Abstract
The transcriptional coactivator OBF-1, which interacts with Oct-1 and Oct-2 and the octamer site DNA, has been shown to be critical for development of a normal immune response and the formation of germinal centers in secondary lymphoid organs. Here we have identified the RING finger protein Siah-1 as a protein interacting specifically with OBF-1. This interaction is mediated by the C-terminal part of Siah-1 and by residues in the N-terminus of OBF-1, partly distinct from the residues required for formation of a complex with the Oct POU domains and the DNA. Interaction between Siah-1 and OBF-1 leads to downregulation of OBF-1 protein level but not mRNA, and to a corresponding reduction in octamer site-dependent transcription activation. Inhibition of the ubiquitin-proteasome pathway in B cells leads to elevated levels of OBF-1 protein. Furthermore, in immunized mice, OBF-1 protein amounts are dramatically increased in primary activated B cells, without concomitant increase in OBF-1 mRNA. These data suggest that Siah-1 is part of a novel regulatory loop controlling the level of OBF-1 protein in B cells.
Keywords: B cell transcription/OBF-1/RING finger proteins/ubiquitin-proteasome pathway
Introduction
OBF-1 (also known as Bob1 or OCA-B) is a B-cell-specific transcription coactivator that binds to conserved octamer elements (ATGCAAAT or reverse) in the DNA together with the POU domain transcription factors Oct-1 or Oct-2 (Gstaiger et al., 1995; Luo and Roeder, 1995; Strubin et al., 1995). OBF-1 is a proline-rich protein consisting of an N-terminal part that mediates binding to DNA and Oct factors, and a C-terminal activation domain (Pfisterer et al., 1995; Gstaiger et al., 1996; Luo et al., 1998). In transfection assays as well as in vitro, OBF-1 strongly potentiates transcription from octamer-containing promoters such as the immunoglobulin κ light chain promoter (Luo and Roeder, 1995; Strubin et al., 1995; Schubart et al., 1996b). In B cells derived from OBF-1 knockout mice but containing an inducible OBF-1 allele it was found that activity of strictly octamer-dependent reporters was also dependent on OBF-1 (Laumen et al., 2000). However, gene targeting in mice suggests that one of the major roles of OBF-1 resides in the germinal center reaction during the immune response, which is strongly impaired in OBF-1–/– mice (Kim et al., 1996; Nielsen et al., 1996; Schubart et al., 1996a). In contrast, transcription of octamer-dependent genes at earlier stages of B-cell development, most prominently immunoglobulins, was largely unaffected in these mice (Schubart et al., 2001). Interestingly, recent structural and functional data have demonstrated that, at least on some of the Ig heavy chain promoters, Oct-1 binds to the octamer site in such a way that OBF-1 can not be recruited to the DNA (Tomilin et al., 2000).
Two recent studies reported that OBF-1 can be specifically upregulated at the level of protein and mRNA when primary splenic B cells are stimulated with either bacterial lipopolysaccharide or a combination of CD40 ligand and interleukin 4 (IL-4) (Qin et al., 1998; Greiner et al., 2000). In addition, a strong upregulation of OBF-1 protein was also observed in activated murine splenic B cells after immunization (Qin et al., 1998). Furthermore, in a variety of human lymphoid tumor samples, OBF-1 was found to be specifically expressed in germinal center-derived tumors (Greiner et al., 2000). All these findings suggest that OBF-1 is strongly induced during the germinal center reaction. While this effect seemed to be mainly at the level of transcription in in vitro stimulation experiments (Qin et al., 1998), the mode of regulation in a physiological context has remained unclear. Here we provide evidence that in vivo this regulation occurs mainly at the post-transcriptional level.
Controlled degradation of proteins through the ubiquitin-proteasome pathway has recently emerged as a major biological regulatory pathway in eukaryotes (reviewed in Ciechanover, 1998). Ubiquitin ligases (E3s) play a key role in this process, by interacting with specific proteins and thereby targeting them for degradation. The RING finger domain is a characteristic feature of a subset of ubiquitin ligases and it interacts with ubiquitin conjugating enzymes (Lorick et al., 1999; Freemont, 2000; Jackson et al., 2000; Joazeiro and Weissman, 2000).
The Drosophila RING finger protein Seven in Absentia (Sina) has been shown to be critical for development of R7 photoreceptor, through interaction with the protein Tramtrack, a repressor of cell-fate determination (Li et al., 1997; Dickson, 1998). The human homologs Siah-1 and Siah-2 (Hu et al., 1997a), and the mouse homologs Siah-1a and -1b and Siah-2 (Della et al., 1993; Holloway et al., 1997), are highly conserved, in particular in their C-terminal portion, where they show >85% homology. Recent work has revealed that mammalian Siah proteins are involved in ubiquitin-mediated degradation of proteins with which they interact, such as DCC (deleted in colorectal cancer) and N-CoR, a nuclear receptor corepressor (Hu et al., 1997b; Zhang et al., 1998; Hu and Fearon, 1999). For this process, the RING finger region of Siah is essential. Here we identify OBF-1 as a novel interactor of human Siah-1 and show that OBF-1 protein level is downregulated by Siah-1 in a manner that is dependent on this interaction. Furthermore, we provide evidence that endogenous OBF-1 is degraded in B cell lines via the ubiquitin-proteasome pathway. Taken together with our observation that OBF-1 is post-transcriptionally upregulated during the immune response, these data suggest a novel mechanism of OBF-1 regulation in vivo.
Results
Identification of Siah-1 as a protein interacting specifically with OBF-1
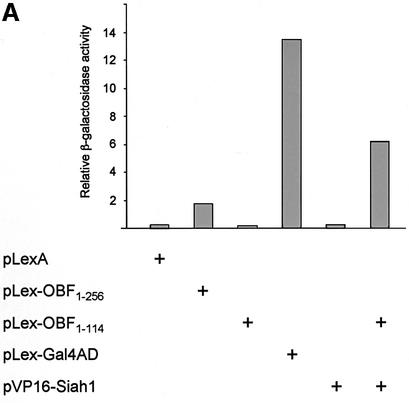
We sought to identify proteins that interact specifically with OBF-1 and that might help explain the function and regulation of this factor in B cells. To this end we set up a yeast two-hybrid screen (Fields and Song, 1989; Gyuris et al., 1993) with OBF-1 as a bait and screened an activation domain-tagged cDNA library derived from the human B-cell line Namalwa. Since full-length OBF-1 activates transcription efficiently in yeast when tethered to DNA via the LexA DNA binding domain (Figure 1), we used the N-terminal portion of OBF-1 (aa 1–114) as a bait, which lacks transcription activation capacity on its own (Figure 1). From a screen of ∼3 × 106 colonies, four cDNAs were identified that allowed growth of the yeast strain on selective medium. These cDNAs were found to be independent full-length isolates of Siah-1, the human homolog of the Drosophila protein Sina. When re-transformed into the parental yeast strain containing Lex-OBF1–114 they all allowed growth on selective medium and also efficiently stimulated expression of a lacZ reporter in an OBF-1-dependent manner (Figure 1A).
