Abstract
In tobacco cells, osmotic stress induced the rapid activation of two protein kinases that phosphorylate myelin basic protein. Immunological studies demonstrated that the 48-kD kinase is the salicylic acid–induced protein kinase (SIPK), a member of the mitogen-activated protein kinase family. SIPK was activated 5 to 10 min after the cells were exposed to osmotic stresses, and its activity persisted for ∼30 min. In contrast, the 42-kD kinase was activated within 1 min after osmotic stress, and its activity was maintained for ∼2 hr. Moreover, in addition to myelin basic protein, the 42-kD kinase phosphorylated casein and two transcription factors, c-Jun and ATF-2. This latter enzyme was inactivated by a serine/threonine–specific phosphatase but, unlike SIPK, was not affected by a tyrosine-specific phosphatase. After the 42-kD kinase was purified to apparent homogeneity, tryptic peptide analysis indicated that it is a homolog of Arabidopsis serine/threonine kinase1 (ASK1).
INTRODUCTION
In nature, plants are exposed to ever-changing environmental conditions. They frequently must withstand drought, temperature extremes, and high salinity. To survive these challenges, plants have developed adaptive mechanisms that manifest themselves in morphological, physiological, developmental, and molecular changes (reviewed in Cheeseman, 1988; Bohnert et al., 1995; Bray, 1997). To date, intracellular pathways leading to these adaptive changes have been poorly understood, although reversible protein phosphorylation is likely to play a crucial role in mediating intracellular response to external stimuli in plants, as has been amply demonstrated in animal and yeast systems. Indeed, changes in protein phosphorylation have been observed when plants are exposed to hormones (Reddy et al., 1987; Koritsas, 1988; Mizoguchi et al., 1994), light (Short and Briggs, 1990; Fallon and Trewavas, 1994), pathogens (Dietrich et al., 1990; Felix et al., 1991; Vriard et al., 1994; Suzuki and Shinshi, 1995), cold (Monroy et al., 1993), and water deficit (Conley et al., 1997).
In mammalian and yeast cells, one of the main pathways that transduces signal from the cell membrane to the nucleus, and leads to expression of the appropriate response genes, is the mitogen-activated protein kinase (MAPK) cascade (reviewed in Errede and Levin, 1993; Karin, 1994; Cano and Mahadevan, 1995; Herskowitz, 1995; Seger and Krebs, 1995; Kyriakis and Avruch, 1996; Treisman, 1996). In general, mammalian growth factors activate the extracellular signal-regulated kinases 1 and 2 (ERK1/ERK2) subfamily of MAPKs (reviewed in Robinson and Cobb, 1997). In contrast, stresses such as osmotic and heat shock, UV irradiation, or exposure to cytokines activate two other MAPK subfamilies, the stress-activated protein kinase or c-Jun N-terminal kinase (SAPK/JNK) and the p38 subfamilies (reviewed in Canman and Kastan, 1996; Kyriakis and Avruch, 1996).
Numerous protein kinases with close sequence similarities to MAPKs and other kinases belonging to the MAPK cascade have been identified in plants (reviewed in Jonak et al., 1994; Nishihama et al., 1995; Stone and Walker, 1995; Hirt, 1997; Mizoguchi et al., 1997). Activation of MAPKs has been observed in barley aleurone protoplasts treated with abscisic acid (Knetsch et al., 1996) as well as in plants exposed to pathogens (Suzuki and Shinshi, 1995; Adam et al., 1997; Ligterink et al., 1997; Zhang and Klessig, 1997, 1998b; Zhang et al., 1998), cold (Jonak et al., 1996), drought (Jonak et al., 1996), and wounding (Seo et al., 1995; Usami et al., 1995; Bögre et al., 1996, 1997; Zhang and Klessig, 1998a; Seo et al., 1999). These results suggest that the MAPK cascade is involved in stress signaling in plants and that many general mechanisms of signal transduction are common among all eukaryotic cells. However, distinct differences between the signaling pathways in plants and mammals or yeast preclude drawing strict analogies between these groups. For example, marked increases of mRNAs encoding several of the MAPKs and their upstream kinases in this cascade have been shown to occur in plants under environmental stress (Seo et al., 1995; Jonak et al., 1996; Mizoguchi et al., 1996; Shinozaki and Yamaguchi-Shinozaki, 1997; Zhang and Klessig, 1998a, 1998b). Moreover, whereas the three subfamilies of MAPKs in animals can be distinguished by the X (an unspecified amino acid) in the TXY motif (ERK1 and ERK2, TEY; SAPK/JNK, TPY; and p38, TGY), all plant MAPKs reported to date possess the TEY motif. In addition, some of the protein kinases involved in signaling appear to be unique to plants (reviewed in Stone and Walker, 1995). The most prevalent of these are the calcium-dependent protein kinases (Harper et al., 1991), which participate in transduction of stress signals in plants (Sheen, 1996).
