The current matching of GenePair to gene can be found in WormBase (www.wormbase.org). About 95% of GenePairs have a one-to-one match with a currently predicted gene. Of the 226 genes found previously to have an embryonic lethal RNAi phenotype, 131 gave >50% embryonic lethality with no maternal sterility. We analysed 113 (86%) of these. In addition, we were able to analyse embryos after RNAi of 34 of 73 (46%) genes where RNAi induced partial or complete sterility in the mother.
The number of genes expected to be found by RNAi was calculated as follows. We identified 226 embryonic lethal genes after screening 13% of the genome (Fraser et al., 2000). We completed assays on 147 here; we identified a further three in the Slow Development and 11 in the Variable classes for which we only have one observation. These latter genes were included in the calculation for the expected number of genes in the Slow Development and Variable classes in Figure 2. Therefore, for each phenotypic class, RNAi could identify [(number of genes in class) × 226]/(147 + 11 + 3)/0.13 genes in the entire genome.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to R.Durbin and M.Gotta for helpful comments on the manuscript. P.Z. was supported by a Wellcome Trust Studentship and is a Junior Research Fellow at Hughes Hall College, A.G.F. by a US Army Breast Cancer Research Fellowship, R.S.K. by a Howard Hughes Medical Institute Predoctoral Fellowship, M.M.-C. by an EC-TMR Network Grant, and J.A. by a Wellcome Trust Senior Research Fellowship (054523).
References
- Clandinin T.R. and Mains,P.E.. (1993) Genetic studies of mei-1 gene activity during the transition from meiosis to mitosis in Caenorhabditis elegans. Genetics, 134, 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Raich,W., Agbunag,C., Leung,B., Hardin,J. and Priess,J.R. (1998) A putative catenin–cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol., 141, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow M.R. and Mains,P.E. (1998) Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics, 150, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper B.W., Mello,C.C., Bowerman,B., Hardin,J. and Priess,J.R. (1996) MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell, 87, 205–216. [DOI] [PubMed] [Google Scholar]
- Edgar L.G., Wolf,N. and Wood,W.B. (1994) Early transcription in Caenorhabditis elegans embryos. Development, 120, 443–451. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu,S, Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Furuta T., Tuck,S., Kirchner,J., Koch,B., Auty,R., Kitagawa,R., Rose,A.M. and Greenstein,D. (2000) EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell, 11, 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S., Siegers,K. and Schiebel,E. (1998) A novel protein complex promoting formation of functional α- and γ-tubulin. EMBO J., 17, 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Schnabel,H., Kaletta,T., Amores,A.D., Hyman,T. and Schnabel,R. (1999) Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol., 144, 927–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. et al. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- Guo S. and Kemphues,K.J. (1996) A non-muscle myosin required for embryonic polarity in Caenorhabditis elegansNature, 382, 455–458. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V. and Glotzer,M. (1999) Depletion of syntaxins in the early Caenorhabditis elegans embryo reveals a role for membrane fusion events in cytokinesis. Curr. Biol., 9, 738–745. [DOI] [PubMed] [Google Scholar]
- Kaitna S., Mendoza,M., Jantsch-Plunger,V. and Glotzer,M. (2000) Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol., 10, 1172–1181. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos,M., Zipperlen,P., Fraser,A.G. and Ahringer,J. (2000) Effectiveness of specific RNA-mediated inter ference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol., 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K.J. and Strome,S. (1997) Fertilisation and establishment of polarity in the embryo. In Riddle,D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R. (eds), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 335–359. [PubMed]
- Krause M. et al. (1997) A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development, 124, 2179–2189. [DOI] [PubMed] [Google Scholar]
- Lin R., Thompson,S. and Priess,J.R. (1995) pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell, 83, 599–609. [DOI] [PubMed] [Google Scholar]
- Montgomery M.K., Xu,S. and Fire,A. (1998) RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 95, 15502–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K.K., Holmer,M. and Lemmon,S.K. (1996) SCD5, a suppressor of clathrin deficiency, encodes a novel protein with a late secretory function in yeast. Mol. Biol. Cell, 7, 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrad A., Meir,J.Y., Barnes,T.M. and Miller,D.M.,III (1997) The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development, 124, 1699–1709. [DOI] [PubMed] [Google Scholar]
- Piano F., Schetterdagger,A.J., Mangone,M., Stein,L. and Kemphues,K.J. (2000) RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol., 10, 1619–1622. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman J.A., Knight,J. and Wood,W.B. (1996) Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev. Biol., 178, 472–483. [DOI] [PubMed] [Google Scholar]
- Reinke V. et al. (2000) A global profile of germline gene expression in C. elegans. Mol. Cell, 6, 605–616. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Downs,W.D., Lin,R., Wittman,C., Bei,Y., Cha,Y.H., Ali,M., Priess,J.R. and Mello,C.C.. (1997) Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell, 90, 707–716. [DOI] [PubMed] [Google Scholar]
- Schlesinger A., Shelton,C.A., Maloof,J.N., Meneghini,M. and Bowerman,B. (1999) Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes Dev., 13, 2028–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. and Mello,C. (1998) A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev., 12, 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K., Lahaye,T., Tan,M.W., Zhou,F., Azevedo,C. and Schulze-Lefert,P. (1999) A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell, 99, 355–366. [DOI] [PubMed] [Google Scholar]
- Sluder A.E., Lindblom,T. and Ruvkun,G. (1997) The Caenorhabditis elegans orphan nuclear hormone receptor gene nhr-2 functions in early embryonic development. Dev. Biol., 184, 303–319. [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke,J.M., Stacey,S.C., Klein,M.E., Mackin,N. and Maine,E.M. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germline development and RNA interference in C. elegans. Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Srayko M., Buster,D.W., Bazirgan,O.A., McNally,F.J. and Mains,P.E. (2000) MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev., 14, 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Steven R., Kubiseski,T.J., Zheng,H., Kulkarni,S., Mancillas,J., Ruiz Morales,A., Hogue,C.W., Pawson,T. and Culotti,J. (1998) UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell, 92, 785–795. [DOI] [PubMed] [Google Scholar]
- Tax F.E., Thomas,J.H., Ferguson,E.L. and Horvitz,H.R. (1997) Identification and characterization of genes that interact with lin-12 in Caenorhabditis elegans. Genetics, 147, 1675–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- Thorpe C.J., Schlesinger,A., Carter,J.C. and Bowerman,B. (1997) Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell, 90, 695–705. [DOI] [PubMed] [Google Scholar]
- Timmons L. and Fire,A. (1998) Specific interference by ingested dsRNA. Nature, 395, 854. [DOI] [PubMed] [Google Scholar]
- Treisman J.E., Luk,A., Rubin,G.M. and Heberlein,U. (1997) eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev., 11, 1949–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J.L., Etemad-Moghadam,B., Guo,S., Boyd,L., Draper,B.W., Mello,C.C., Priess,J.R. and Kemphues,K.J. (1996) par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development, 122, 3133–3140. [DOI] [PubMed] [Google Scholar]
- Williams B.D. and Waterston,R.H. (1994) Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol., 124, 475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A., Ingles,J., McGhee,J.D. and Mains,P.E. (1997) Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev., 11, 409–422. [DOI] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]