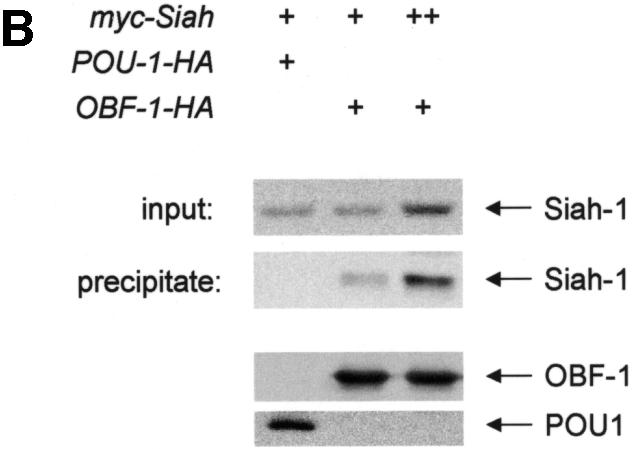
Fig. 1. Interaction between OBF-1 and Siah-1 in yeast and in mammalian cells. (A) β-galactosidase activity in the yeast strain EGY48 containing a lacZ reporter under the transcriptional control of multimerized lexA binding sites and different combinations of expression plasmids as indicated. Importantly, VP16-Siah-1 only activates transcription in cells also expressing Lex-OBF1–114. The relative β-galactosidase activity of the different strains was normalized for protein level. (B) Co-immunoprecipitation assay. 293T cells were transfected with an expression vector for myc-Siah-1 (500 ng or 1 µg), together with expression vectors for POU-1-HA or OBF-1-HA, as indicated. Forty-eight hours after transfection cells were treated for 6 h with 500 nM MG262 in order to stabilize Siah-1. After cell extract preparation, HA-tagged proteins were immunoprecipitated with the 12CA5 mAb. The precipitate was analyzed by SDS–PAGE and blotted with the 9E10 mAb to detect potential association of Siah-1. Expression of POU-1 or OBF-1 was detected by reprobing the membrane with the 12CA5 mAb (lower part).
To test whether OBF-1 and Siah-1 can also interact in mammalian cells, a co-immunoprecipitation assay was performed. For this, the Siah-1 cDNA was recloned in a mammalian expression vector so as to express a protein tagged with a myc epitope. Human 293T cells were transfected with myc-Siah together with a plasmid encoding a hemagglutinin (HA)-tagged version of OBF-1 or, as a control, an HA-tagged Oct-1 POU domain. Cell extracts were prepared from the transfected cells and OBF-1 or POU-1 was immunoprecipitated with an anti-HA antibody. The precipitated material was resolved by SDS–PAGE, and subsequently the presence of Siah-1 was detected by western blotting using an anti-myc antibody. As seen in Figure 1B, immunoprecipitation of OBF-1, but not of POU-1, led to co-precipitation of Siah-1. This indicates that the two proteins OBF-1 and Siah-1 are also able to interact in mammalian cells, and further indicates that this interaction is specific since no association was detected with the Oct-1 POU domain.
The N-terminal part of OBF-1 interacts with Siah-1 through residues distinct from those mediating interaction with the POU domain
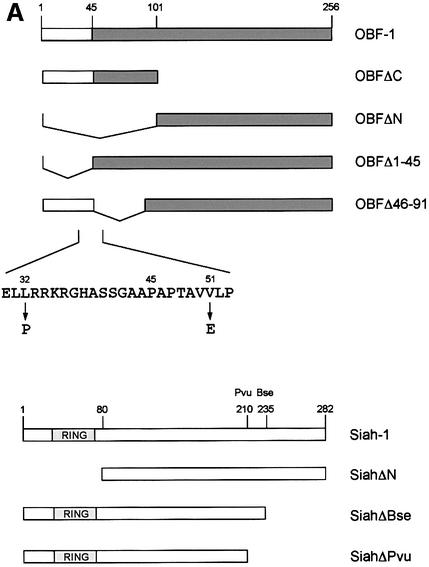
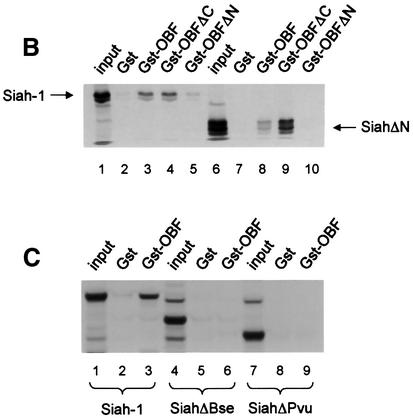
To confirm the interaction between OBF-1 and Siah-1, an in vitro binding assay was used. Glutathione S-transferase (GST) fusion proteins containing different parts of OBF-1 (Figure 2A) were expressed in Escherichia coli and affinity purified on glutathione–Sepharose beads. These proteins were then incubated with radiolabeled Siah-1 protein, either full-length or lacking the N-terminal RING finger domain (SiahΔN). After extensive washing, bound protein was separated on an SDS–polyacrylamide gel and visualized by autoradiography. As shown in Figure 2B, full-length Siah-1 bound efficiently to OBF-1 or to its N-terminal portion (GST–OBFΔC, lane 4), but not to its C-terminus (GST–OBFΔN, lane 5). When a Siah-1 truncation was used that lacked the first 80 aa including the RING finger (SiahΔN), the same binding specificity to OBF-1 was observed, but the binding was somewhat stronger, especially to the N-terminus of OBF-1 (Figure 2B, lane 9). To further define the residues within Siah-1 necessary for interaction with OBF-1, two C-terminal deletions were tested (Figure 2A). As shown in Figure 2C, removal of the last 47 aa (SiahΔBse, lanes 4–6) dramatically reduced binding to full-length OBF-1; however, very weak binding to the N-terminus of OBF-1 was still detected (not shown). Elimination of an additional 25 aa of Siah-1 completely abolished its interaction (SiahΔPvu, lanes 7–9). Thus, binding to OBF-1 is mediated by residues comprised within the last 72 aa of Siah-1.
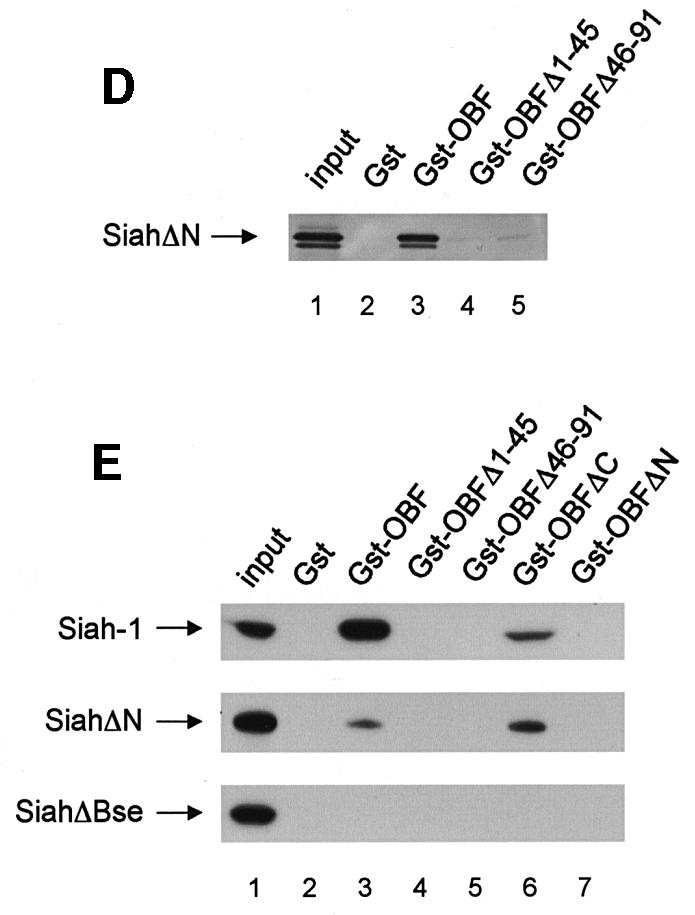
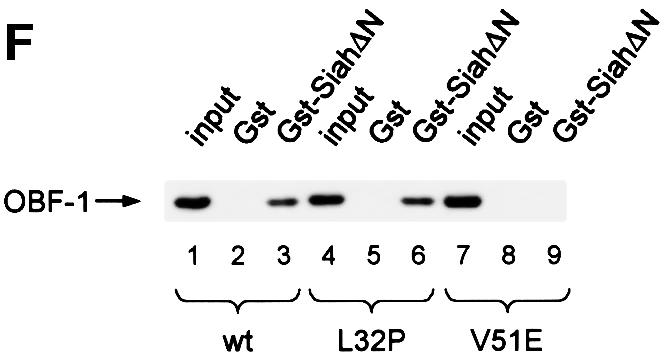
Fig. 2. OBF-1 and Siah-1 interact in vitro. (A) Schematic representation of the cDNAs used in this study. Depending on the experiment and as indicated in the relevant figure legends, the different proteins were expressed in bacteria as GST fusion proteins, made in reticulocyte lysate or expressed in eukaryotic cells from an appropriate expression vector. For OBF-1, the open rectangle represents the region that is necessary for interaction with the POU domain and DNA (Gstaiger et al., 1996; Chasman et al., 1999). (B–D) In vitro pull-down assays. Equal amounts of GST–OBF-1 fusion proteins as indicated were incubated with 35S-labeled Siah-1 proteins. After extensive washing, radioactive protein that was retained was visualized by SDS–PAGE followed by autoradiography. (E) Equal amounts of extracts from 293T cells transiently transfected with expression vectors for HA-tagged Siah-1, SiahΔN or SiahΔBse were incubated with beads loaded with GST–OBF-1 fusion proteins, as indicated. After extensive washing, bound proteins were run on SDS–PAGE and retained Siah was detected by western blotting using an anti-HA antibody. (F) Extracts from 293T cells transfected with Myc-tagged OBF-1 constructs as indicated were incubated with GST alone or GST–SiahΔN on glutathione beads. Subsequent analysis was carried out as in (E).