The main goal of this study was to identify protein kinases involved in immediate responses to salinity stress in plants. In tobacco cells exposed to 250 mM NaCl, rapid activation of two protein kinases was observed. One of these kinases belongs to the MAPK family, whereas the second is a homolog of Arabidopsis serine/threonine kinase 1 (ASK1), a member of the sucrose nonfermenting 1 (SNF1) kinase family. These two enzymes are also activated by high concentrations of sorbitol, which suggests their involvement in the response to osmotic stress in plant cells.
RESULTS
Salinity Stress Transiently Activates the 48- and 42-kD Protein Kinases in Tobacco Cells
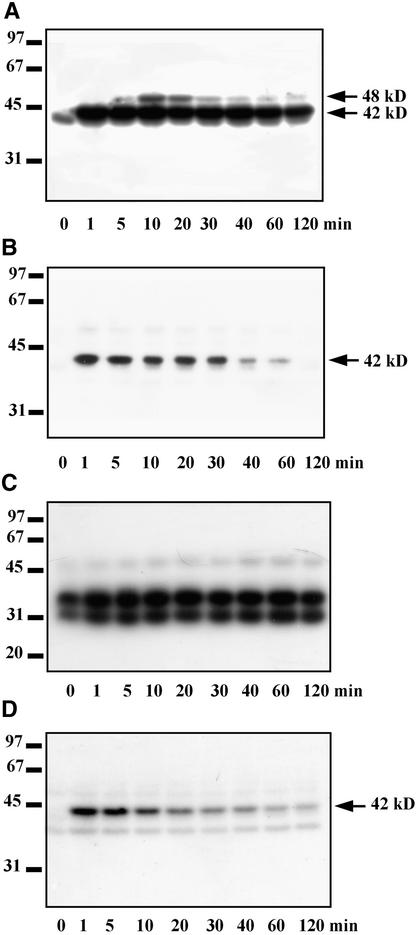
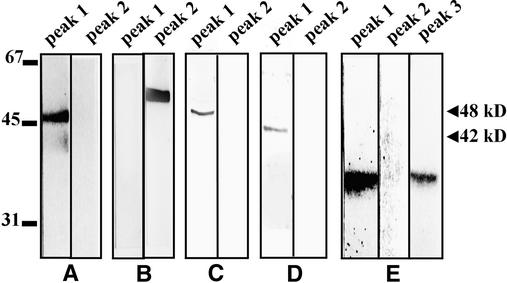
To identify protein kinases involved in response to salinity stress, we monitored the activity of protein kinases present in tobacco cells exposed to 250 mM NaCl by using in-gel kinase assays with myelin basic protein (MBP), ATF-2, and casein as substrates. Salt stress caused a rapid and transient activation of two protein kinases that phosphorylate MBP. The molecular mass of one of these kinases is ∼48 kD, whereas that of the other is 42 kD (Figure 1A). Activity of the 48-kD kinase appeared ∼5 min after the addition of salt and reached a maximum by 10 min, returning to the control value within 30 min. The 42-kD kinase was activated even more rapidly, responding strongly within 1 min of salt treatment and maintaining this activity for >2 hr (Figure 1A).
Figure 1.
Identification of Protein Kinases Activated by Salinity Stress.
Tobacco suspension culture cells were treated with 250 mM NaCl. Aliquots of the culture were taken at the indicated times, and kinase activities were analyzed in cell cytosolic extracts by the in-gel kinase assays using MBP, recombinant GST–ATF-2, or casein as the substrates. Molecular mass markers are given at left in kilodaltons. The 42 and 48 kD at right indicate molecular masses of the protein kinases activated by salinity stress.
(A) Identification of protein kinases that phosphorylate MBP.
(B) Identification of protein kinases that phosphorylate ATF-2.
(C) and (D) Identification of protein kinases that phosphorylate casein. The assay shown in (D) was performed in the presence of 100 μg/mL heparin.
The in-gel kinase assays using immobilized ATF-2 or casein also indicated the activation of a protein kinase(s) with molecular mass(es) of ∼42 kD (Figures 1B and 1D); the 48-kD kinase showed no activity with these substrates. The 42-kD protein kinase(s) became fully active 1 min after the addition of salt; during the next 2 hr, its (their) activity gradually declined to a basal level. When casein was the substrate, the 42-kD kinase activated by salt stress was detected only when casein kinases CK1 and CK2 were inhibited by heparin, a potent casein kinase inhibitor (Pinna, 1990) (Figures 1C and 1D).