OBF-1 forms a ternary complex with the POU domain of Oct-1 or Oct-2, and the octamer DNA. Based on mutagenesis as well as crystallographic studies, a minimal domain has been defined in the N-terminal moiety of OBF-1, which encompasses residues 1–44 and which is necessary for specific interaction with the POU domain and DNA (Gstaiger et al., 1996; Chasman et al., 1999). Since Siah-1 was also found to interact with the N-terminal half of OBF-1, it was of interest to determine whether the same OBF-1 residues might be involved in the two types of interaction. To investigate this, deletions as well as point mutants of OBF-1 were made. As shown in Figure 2D, GST fusion proteins lacking either residues 1–45 or residues 46–91 of OBF-1 failed to interact with radiolabeled in vitro translated SiahΔN. Similar results were obtained with Siah-1 full-length (data not shown). To verify these results we next used an alternative binding assay, in which the GST–OBF fusion proteins were incubated with extracts from transiently transfected 293T cells expressing different forms of Siah proteins. In several instances we have found this assay to be more stringent and to give cleaner results. In this case, as shown in Figure 2E, no significant binding was observed between OBFΔ1–45 or OBFΔ46–91, and Siah-1 or SiahΔN. Together these results therefore suggest that the region of OBF-1 required for interaction with Siah-1 is partly overlapping with the region needed for formation of a complex with the POU domain and DNA, but extends further towards the C-terminus of OBF-1.
This analysis was extended by testing the binding capacity of two previously characterized OBF-1 point mutants, OBF-L32P and OBF-V51E (Gstaiger et al., 1996). The mutant protein OBF-L32P has been shown to have a weakened interaction with the POU domain and consequently a poor transcription activation capacity (Gstaiger et al., 1996). Crystallographic studies have revealed that residue L32 of OBF-1 is in direct contact with the POU-specific domain of Oct-1 (Chasman et al., 1999); however, this mutant is bound in vitro by SiahΔN like wild-type (WT) OBF-1 (Figure 2F; see also Figure 5C).
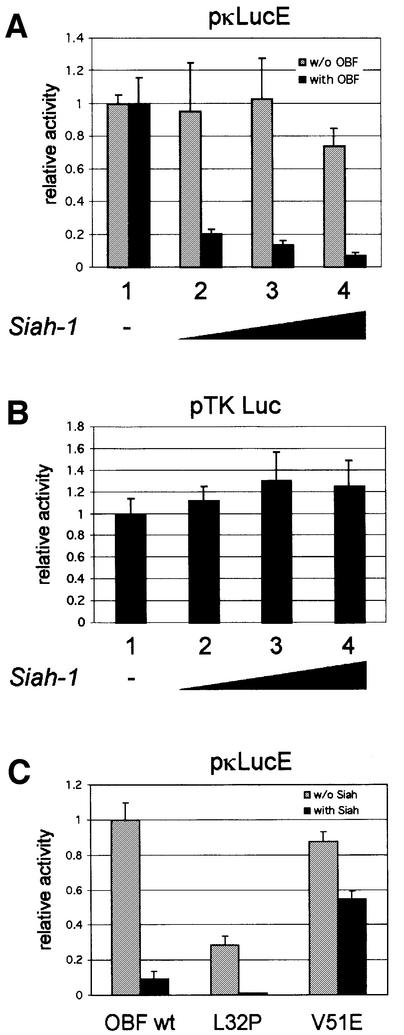
Fig. 5. Siah-1 antagonizes transcription activation mediated by OBF-1. (A) 293T cells were transiently transfected with 500 ng of the reporter pκLucE, together with an HA-Siah-1 expression vector in increasing amounts (lanes 1–4; 0, 100, 250 and 500 ng, respectively; filled up to 500 ng with empty vector). This was done without (gray bars) or with (black bars) 500 ng of a Myc-OBF-1 expression vector and the first value (lane 1, without Siah) of each series was set to 1. When looking at absolute values, OBF-1 stimulated the reporter activity >50-fold (not shown). (B) Cells were transfected with the reporter pTK-Luc and with a Siah-1 expression vector as in (A). (C) Cells were transfected with the reporter pκLucE and Myc-OBF-1 WT, L32P or V51E, together with an empty expression vector (gray bars) or an HA-Siah-1 expression vector (black bars). The data represent the average of three independent experiments, and each value was measured in duplicate. Values were normalized to the protein concentration of each sample, determined by the Bradford method.
In contrast, the OBF-1 mutant V51E has been found earlier to still interact efficiently with the Oct-1 POU domain, and to activate transcription like WT (Gstaiger et al., 1996). In the crystal structure, this residue is not in contact with the POU domain. As shown here, however, this mutant is strongly impaired in its interaction with Siah-1 (Figure 2F, lane 9). Together, these results show that the regions of OBF-1 required for interaction with Siah-1 and the Oct POU domain are overlapping, but distinct contacts are made.
OBF-1 protein level is specifically downregulated through interaction with Siah-1
Siah-1 has been shown to specifically mediate degradation of DCC (Hu et al., 1997b; Hu and Fearon, 1999), NCoR (Zhang et al., 1998), Kid (Germani et al., 2000) and Bag-1 (Sourisseau et al., 2001). We therefore tested whether the interaction identified here might regulate OBF-1 expression, perhaps by promoting its degradation. For this, 293T cells were transfected with expression vectors for OBF-1 in the absence or presence of a Siah-1 expression vector, and 2 days later the levels of the different proteins were examined by western blotting. As seen in Figure 3A, addition of increasing amounts of the Siah-1 expression vector led to a dose-dependent reduction of OBF-1 protein (lanes 3–5). Siah-1 itself is degraded through the proteasome in a RING finger-dependent manner (Hu and Fearon, 1999) and was expressed only at low level. When SiahΔN was used, however, no reduction in OBF-1 level was observed and the amount of OBF-1 was in fact reproducibly increased (lanes 8–10). This may reflect competition between transfected SiahΔN and endogenous Siah-1 protein, leading to protection of OBF-1 from degradation. In addition, SiahΔN was expressed at much higher levels than WT Siah, consistent with the fact that the former protein is no longer degraded through the proteasome (Hu and Fearon, 1999). In contrast, the presence of Siah-1 or SiahΔN had no impact on the amount of TATA binding protein (TBP), indicating that this effect is specific for OBF-1. Importantly, there was no influence of Siah-1 or SiahΔN on the amount of OBF-1 mRNA expressed from the transfected plasmid (Figure 3A).
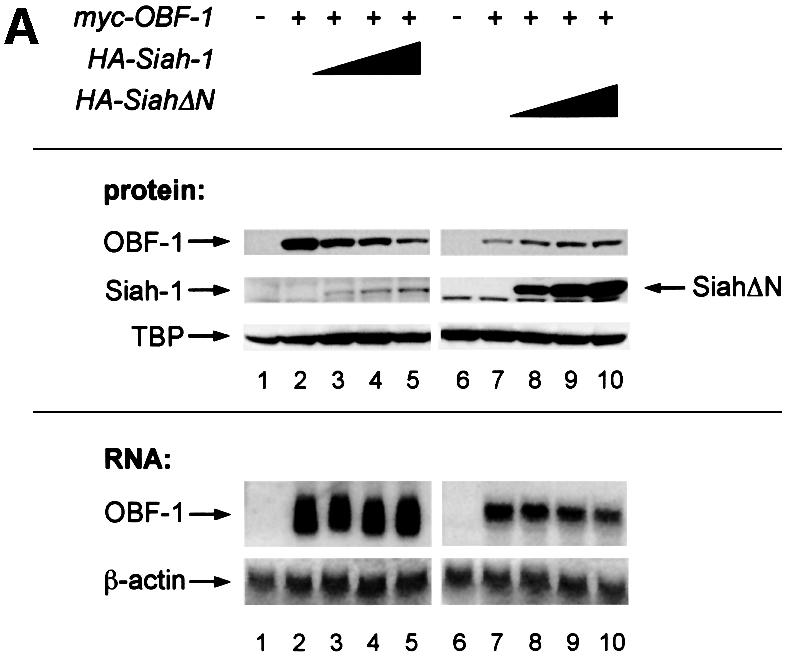
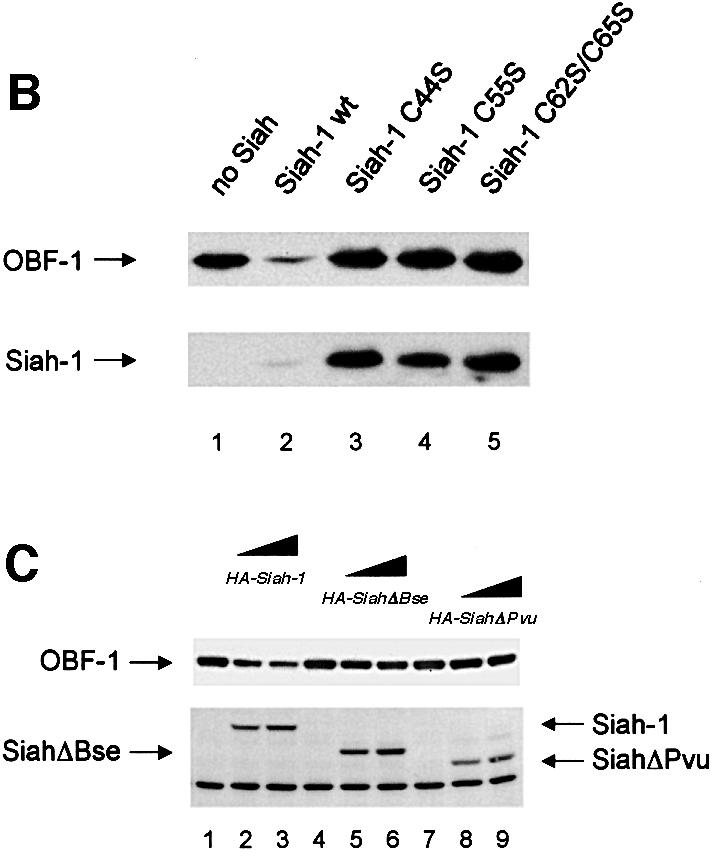
Fig. 3. Interaction between Siah-1 and OBF-1 reduces OBF-1 protein expression. (A) 293T cells were transiently transfected with expression vectors for HA-tagged Siah-1 or Siah-ΔN, and Myc-tagged OBF-1, as indicated. Control transfections in lanes 1 and 6 contained empty expression vectors. Protein extracts and RNA were prepared and equal amounts were analyzed by western (upper panels) or northern blotting (lower panels). OBF-1 and Siah proteins were detected with monoclonal antibodies directed against the respective tags (anti-Myc, 9E10; anti-HA, 12CA5). As a control for endogenous protein, TBP was detected with the specific monoclonal antibody SL-39. For the experiment shown on the right of the figure, less OBF-1 expression vector was used, explaining the weaker signal for OBF-1 protein and RNA. (B) Cells were transfected with Myc-tagged OBF-1 (lanes 1–5) and WT or point mutated Siah (HA-tagged at the N-terminus), as indicated. After protein extract preparation, the respective proteins were detected as described above. Because the Siah-1 point mutant was expressed at a much higher level than WT Siah-1, less expression vector was used for the transfection. (C) Cells were transfected with Myc-tagged OBF-1 (lanes 1–9) and the indicated HA-Siah expression plasmids. OBF-1 and Siah proteins were detected as in (A). The band of equal intensity visible across the lower part of the picture corresponds to an endogenous protein cross-reacting with the anti-HA antibody. (D) Cells were transfected with Myc-tagged expression vectors for WT OBF-1 or the two point mutants indicated, together with a plasmid expressing Siah-1 (lanes 2, 4 and 6). Protein extracts were prepared and OBF-1 protein was detected by western blotting as above. (E) Cells were transfected with Myc-tagged expression vectors for WT OBF-1 or the two deletion mutants indicated, together with a plasmid expressing Siah-1 or SiahΔBse. The level of OBF-1 expression was monitored by western blotting, as above.
The function of the RING finger has been shown to depend on several conserved cysteine residues (Freemont, 2000; Jackson et al., 2000; Joazeiro and Weissman, 2000). To confirm the importance of the RING finger of Siah-1 for downregulating OBF-1 expression, we generated cysteine-to-serine point mutants in the Siah-1 RING finger (Hu and Fearon, 1999) and tested them in the transfection assay described above. As seen in Figure 3B, while WT Siah-1 downregulated OBF-1 efficiently, the point mutants Siah-C44S, Siah-C55S and the double mutant Siah-C62S/C65S all failed to degrade OBF-1. As expected, these three point mutants were expressed at significantly higher levels, since they are no longer degraded by the proteasome.
We next examined whether downregulation of OBF-1 expression indeed required a direct interaction with Siah-1. For this, we used the C-terminally truncated Siah proteins SiahΔBse and SiahΔPvu, which, as we have shown above, are no longer able to interact with OBF-1 (Figure 2C). As shown in Figure 3C, SiahΔBse and SiahΔPvu both failed to downregulate OBF-1 expression; however, these two Siah-1 mutants are still degraded normally through the proteasome pathway and can be stabilized efficiently by proteasome inhibitors (data not shown and see below). Finally, the point mutated protein OBF-V51E, which no longer interacts with Siah in vitro (Figure 2F), was also not subject to downregulation by Siah-1 in vivo, whereas OBF-L32P was still efficiently downregulated (Figure 3D). Likewise, the two deletion mutants OBFΔ1–45 and OBFΔ46–91 were both resistant to downregulation by Siah-1 (Figure 3E). Together, these results show that OBF-1 expression is regulated by Siah-1 in a manner requiring specific interaction between these two proteins.
Inhibition of the proteasome can rescue Siah-1-mediated downregulation of OBF-1 protein expression
To test whether Siah-1-mediated reduction in OBF-1 expression requires the ubiquitin-proteasome pathway, transiently transfected cells were treated with the highly specific proteasome inhibitor MG262 (McCormack et al., 1997). As shown in Figure 4, addition of 500 nM MG262 for 12 h rescued almost completely the downregulation of OBF-1 by Siah-1, suggesting that Siah-1 indeed mediates proteasomal degradation of OBF-1. Under these conditions, Siah-1, which is normally expressed only at very low levels, owing to constant degradation, was found to be stabilized efficiently and no effect was observed on the TBP level. Perhaps surprisingly, MG262 was found to modestly increase the amount of OBF-1 even in the absence of transfected Siah-1 (Figure 4, lane 3). This effect might be explained by ‘basal’ degradation of OBF-1 through endogenous Siah-1 protein in the absence of proteasome inhibitor. This observation is also in good agreement with the results obtained with SiahΔN, in the presence of which an upregulation of OBF-1 was observed (see Figure 3A).