Activation of the 48- and 42-kD Protein Kinases Is Also Induced by Hyperosmotic Stress
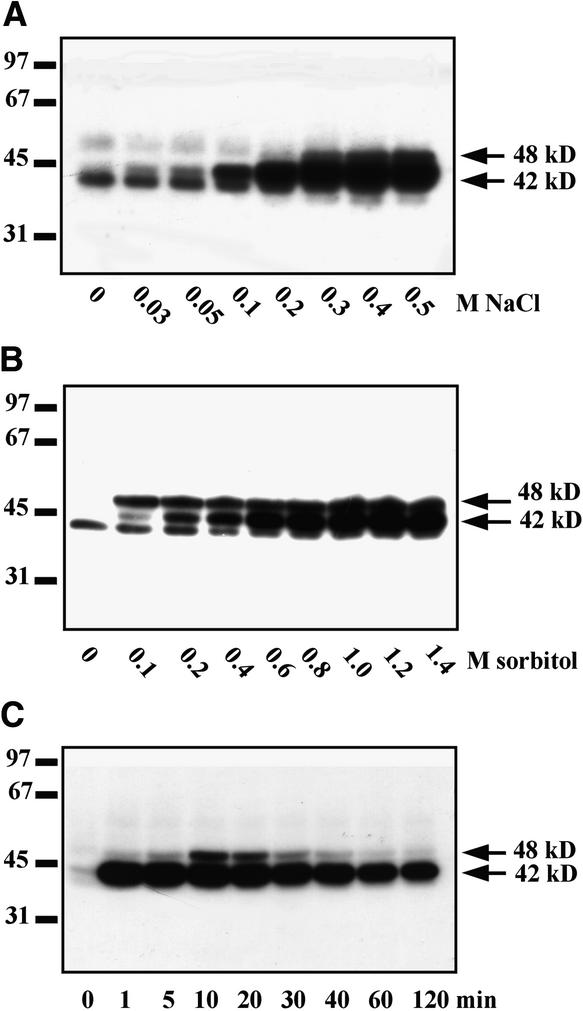
To determine whether activation of these protein kinases was associated with the salinity stress exclusively or with a more general response to osmotic stress, we analyzed the effect of various concentrations of NaCl and sorbitol on the activities of the two kinases, using MBP as a substrate (Figures 2A and 2B). Both stresses caused the activation of the 48- and 42-kD kinases. A third kinase of slightly lower molecular mass was also evident in Figures 2A and 2B, but its activity was not increased by either NaCl or sorbitol and may even have been suppressed.
Figure 2.
Activation of Protein Kinases in Tobacco Cells Exposed to NaCl and Sorbitol Stress.
(A) and (B) Dose–response. Tobacco cells were treated with various concentrations of NaCl (A) or sorbitol (B) for 10 min, and protein kinase activities were monitored in cell extracts by using an in-gel kinase assay with MBP as substrate.
(C) Time course. Tobacco cells were treated with 900 mM sorbitol. Aliquots of the culture were taken at the indicated times, and kinase activities were analyzed in cell extracts by the in-gel kinase assay with MBP as substrate.
Molecular mass markers are given at left in kilodaltons. The 42 and 48 kD at right indicate molecular masses of the protein kinases activated by salinity stress.
We also compared the time courses of kinase activation by 250 mM NaCl and 900 mM sorbitol. The results showed a very similar pattern of protein kinase activation for both stressors (Figures 1A and 2C), suggesting that the 48- and 42-kD kinases can indeed be classified as osmotic stress–responsive kinases. Based on the dose response and time course of activation (Figures 2A and 1A, respectively), tobacco cells treated for 10 min with 250 mM NaCl or 900 mM sorbitol were chosen as the material for further characterization of osmotic stress–activated protein kinases.
Separation of Osmotic Stress–Activated Protein Kinases on a Mono Q Column
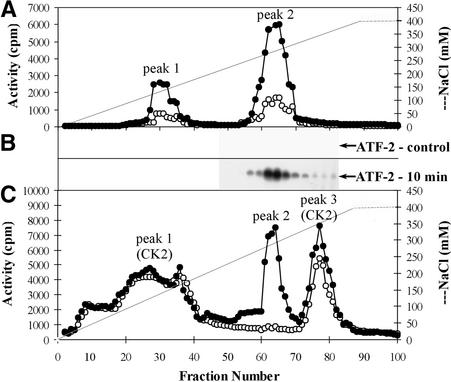
The osmotic stress–activated kinases from cytosolic extracts of tobacco cells treated for 10 min with 250 mM NaCl were separated by chromatography on a Mono Q column at the same time as extracts from untreated (control) cells. Each eluted fraction was independently tested for kinase activity with one of the following substrates: MBP, recombinant ATF-2, or casein (Figures 3A to 3C). The elution profile showed two main peaks of activity with MBP as substrate. Peak 1 eluted at 150 mM NaCl, and peak 2 eluted at 300 mM NaCl (Figure 3A). Protein kinase activity in both peaks was three to four times greater in extracts from cells exposed to salt than in extracts from control cells. The kinase activity that phosphorylated ATF-2 eluted at 300 mM NaCl in a sharp peak located at the same position as peak 2 from the samples with MBP-phosphorylating activity (Figure 3B). In control cells, no ATF-2–phosphorylating activity was detected.
Figure 3.
Elution Profile of Protein Kinases from the Mono Q HR 5/5 Column.