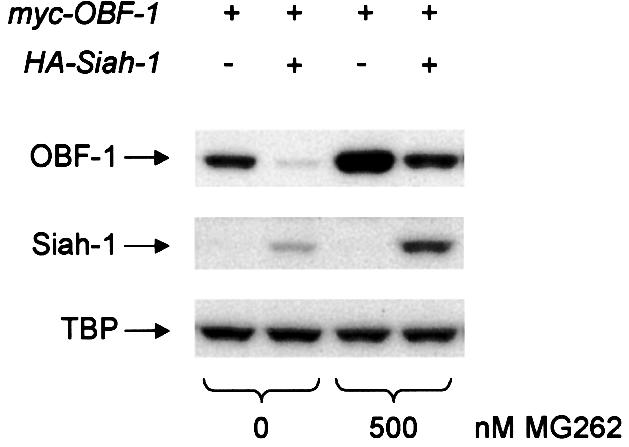
Fig. 4. Siah-1 promotes OBF-1 degradation by the proteasome. L929 cells were transfected with Myc-tagged OBF-1 and HA-tagged Siah-1 expression vectors, as indicated. After treatment for 12 h with the proteasome inhibitor MG262, or with H2O as control, extracts were prepared and proteins detected by western blotting.
Siah-1 specifically antagonizes transcription activation mediated by OBF-1
The interaction identified here between Siah-1 and the transcriptional coactivator OBF-1 suggested that in vivo OBF-1-dependent transcription might be modulated by Siah-1. To test this, we set up reporter assays in which a luciferase gene under the control of an octamer site-containing promoter was transfected into 293T cells, together with the expression vectors indicated in Figure 5. When increasing amounts of Siah-1 expression vector were included in the transfection, a dose-dependent decrease was observed in OBF-1-stimulated luciferase activity (Figure 5A, lanes 2–4, black bars). This effect was dependent specifically on OBF-1, since in its absence Siah-1 did not repress the basal level of luciferase expression (lanes 2–4, gray bars). The same effect was also observed on several other octamer site-containing promoters (data not shown), and Siah-1 cotransfection did not repress a luciferase reporter under the control of the proximal thymidine kinase (TK) promoter, which does not contain an octamer site (Figure 5B).
Inhibition of OBF-1-mediated transcription activation was specifically dependent on its interaction with Siah-1 as well as on the ubiquitin-proteasome pathway, since SiahΔBse, SiahΔPvu and SiahΔN did not have any repressive effect (data not shown). The two OBF-1 point mutants mentioned above also behaved in a manner expected from their binding behavior; OBF-L32P is impaired in interaction with the Oct-1 POU domain and therefore poorly activates transcription. However, this mutant interacted well with Siah-1, with the consequence that transcription activation was antagonized by Siah-1, as seen in Figure 5C. In contrast, OBF-V51E activates transcription efficiently, but was not targeted for degradation by Siah-1. Accordingly, transcription activation by this mutant was only little affected by Siah-1. Thus, Siah-1 specifically antagonizes transcription activation by OBF-1 through promoting its degradation.
In B cells, endogenous OBF-1 protein is degraded via the ubiquitin-proteasome pathway
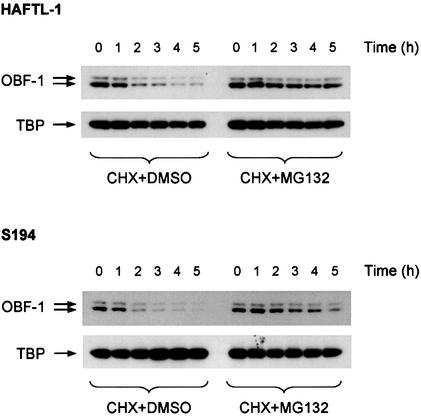
The data presented so far demonstrate that Siah-1 and OBF-1 interact specifically and that this interaction results in downregulation of OBF-1 protein level, which can be prevented by inhibition of the ubiquitin-proteasome pathway. As a consequence, transcriptional activation of target promoters is reduced. To test whether in B cells OBF-1 protein levels might indeed be controlled through the ubiquitin-proteasome pathway, we measured the amount of OBF-1 protein present in several B-cell lines after treatment with the proteasome inhibitor MG132 (Palombella et al., 1994; Rock et al., 1994). For these experiments cells were treated with cycloheximide in order to inhibit protein synthesis. As shown in Figure 6, in the mouse B-cell lines HAFTL-1sc1 and S194 the amount of endogenous OBF-1 decreased rapidly when protein synthesis was blocked (left half), showing a half-life of ∼2 h. However, when the proteasome inhibitor was added, a rescue of OBF-1 level was obtained (right half). In contrast, under the same conditions TBP was found to be stable and was not affected by inhibition of the proteasome. Similar results were also obtained in the B-cell line P3X63-Ag8.653 (not shown). Together, these data show that endogenous OBF-1 is subject to degradation through the proteasome pathway, which is likely to involve Siah-1.
Fig. 6. Inhibition of the ubiquitin-proteasome pathway in B cells leads to upregulation of endogenous OBF-1 levels. The B-cell lines HAFTL-1 and S194 were treated with 20 µg/ml cycloheximide together with 25 µM proteasome inhibitor MG132 or dimethylsulfoxide as a control. At the indicated time points cell extracts were prepared and analyzed by western blotting with an anti-OBF-1 antibody. After stripping, the filters were reprobed with an anti-TBP antibody.
In activated primary B cells OBF-1 is upregulated post-transcriptionally, consistent with a role for Siah-1
Previous work has shown that in splenic B cells OBF-1 expression can be induced, both at the RNA and protein level, by stimulation with LPS or CD40/IL-4 (Qin et al., 1998; Greiner et al., 2000). It has also been found that OBF-1 protein levels are particularly high in germinal center (GC) B cells, where OBF-1 plays a critical role (Qin et al., 1998; Greiner et al., 2000). Here we sought to determine whether the dramatic upregulation of OBF-1 protein observed in GC B cells is controlled at the level of transcription or whether it reflects a post-transcriptional regulation mechanism.
For this, mice were immunized with DNP-KLH and 10 days later splenic B cells were isolated and separated by centrifugation into high density and low density B cells, the latter corresponding mostly to activated GC cells (Thompson et al., 1984). In these two B-cell fractions the levels of OBF-1 protein and RNA were examined and correlated to the level of Siah-1 RNA, since no antibody against Siah was available. As shown in Figure 7, the amount of OBF-1 protein was dramatically increased in low density B cells, consistent with the reported upregulation of this factor in GC B cells. Strikingly, however, the amount of OBF-1 mRNA was unchanged, or even slightly less in the low density fraction, indicating that this upregulation takes place almost entirely at the post-transcriptional level. Furthermore, the level of Siah-1 mRNA was found to be moderately, but significantly, decreased in activated (low density) B cells, as would be expected if downregulation of Siah-1 were part of the mechanism controlling the amount of OBF-1 in GC B cells.
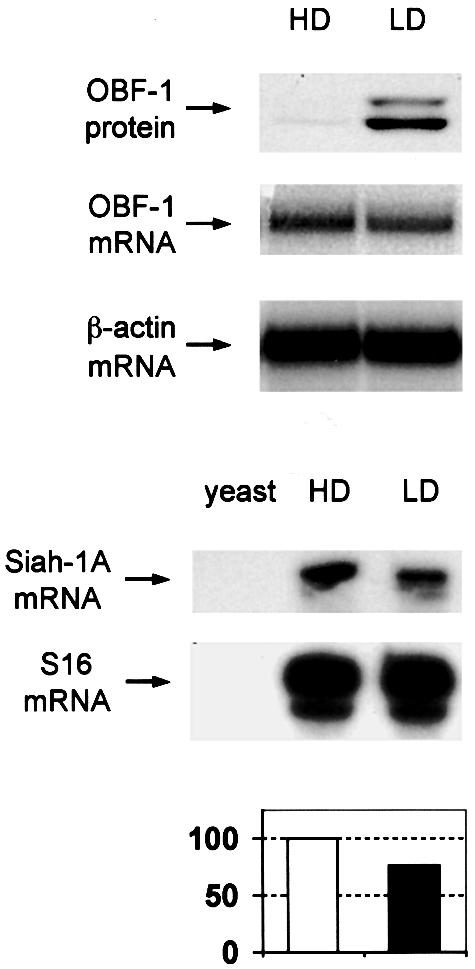
Fig. 7. In activated primary B cells, OBF-1 protein levels are elevated at the post-transcriptional level. Mice (C57Bl6) were immunized with 150 µg of DNP-KLH adsorbed to aluminium hydroxide, and 10 days later splenic B cells were isolated and centrifuged over Percoll step gradients. High density (HD) and low density (LD) B cells were isolated, and protein and RNA prepared simultaneously. Levels of OBF-1 protein were determined by western blot analysis with an anti-OBF-1 antibody. OBF-1 RNA was measured by northern blotting, also using a probe for β-actin as control. The level of Siah-1 RNA was determined by RNase protection assay, using a probe for the ribosomal protein S16 as control. A control hybridization with yeast RNA is also shown. In the lower part, a Phosphorimager quantification of these data is presented, using the level of S16 RNA for normalization.
Discussion
Siah-1, a novel partner of OBF-1
Here we have identified the RING finger protein Siah-1 as a binding partner for the B-cell-specific transcriptional coactivator OBF-1. Using the N-terminal half of OBF-1 as a bait to screen a B-cell-derived cDNA library, several independent isolates of Siah-1 were obtained and specific interaction between these two proteins was confirmed in vitro and in vivo. We found that in transfected cells the Siah–OBF-1 interaction leads to downregulation of OBF-1 expression at the protein level and a concomitant decrease in transcription activation of octamer site-containing reporter plasmids. OBF-1 downregulation was dependent on interactions between the two proteins, and required an intact RING finger within Siah-1. Furthermore, it could be prevented to a large extent by inhibition of the proteasome, consistent with the known mode of action of RING finger proteins. The significant but not complete rescue by proteasome inhibition may suggest that part of the downregulation observed here is independent of the proteasome. Precisely this was found to be the case for the observed inhibition of the Vav pathway by Siah-2 (Germani et al., 1999); yet treatment of several B-cell lines with a proteasome inhibitor resulted in elevated OBF-1 protein levels, indicating that in B cells OBF-1 turnover indeed involves degradation by the ubiquitin-proteasome pathway. Finally, we found that in primary activated B cells, the OBF-1 protein is dramatically upregulated, however without increase in RNA level. Given the high conservation between Siah-1 (in the mouse, Siah-1a and -1b) and Siah-2, it appears likely that Siah-2 is also able to interact with OBF-1 and in fact such an interaction has been shown (T.Wirth, personal communication). Siah genes are expressed in a broad range of organs and cells (Della et al., 1993), and from RT–PCR analysis, mouse Siah-1a, the homolog of the protein we identified, appears to be the most expressed Siah family member in murine splenic B cells (data not shown).
Structural aspects of the interaction between Siah-1 and OBF-1
Previous work has shown that OBF-1 makes a ternary complex with the POU domain of Oct-1 and Oct-2 and the octamer site DNA. Genetic and biochemical studies demonstrated that OBF-1 interacts with both moieties of the POU domain, the POUS and the POUH subdomains, as well as with the DNA (Babb et al., 1997; Sauter and Matthias, 1998). These multiple interactions may lead to stabilization of the Oct protein on its cognate site. A recent crystal structure of the Oct-1 POU domain in complex with the octamer site and a peptide comprising residues 1–44 of OBF-1 has confirmed these observations and defined the direct contacts between OBF-1 and residues in the POUS and POUH subdomains and in the DNA (Chasman et al., 1999). Interestingly, the novel interaction identified here with Siah-1 also takes place through amino acids located in the N-terminus of OBF-1, in a region that is broadly overlapping with the segment required for binding to the POU domain. Two deletion mutants of OBF-1, lacking residues 1–45 or 46–91, respectively, both failed to interact with Siah-1 in vitro (Figure 2D and E; or in vivo, data not shown). In addition, two OBF-1 point mutants gave additional useful information: OBF-L32P, which fails to interact efficiently with the POU domain (Gstaiger et al., 1996), still bound well to Siah-1. On the contrary, OBF-V51E, which does not impair interaction with Oct proteins, disrupted binding to Siah-1. Thus, the regions required for interaction with Oct and Siah-1 are overlapping, but distinct contacts are made. It remains to be determined whether, for example, Siah-1 may be able to bind to OBF-1 complexed with the POU domain and the DNA, and if so, whether it might also indirectly target the Oct factors for degradation. In Siah-1, residues within the C-terminal 72 amino acids are necessary for binding to OBF-1. Although the precise Siah amino acids involved in interaction have not been defined in any of the published studies, in each case (N-CoR, DCC or the kinesin Kid) the interaction involved the C-terminal half of Siah (Hu et al., 1997b; Germani et al., 2000).
The binding of Siah-1 to OBF-1 identifies a novel regulatory mechanism operating in B cells
One of the most striking defects in OBF-1-deficient mice is their dramatically reduced immune response accompanied by a total absence of germinal center development. Thus, in the absence of OBF-1 a very late stage of B-cell maturation is affected, which takes place in secondary lymphoid organs in response to external stimuli, in particular CD40 signaling. This late phenotype had been somewhat unexpected because OBF-1 expression at the RNA level had been found to be rather constant in B-cell lines representing different stages of B-cell differentiation. A partial explanation came from the realization that the OBF-1 protein is upregulated in GC B cells (Qin et al., 1998) or in cell lines derived thereof (Greiner et al., 2000), and that in purified splenic B cells, treated in vitro with CD40L and IL-4 or CD40L and anti-IgM, OBF-1 mRNA and protein expression can be induced (Qin et al., 1998; our unpublished results). Because CD40 signaling is essential for the development of GCs, it has been assumed that upregulation of OBF-1 RNA expression by CD40L leads to the increased OBF-1 protein amount observed in GC cells (Qin et al., 1998). Here we found that in primary activated B cells that had been stimulated in vivo (i.e. in cells isolated from immunized mice) there is indeed a dramatic upregulation of OBF-1 protein, but there is no increase in OBF-1 RNA (Figure 7). Thus, the upregulation of OBF-1 protein level observed in vivo must take place, largely if not exclusively, at the post-transcriptional level. The identification of an interaction between OBF-1 and a RING finger protein known to target other proteins to proteasomal degradation offers an attractive solution to this paradox. A central tenet of the model proposed here is that Siah protein level (or activity) becomes downregulated as cells are activated in vivo. Since antibodies against Siah-1 were not available, we could not directly test the hypothesis that downregulation of Siah-1 protein level correlates with the upregulation of OBF-1 observed in GC cells. However, analysis of Siah-1 RNA showed that its level decreases moderately upon in vivo B-cell activation, in good agreement with the proposed model. In addition, in B cells stimulated in vitro with anti-CD40 or anti-CD40 and IL-4 the Siah-1 mRNA level was found to decrease moderately (data not shown). In transfected cells we have observed that small differences in the respective amounts of Siah-1 and OBF-1 can have important consequences regarding the magnitude of OBF-1 protein downregulation. Therefore, it seems reasonable to assume that in B cells, even relatively subtle changes in the amounts (or activity) of Siah-1 may lead to rather large differences in OBF-1 protein levels. Furthermore, in multiple instances it has been shown that degradation of proteins by the proteasome is ultimately modulated by post-translational modification, such as phosphorylation, either of the target protein itself or of the RING finger protein. For example, the tumor suppressor protein p53 is normally degraded by the ubiquitin-proteasome pathway, and for this, interaction with the RING finger protein MDM2 is critical. In response to DNA damage, p53 becomes phosphorylated by the kinase Chk2 on Ser20; this then leads to disruption of the p53–MDM2 interaction and p53 stabilization (Caspari, 2000). In this respect it is worth mentioning that OBF-1 has been found to be phosphorylated in its C-terminal portion (Zwilling et al., 1997). Mutation of the previously described phosphorylation site S184 to either A, D or E, however, had no impact on OBF-1 downregulation in transfection assays (data not shown).