Chromatography of cytosolic extracts of tobacco cells treated for 10 min with 250 mM NaCl (filled circles) and untreated (control) cells (open circles) on a Mono Q column was performed. Protein kinase activities were determined by an in-solution kinase assay (see Methods).
(A) Analysis of protein kinase activities that phosphorylate MBP.
(B) Analysis of protein kinase activities that phosphorylate ATF-2.
(C) Analysis of protein kinase activities that phosphorylate casein.
When casein was used as the substrate, three peaks of activity were found, eluting at 100 to 175, 300, and 350 mM NaCl (Figure 3C, peaks 1 to 3). Because the cell extracts also contained casein kinases (see Figures 1C and 1D), several approaches were used to determine whether any of the peaks corresponded to those kinases. Use of substrates specific for CK1 (RRKDLHDDEEDEAMSITA) and CK2 (RRRDDDSDDD) (Marin et al., 1994) showed that CK1 washed out in the flow-through of the Mono Q column (data not shown), whereas CK2 eluted in peak 1 (see Table 1) and peak 3. The presence of CK2 in peaks 1 and 3 was also demonstrated by using protein gel blot analysis with antibodies to the α subunit of CK2 (see below). Gel filtration confirmed that the monomeric form of CK2 (molecular mass ∼40 kD) was in peak 1, whereas the oligomeric form (molecular mass ∼135 kD) was responsible for the kinase activity in peak 3 (data not shown). With casein as substrate, the amounts of activity in peaks 1 and 3 were similar in extracts from the control and from NaCl-stressed tobacco, whereas the kinase activity in peak 2 was greatly increased in extracts from NaCl-treated cells (Figure 3C). Peak 2 eluted at the same salt concentration (300 mM) as the activity that phosphorylates MBP and ATF-2 (Figures 3A and 3B). Thus, this kinase activity, which utilizes casein as well as MBP and ATF-2, can be separated from typical casein kinases, which have more restrictive substrate specificities.
Table 1.
Substrate Specificity of the Osmotic Stress–Activated Protein Kinases
| Phosphorylation Ratea,b(pmol mL−1 min−1)
|
||
|---|---|---|
| Substrate | Peak 1 | Peak 2 |
| MBP | 25 | 32 |
| GST–ATF-2 (96) | NDc | 6 |
| GST–c-Jun (79) | ND | 5 |
| Casein | 28 | 16 |
| Casein + heparind | ND | 15 |
| RRRDDDSDDD | 42 | ND |
| RRRDDDSDDD + heparind | ND | ND |
Phosphorylation of the listed substrates by protein kinases eluted from the Mono Q column in peaks 1 and 2 was performed as described in Methods.
Phosphorylation rates represent the mean values from five independent experiments. Standard deviation was ∼10 to 15%.
ND, not detectable.
Heparin, an inhibitor of typical casein kinases, was introduced at a concentration of 100 μg/mL.
Estimation of Molecular Masses of the Osmotic Stress–Activated Protein Kinases
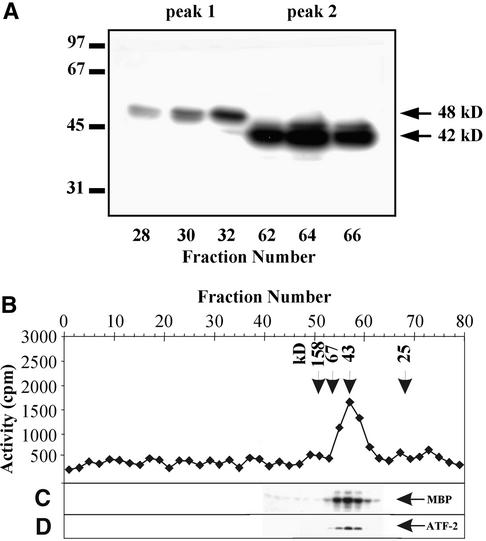
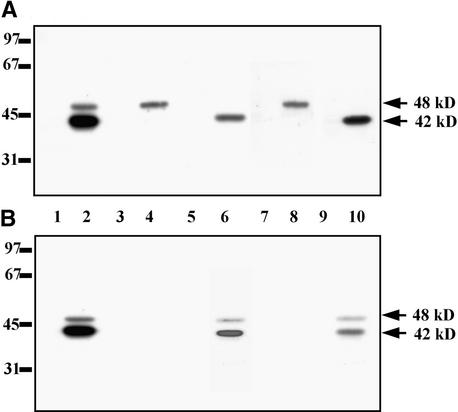
The enzymes that eluted in peaks 1 and 2 from the Mono Q column were subjected to the in-gel kinase assay with MBP as the substrate (Figure 4A). In peak 1, a kinase with a molecular mass of 48 kD was detected, whereas peak 2 contained a 42-kD kinase. In addition, the molecular mass of the kinase(s) that eluted in peak 2 was estimated by gel filtration on the Superose 12 column attached to a fast-performance liquid chromatography system. The kinase activity that eluted from the Superose 12 column was analyzed by using MBP (Figure 4B), casein (Figure 4C), or ATF-2 (Figure 4D) as substrates. All three substrates were phosphorylated by the kinase(s) that eluted in fractions 54 to 60 from the Superose 12 column. The molecular mass of the kinase(s) was estimated at 40 to 42 kD. Similar results were obtained by the in-gel kinase assay analysis (Figure 4A), indicating that osmotic stress–activated protein kinase(s) eluting in peak 2 from the Mono Q column is a monomeric 42-kD enzyme.
Figure 4.
Estimation of Molecular Masses of Protein Kinases Activated by Osmotic Stress.
(A) Enzymes eluted from the Mono Q column in peak 1 (fractions 28, 30, and 32) and peak 2 (fractions 62, 64, and 66) were analyzed by using an in-gel kinase assay with the MBP. Molecular mass markers are indicated in kilodaltons at left. The 42 and 48 kD at right indicate molecular masses of the protein kinases activated by salinity stress.
(B) to (D) The protein kinases eluted in peak 2 were chromatographed on the Superose 12 column. Protein kinase activities were assayed by using casein (B), MBP (C), or ATF-2 (D) as substrate. Arrowheads and numbers in kilodaltons at top indicate molecular mass markers.
Immunoblot of the Osmotic Stress–Activated Protein Kinases
The kinases isolated from salt-treated tobacco cells and eluted from the Mono Q column were subjected to immunoblot analysis with anti–ERK1/ERK2, anti-p38, anti-JNK1, anti–salicylic acid–induced protein kinase (SIPK), anti–wound-induced protein kinase (WIPK), and anti-CK2α antibodies. Anti–ERK1/ERK2 antibodies recognized a protein with a molecular mass of ∼48 kD in the pooled fractions of peak 1 (Figure 5A) but reacted with none of the proteins in peak 2.
Figure 5.
Immunological Analysis of Tobacco Protein Kinases Activated by Osmotic Stress.
For protein gel blot analysis of proteins eluted from the Mono Q column in peaks 1 and 2 and occasionally in peak 3, the blots were probed with the following antibodies:
(A) Rabbit polyclonal anti–ERK1/ERK2 antibodies.
(B) Rabbit polyclonal anti-JNK1 antibodies.
(C) Rabbit polyclonal anti-SIPK antibodies.
(D) Rabbit polyclonal anti-WIPK antibodies.
(E) Rabbit polyclonal anti-CK2α antibodies.
Molecular mass markers are given in kilodaltons at left; 42 and 48 kD at right indicate molecular masses of the protein kinases activated by salinity stress.
Anti-JNK1 antibodies recognized one protein band in peak 2 with a molecular mass of 52 kD (Figure 5B). However, the in-gel kinase assay with ATF-2 as substrate detected no active kinase of this size (Figure 1B), indicating that the 52-kD MAPK family member was not activated by osmotic stress. Anti-p38 antibodies did not recognize any proteins in peaks 1 and 2 (data not shown). Although this might suggest that a mammalian p38 ortholog does not exist in plants, it is at least as likely that the antiserum used, which was raised against N- and C-terminal peptides of human p38, does not cross-react with the tobacco p38 ortholog. Anti-SIPK antibodies recognized the 48-kD protein from peak 1 (Figure 5C), suggesting that the osmotic stress–induced 48-kD kinase is SIPK. Anti-WIPK antibodies reacted with a 44-kD protein from peak 1 (Figure 5D). Neither anti-SIPK nor anti-WIPK antibodies recognized any proteins that eluted in peak 2 (Figures 5C and 5D). Anti-CK2α antibodies recognized casein kinases type II, which are ubiquitously distributed in the plant kingdom. These antibodies recognized both a 38-kD protein from peak 1, which coeluted with the salt-activated 48-kD protein kinase, and a protein of similar size from peak 3. In contrast, none of the protein eluting in peak 2 was recognized by the anti-CK2α antibodies (Figure 5E).
To demonstrate rigorously that the salinity-activated 48-kD kinase was SIPK, we immunoprecipitated cell extracts from untreated and 250 mM NaCl–treated tobacco cells with anti-SIPK antibodies (and anti-WIPK antibodies as control) and analyzed the resulting immunocomplexes with the in-gel kinase assay, using MBP (Figures 6A and 6B). The activated 48-kD kinase was immunoprecipitated by the anti-SIPK antibodies, thus confirming its identity. To identify the 48-kD kinase activated by another osmotic stress, we performed a similar immunocomplex kinase assay with extracts prepared from untreated or 900 mM sorbitol–treated cells. This kinase was recognized by the anti-SIPK but not the anti-WIPK antibodies, again demonstrating that it is SIPK (Figures 6A and 6B).
Figure 6.
SIPK-Specific Antibodies Immunoprecipitate the 48-kD Osmotic Stress–Activated Protein Kinase.