Finally, our finding that treatment of several B-cell lines with a proteasome inhibitor leads to a significant upregulation of OBF-1 protein lends additional support to the notion that in vivo OBF-1 is, at least partly, regulated at the post-transcriptional level.
Materials and methods
Yeast two-hybrid screen
The yeast two-hybrid screen (Fields and Song, 1989) in this study was conducted using the ‘interaction trap’ version (Gyuris et al., 1993). In brief, the yeast strain EGY48, which contains a LEU2 gene under the transcriptional control of multimerized lexA operator elements, was transformed with a bait vector expressing a fusion of lexA and the first 114 aa of human OBF-1, and with a library derived from the human B-cell line Namalwa, which expressed the containing cDNAs as fusions to the VP16 activation domain (Strubin et al., 1995).
Primary transformants (3 × 106) were screened on medium lacking leucine, and from this Siah-1 was isolated independently four times.
To confirm interaction in yeast, a second lexA-dependent reporter bearing a lacZ gene was transfomed together with the respective expression plasmids, and β-galactosidase activity was measured by standard protocols.
Bacterial and in vitro protein expression
To generate GST fusion proteins, the relevant DNA fragments were cloned into pGex2T (Pharmacia) and transformed into the bacterial strains BL21 or TOPP (Stratagene). Bacteria were grown at room temperature and induced for 2 h with 0.4 mM isopropyl-β-d-thiogalactopyranoside before lysis. Lysates were prepared by treatment with lysozyme and repeated freeze and thaw followed by ultracentrifugation to remove cell debris. GST fusion proteins were purified on glutathione–Sepharose beads.
In vitro translation was performed with clones in the plasmid pBGO-ATG using the ‘TnT’ system (Promega). Proteins were radiolabeled during synthesis with [35S]methionine (Amersham).
Cell culture and protein expression in eukaryotic cells
Adherent cells were maintained in Dulbecco’s modified Eagle’s medium plus 10% fetal calf serum (FCS). All B-cell lines were grown in RPMI + 10% FCS + 50 µM β-mercaptoethanol.
For protein expression, OBF-1 fragments were PCR-cloned into the vector pBact (kindly provided by A.Matus; Cravchik and Matus, 1993) to carry a myc tag at the N-terminus. Template plasmids containing OBF-1 point mutations were a gift from W.Schaffner (Gstaiger et al., 1996); their relevant portion was subcloned into pBact by PCR.
Siah-1 derivatives were cloned into pCDNA3 and carry an HA tag at the N-terminus. SiahΔN was generated using the internal BamHI site and C-terminal truncations were made using the enzymes indicated in the name. Further details about the clones used in this study are available on request.
HEK 293T cells were transfected in six-well plates using the calcium phosphate coprecipitation method. Generally and if not indicated otherwise in the respective figure, 500 ng of each construct were transfected and samples were filled up to have equal amounts of each promoter.
L929 cells were transfected using Lipofectamine 2000 (Gibco) according to the manufacturer’s instructions. One microgram of each construct was used.
Protein extract, western blotting and luciferase assay
Adherent cells were lysed in 1% Triton X-100, 10% glycerol, 25 mM Tris–HCl pH 7.8, 2 mM dithiothreitol (DTT), 2 mM EDTA and ‘complete’ protease inhibitor cocktail (Roche). Protein concentrations were determined with Bradford reagent (Bio-Rad) and 30 µg of protein for each sample were resolved by SDS–PAGE with subsequent transfer onto Immobilon membrane (Millipore). Myc-tagged proteins were detected using the monoclonal antibody 9E10, HA-tagged proteins with the monoclonal antibody 12CA5. Endogenous TBP was detected using the monoclonal antibody SL39 (kindly provided by N.Hernandez). Bands were visualized with the ECL system (Amersham).
B cells were lysed by repeated cycles of freeze and thaw in 20 mM HEPES pH 7.9, 20% glycerol, 400 mM NaCl, 0.5 mM EDTA, 0.25 mM EGTA, 0.025% NP-40, 1 mM DTT, 0.5 mM NaF and protease inhibitor cocktail. After western blotting as above, endogenous OBF-1 was detected using a polyclonal antibody that was raised in rabbits against the C-terminal 154 amino acids of murine OBF-1.
To measure luciferase activity, reporter plasmids were cotransfected as indicated in Figure 5, and cell lysates generated as described above were measured according to standard protocols.
Co-immunoprecipitation and in vitro binding assays
For co-immunoprecipitation, HEK 293T cells were transfected by the calcium phosphate method in 60-mm dishes with the constructs indicated in Figure 1B. After 48 and 6 h treatment with MG262, cells were lysed as described above and ∼500 µg of extract were incubated for 1 h on ice with 100 µl of 12CA5 hybridoma supernatant in 1 ml of IP buffer. After this, 20 µl of protein A–Sepharose slurry were added and samples were incubated overnight at 4°C with gentle agitation. Beads were washed three times with IP buffer and subsequently resuspended and boiled in 20 µl of loading buffer for SDS–PAGE.
IP buffer was 20 mM HEPES pH 7.4, 0.5 mM EDTA, 150 mM NaCl and 0.1% Triton X-100.
For the in vitro binding assay, GST fusion proteins on glutathione beads were incubated with either in vitro translated radiolabeled protein or with extract from transfected cells. The binding and washing of the beads was done in 100 mM NaCl, 40 mM HEPES pH 7.9, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride and 0.05% NP-40. Washed beads were boiled in loading buffer and bound proteins were run on denaturing gels followed by either autoradiography or western blotting. Generally, in lanes labeled ‘input’ 1/10 of the material used for the binding reaction was loaded.
Northern blotting
RNA was prepared by Trizol (Gibco) according to the manufacturer’s instructions. Total RNA (15 µg) was analyzed by northern blotting as described in Qiagen News (Issue 2/3, 2000) using Hybond-XL membranes (Amersham Pharmacia). OBF-1 was visualized with a radiolabeled coding region probe.
Proteasome inhibitor and cycloheximide treatment
For L929 cells, the highly potent proteasome inhibitor MG262 (Affiniti; McCormack et al., 1997) was used at a concentration of 500 nM and treatment lasted 12 h, unless indicated otherwise.
B cells were treated with 25 µM proteasome inhibitor MG132 (Sigma; Palombella et al., 1994; Rock et al., 1994) and 20 µg/ml cycloheximide for the times indicated in the respective figure.
Immunization and splenic cell fractionation
Mice were injected intraperitoneally with 150 µg of aluminium hydroxide-adsorbed DNP-KLH. After 10 days, mice were killed, splenic cells were isolated and depleted for T cells by magnetic beads coupled to the pan-T-cell marker Thy1.2 (Dynal). The remaining cells (mainly B cells) were separated into high density (largely resting) and low density (largely activated) B cells by Percoll gradient centrifugation as described in Coligan (1992). Protein and RNA were prepared as described above.
RNase protection assay
RNase protection was carried out as described earlier (Schubart et al., 1996b). In brief, a probe was in vitro transcribed from an IMAGE clone (clone ID 523455) that contained part of the murine Siah-1A cDNA. The gel-purified radiolabeled probe was afterwards hybridized with 20 µg of total RNA at 50°C over night. After RNase digestion, RNA was resolved on a 5% polyacrylamide gel and autoradiographed. The protected part of the murine Siah-1A cDNA covered the region from approximately position +90 to +370 to give a band of 280 bases.
Acknowledgments
Acknowledgements
We wish to thank O.Georgiev, M.Gstaiger and W.Schaffner for providing plasmids with OBF-1 point mutations, Y.Nagamine, I.Galetic and M.Hill for critical comments on the manuscript, M.Gstaiger, A.Hergovich and J.Lisztwan for technical help, and laboratory members for discussions. This work was supported by the Novartis Research Foundation.
Note added in proof
Independently, similar results are reported by Boehm et al. on pages 4153–4162 of this issue.