Protein extracts (50 μg) from untreated cells (lanes 1, 3, 5, 7, and 9), from cells exposed to 900 mM sorbitol for 10 min (lanes 2, 4, and 6), or to 250 mM NaCl (lanes 8 and 10) were analyzed by using an in-gel kinase assay with MBP as substrate, as described in Methods, before and after immunoprecipitation with anti-SIPK or anti-WIPK antibodies.
(A) Immunoprecipitation with anti-SIPK antibodies.
(B) Immunoprecipitation with anti-WIPK antibodies.
Lanes 1 and 2 show kinase activity of protein extracts not incubated with antibodies; lanes 3, 4, 7, and 8 show kinase activity of immunocomplexes; and lanes 5, 6, 9, and 10 show proteins possessing kinase activity that were not bound to antibodies, remaining in the supernatant after immunoprecipitation. Molecular mass markers are given in kilodaltons at left; 42 and 48 kD indicate molecular masses of the osmotic stress–activated protein kinases.
Substrate Specificity of Protein Kinases Activated by Osmotic Stress
The activities of the protein kinases separated on the Mono Q column as peak 1 and peak 2 were analyzed with various protein and peptide substrates to determine their specificity (Table 2). The kinases that eluted in peak 1 were able to phosphorylate MBP and two typical CK2 substrates, casein and RRRDDDSDDD, but this phosphorylation was totally suppressed by the CK2 and CK1 inhibitor heparin. The MBP-phosphorylating activity in this peak increased after salt stress (Figure 3A), but the casein-phosphorylating activity was unaffected (Figure 3C). These results are consistent with the presence in this peak of the 48-kD SIPK, which preferentially uses MBP as substrate, and a 38-kD CK2, which is specific for casein (Figures 5 and 6). The kinase(s) that eluted in peak 2 phosphorylated MBP, casein, ATF-2, and c-Jun, with the phosphorylation rates being greatest for MBP and casein (Table 1). The kinase activity was not affected by heparin at 100 μg/mL, indicating that this kinase does not belong to the CK1 or CK2 family. This conclusion was confirmed by the demonstration that the CK2-specific peptide substrate was not phosphorylated by the kinase activity in peak 2 (Table 1) and by the lack of recognition by anti-CK2α antibodies (Figure 5E).
Table 2.
Effect of the Inhibition of Protein Phosphatases on the Activity of the Osmotic Stress–Activated Protein Kinasesa
| MBP Phosphorylationb(% control)
|
ATF-2 Phosphorylationb(% control)
|
Casein Phosphorylationb(% control)
|
||
|---|---|---|---|---|
| Treatment | Peak 1 | Peak 2 | Peak 2 | Peak 2 |
| PP2A + OA (control) | 100 | 100 | 100 | 100 |
| PP2A | 56 | 30 | 8 | 12 |
| PTP + van (control) | 100 | 100 | 100 | 100 |
| PTP | 40 | 98 | 97 | 105 |
Protein kinases eluted from the Mono Q column in peaks 1 and 2 were incubated with PP2A or PTP in the presence (controls) or absence of specific inhibitors (okadaic acid [OA] for PP2A and orthovanadate [van] for PTP inhibition). The reaction was performed as described in Methods. Protein kinases were analyzed using MBP (peak 1) or MBP, ATF-2, and casein (peak 2) as the substrates.
Kinase activities represent the mean values from five independent experiments. Standard deviation was ∼ 5 to 10%.
Phosphorylation of Both Tyrosine and Serine/Threonine Residues Is Essential for the 48-kD SIPK Activity, Whereas the Activation of the 42-kD Kinase Requires Phosphorylation of Serine/Threonine but Not Tyrosine
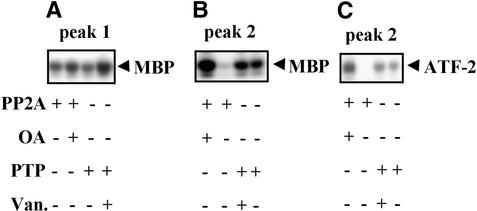
To assess whether phosphorylation was essential for the activation of these stress-responsive protein kinases, we tested the effect of dephosphorylation by protein phosphatases on the activities of the kinases that eluted in peaks 1 and 2 from the Mono Q column. A serine/threonine protein phosphatase type 2A (PP2A) purified from maize seedlings and commercially available phosphotyrosine phosphatase (PTP) were used. The kinase activity was measured by using MBP as substrate for the enzyme that eluted in peak 1, whereas MBP, casein, and ATF-2 were used as substrates for the kinase that eluted in peak 2. Treatment by both PP2A and PTP inactivated the 48-kD SIPK in peak 1 (Figure 7A and Table 2). Zhang and Klessig (1997) observed similar inactivation of the purified 48-kD SIPK with another serine/threonine protein phosphatase type 1 and PTP. The 42-kD protein kinase that eluted in peak 2 was inactivated by PP2A, whereas PTP had no effect on its activity (Figures 7B and 7C and Table 2). Inactivation by PP2A did not depend on the substrate used for measuring the kinase activity.
Figure 7.
Inhibition by Treatment with Protein Phosphatases of the Protein Kinases Activated by Osmotic Stress.
Kinases eluted in peaks 1 and 2 from the Mono Q column were preincubated for 20 min with PP2A or PTP at 30°C in the presence (controls) or absence of specific phosphatase inhibitors ocadaic acid (OA) for PP2A and sodium vanadate (Van.) for PTP. (+) and (−) indicate the presence and absence, respectively, of protein phosphatase or inhibitor in the reaction mixture.
(A) Effect of dephosphorylation on activities of protein kinase(s) eluted in peak 1 measured with MBP as the substrate.
(B) Effect of dephosphorylation on activities of protein kinase(s) eluted in peak 2 measured with MBP as the substrate.
(C) Effect of dephosphorylation on activities of protein kinase(s) eluted in peak 2 measured with ATF-2 as the substrate. Phosphorylation of MBP and ATF-2 was determined by autoradiography of phosphorylated substrates resolved by SDS-PAGE.
Purification of the 42-kD Kinase
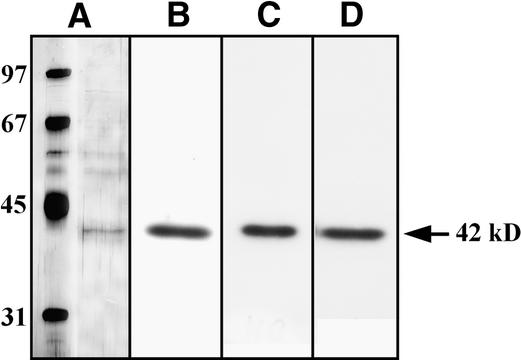
The tobacco 42-kD kinase was purified to near-homogeneity by four-step chromatography on SOURCE 15Q, phenyl-Sepharose, heparin-Sepharose, and, finally, Mono Q columns (see Methods for details). The purification procedure is summarized in Table 3, and typical elution profiles from each of the purification steps are shown in Figures 8A to 8D. The enzyme was purified ∼12,000-fold to near-homogeneity, showing only one protein band (molecular mass of 42 kD) in the 35- to 45-kD range (Figure 9A) after SDS-PAGE and silver staining. To ensure that the 42-kD protein band visible on the polyacrylamide gel after silver staining was the kinase of interest, we performed in-gel kinase assays with casein, MBP, or a mixture of equal weights of casein and MBP after the last step of purification. In each assay, only one band of kinase activity was detected, and its molecular mass was 42 kD (Figures 9B to 9D), which correlated with the protein detected by silver staining. These results indicate that this polypeptide was the 42-kD osmotic stress–activated kinase.
Table 3.
Purification of the 42-kD Kinase
| Fraction | Protein (mg)a |
Total Activity (pmol min −1) |
Specific Activity (pmol min −1 mg−1) |
Recovery (%) |
Purification (fold) |
|---|---|---|---|---|---|
| Total extractb | 3,900 | 135,546 | 34 | 100 | 1 |
| SOURCE 15Q | 38 | 87,150 | 2,305 | 64 | 68 |
| Phenyl-Sepharose | 61 | 49,500 | 8,250 | 36 | 243 |
| Heparin-Sepharose | 0.510 | 34,900 | 68,431 | 26 | 2,013 |
| Mono Q HR 5/5 | 0.025 | 10,150 | 406,000 | 7 | 11,941 |
The cell extract was prepared from ∼500 g of BY-2 cells treated for 5 min with 250 mM NaCl.
The amount of protein was estimated by comparing different amounts of BSA with the 42-kD protein band on a silver-stained SDS–polyacrylamide gel.
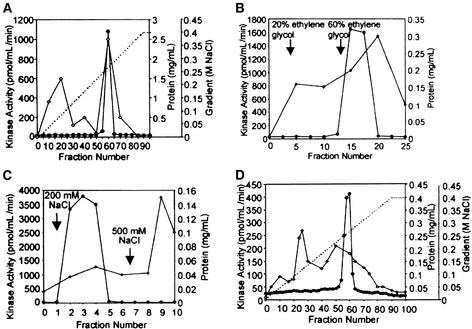
Figure 8.
Elution Profiles of Kinase Activity and Protein from Each Purification Step.
(A) Chromatography on a SOURCE 15Q column.
(B) Chromatography on a phenyl-Sepharose column.
(C) Chromatography on a heparin-Sepharose column.
(D) Chromatography on a Mono Q column.
Kinase activity (filled circles) was determined by the in-solution kinase assay with casein as substrate. For inhibition of casein kinases, heparin was added to the incubation mixtures at a concentration of 100 μg/mL. The dashed lines indicate NaCl gradients, and lines with open diamonds indicate protein concentration.
Figure 9.
The 42-kD Osmotic Stress–Activated Protein Kinase Phosphorylates MBP and Casein Equally Well.