References
- Babb R., Cleary,M.A. and Herr,W. (1997) OCA-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol. Cell. Biol., 17, 7295–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T. (2000) How to activate p53. Curr. Biol., 10, R315–R317. [DOI] [PubMed] [Google Scholar]
- Chasman D., Cepek,K., Sharp,P.A. and Pabo,C.O. (1999) Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein–DNA interface. Genes Dev., 13, 2650–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. (1998) The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J., 17, 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan J.E. (1992) Current Protocols in Immunology. Greene Publishing and Wiley Interscience, New York, NY.
- Cravchik A. and Matus,A. (1993) A novel strategy for the immunological tagging of cDNA constructs. Gene, 137, 139–143. [DOI] [PubMed] [Google Scholar]
- Della N.G., Senior,P.V. and Bowtell,D.D. (1993) Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development, 117, 1333–1343. [DOI] [PubMed] [Google Scholar]
- Dickson B.J. (1998) Photoreceptor development: breaking down the barriers. Curr. Biol., 8, R90–R92. [DOI] [PubMed] [Google Scholar]
- Fields S. and Song,O. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Freemont P.S. (2000) RING for destruction? Curr. Biol., 10, R84–R87. [DOI] [PubMed] [Google Scholar]
- Germani A., Romero,F., Houlard,M., Camonis,J., Gisselbrecht,S., Fischer,S. and Varin-Blank,N. (1999) hSiah2 is a new Vav binding protein which inhibits Vav-mediated signaling pathways. Mol. Cell. Biol., 19, 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germani A., Bruzzoni-Giovanelli,H., Fellous,A., Gisselbrecht,S., Varin-Blank,N. and Calvo,F. (2000) SIAH-1 interacts with α-tubulin and degrades the kinesin Kid by the proteasome pathway during mitosis. Oncogene, 19, 5997–6006. [DOI] [PubMed] [Google Scholar]
- Greiner A., Müller,K.B., Hess,J., Pfeffer,K., Muller-Hermelink,H.K. and Wirth,T. (2000) Up-regulation of BOB.1/OBF.1 expression in normal germinal center B cells and germinal center-derived lymphomas. Am. J. Pathol., 156, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M., Knoepfel,L., Georgiev,O., Schaffner,W. and Hovens,C.M. (1995) A B-cell coactivator of octamer-binding transcription factors. Nature, 373, 360–362. [DOI] [PubMed] [Google Scholar]
- Gstaiger M., Georgiev,O., van Leeuwen,H., van der Vliet,P. and Schaffner,W. (1996) The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J., 15, 2781–2790. [PMC free article] [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Holloway A.J., Della,N.G., Fletcher,C.F., Largespada,D.A., Copeland,N.G., Jenkins,N.A. and Bowtell,D.D. (1997) Chromosomal mapping of five highly conserved murine homologues of the Drosophila RING finger gene seven-in-absentia. Genomics, 41, 160–168. [DOI] [PubMed] [Google Scholar]
- Hu G. and Fearon,E.R. (1999) Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol., 19, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Chung,Y.L., Glover,T., Valentine,V., Look,A.T. and Fearon,E.R. (1997a) Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics, 46, 103–111. [DOI] [PubMed] [Google Scholar]
- Hu G., Zhang,S., Vidal,M., Baer,J.L., Xu,T. and Fearon,E.R. (1997b) Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev., 11, 2701–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P.K., Eldridge,A.G., Freed,E., Furstenthal,L., Hsu,J.Y., Kaiser,B.K. and Reimann,J.D. (2000) The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol., 10, 429–439. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A. and Weissman,A.M. (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell, 102, 549–552. [DOI] [PubMed] [Google Scholar]
- Kim U., Qin,X.F., Gong,S., Stevens,S., Luo,Y., Nussenzweig,M. and Roeder,R.G. (1996) The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature, 383, 542–547. [DOI] [PubMed] [Google Scholar]
- Laumen H., Nielsen,P.J. and Wirth,T. (2000) The BOB.1/OBF.1 co-activator is essential for octamer-dependent transcription in B cells. Eur. J. Immunol., 30, 458–469. [DOI] [PubMed] [Google Scholar]
- Li S., Li,Y., Carthew,R.W. and Lai,Z.C. (1997) Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell, 90, 469–478. [DOI] [PubMed] [Google Scholar]
- Lorick K.L., Jensen,J.P., Fang,S., Ong,A.M., Hatakeyama,S. and Weissman,A.M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA, 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. and Roeder,R.G. (1995) Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol., 15, 4115–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Ge,H., Stevens,S., Xiao,H. and Roeder,R.G. (1998) Coactivation by OCA-B: definition of critical regions and synergism with general cofactors. Mol. Cell. Biol., 18, 3803–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack T. et al. (1997) Active site-directed inhibitors of Rhodococcus 20 S proteasome. Kinetics and mechanism. J. Biol. Chem., 272, 26103–26109. [DOI] [PubMed] [Google Scholar]
- Nielsen P.J., Georgiev,O., Lorenz,B. and Schaffner,W. (1996) B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur. J. Immunol., 26, 3214–3218. [DOI] [PubMed] [Google Scholar]
- Palombella V.J., Rando,O.J., Goldberg,A.L. and Maniatis,T. (1994) The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell, 78, 773–785. [DOI] [PubMed] [Google Scholar]
- Pfisterer P., Zwilling,S., Hess,J. and Wirth,T. (1995) Functional characterization of the murine homolog of the B-cell-specific coactivator BOB.1/OBF.1. J. Biol. Chem., 270, 29870–29880. [DOI] [PubMed] [Google Scholar]
- Qin X.-F., Reichlin,A., Luo,Y., Roeder,R.G. and Nussenzweig,M.C. (1998) OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J., 17, 5066–5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K.L., Gramm,C., Rothstein,L., Clark,K., Stein,R., Dick,L., Hwang,D. and Goldberg,A.L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell, 78, 761–771. [DOI] [PubMed] [Google Scholar]
- Sauter P. and Matthias,P. (1998) Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol. Cell. Biol., 18, 7397–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart D.B., Rolink,A., Kosco-Vilbois,M.H., Botteri,F. and Matthias,P. (1996a) B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature, 383, 538–542. [DOI] [PubMed] [Google Scholar]
- Schubart D.B., Sauter,P., Massa,S., Friedl,E.M., Schwarzenbach,H. and Matthias,P. (1996b) Gene structure and characterization of the murine homologue of the B cell-specific transcriptional coactivator OBF-1. Nucleic Acids Res., 24, 1913–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart K., Massa,S., Schubart,D., Corcoran,L.M., Rolink,A.G. and Matthias,P. (2001) B cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nature Immunol., 2, 69–74. [DOI] [PubMed] [Google Scholar]
- Sourisseau T., Desbois,C., Debure,L., Bowtell,D.D., Cato,A.C., Schneikert,J., Moyse,E. and Michel,D. (2001) Alteration of the stability of Bag-1 protein in the control of olfactory neuronal apoptosis. J. Cell Sci., 114, 1409–1416. [DOI] [PubMed] [Google Scholar]
- Strubin M., Newell,J.W. and Matthias,P. (1995) OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell, 80, 497–506. [DOI] [PubMed] [Google Scholar]
- Thompson C.B., Scher,I., Schaefer,M.E., Lindsten,T., Finkelman,F.D. and Mond,J.J. (1984) Size-dependent B lymphocyte subpopulations: relationship of cell volume to surface phenotype, cell cycle, proliferative response, and requirements for antibody production to TNP-Ficoll and TNP-BA. J. Immunol., 133, 2333–2342. [PubMed] [Google Scholar]
- Tomilin A., Remenyi,A., Lins,K., Bak,H., Leidel,S., Vriend,G., Wilmanns,M. and Scholer,H.R. (2000) Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell, 103, 853–864. [DOI] [PubMed] [Google Scholar]
- Zhang J., Guenther,M.G., Carthew,R.W. and Lazar,M.A. (1998) Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev., 12, 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling S., Dieckmann,A., Pfisterer,P., Angel,P. and Wirth,T. (1997) Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science, 277, 221–225. [DOI] [PubMed] [Google Scholar]