Substrate specificity of the purified 42-kD osmotic stress–activated protein kinase was analyzed by using an in-gel kinase assay.
(A) SDS-PAGE of the purified 42-kD osmotic stress–activated protein kinase. Proteins were visualized by silver staining.
(B) In-gel kinase assay of the 42-kD protein kinase with MBP.
(C) In-gel kinase assay of the 42-kD protein kinase with casein.
(D) In-gel kinase assay of the 42-kD protein kinase with equal weights of MBP and casein.
The 42-kD Kinase Is a Homolog of the Arabidopsis Protein Kinase ASK1
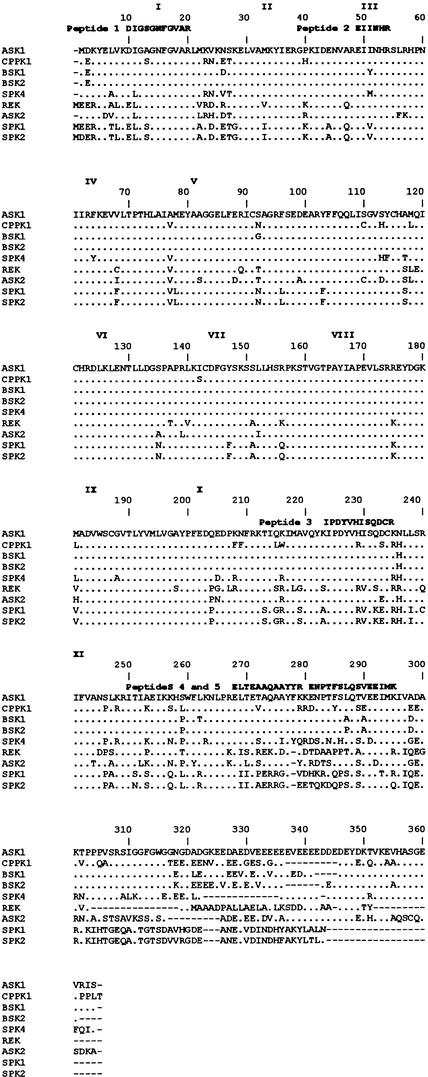
After the final purification step, 42-kD kinase was used in preparative SDS-PAGE. The 42-kD protein band was then excised from the gel for microsequencing, in which partial amino acid sequences of five internal tryptic peptides were obtained (Figure 10). Peptide 1 has 100% identity with the peptide coming from the first serine/threonine kinase subdomain of the Craterorostigma plantagineum protein kinase CPPK1 (P. Heino, M. Nylander, T. Palva, and D. Bartels, unpublished results; GenBank accession number CAA06503) and 91% identity with the analogous peptide of the Arabidopsis serine/threonine kinase ASK1 of unknown function (Park et al., 1993). Peptide 2 exhibits 100% identity with the peptide from the third kinase subdomain of both the CPPK1 and ASK1 kinases. Peptide 3 shares 83 and 92% identity, whereas peptide 4 is 83 and 75% identical to the corresponding peptides of the CPPK1 and ASK1 kinases, respectively. Peptide 5 exhibits 73 and 93% identity, respectively, with the related peptides from both kinases. The third peptide is located between the 10th and 11th kinase subdomains, whereas the fourth and fifth peptides lie beyond the kinase domain. This strong homology of all five peptides with these two kinases indicates that the 42-kD osmotic stress–activated protein kinase is an ASK1 homolog.
Figure 10.
Comparison of Tryptic Peptide Sequences of the 42-kD Kinase with Other Proteins.
Proteins exhibiting the greatest sequence similarities to peptides derived from the 42-kD kinase are shown. Dots represent amino acid residues that are identical to ASK1 (Park et al., 1993). References for the sequences of the other kinases are as follows: CPPK1 (P. Heino, M. Nylander, T. Palva, and D. Bartels, unpublished results; GenBank accession number CAA06503), BSK1 (Park et al., 1995), BSK2 (Park et al., 1995), SPK4 (Yoon et al., 1997), rice endosperm protein kinase (REK; Hotta et al., 1998), ASK2 (Park et al., 1993), soybean protein kinase 1 (SPK1; P.G. Shin, H. Yoon, Y.H. Jeong, J. Bahk, J.C. Hong, and M.J. Cho, unpublished results; GenBank accession number AAA33979), and soybean protein kinase 2 (SPK2; P.G. Shin, H. Yoon, M. Kim, J. Bahk, J.C. Hong, and M.J. Cho, unpublished results; GenBank accession number AAA34017). The conserved subdomains of the serine/threonine kinase domain are indicated by roman numerals. Gaps are marked with dashes and were introduced to maximize alignment. The peptide sequences obtained by microsequencing are shown above the ASK1 sequence.
DISCUSSION
At least two protein kinases were activated in tobacco cells that had been subjected to osmotic stress (250 mM NaCl or 900 mM sorbitol). These enzymes were separated and partially purified by chromatography on a Mono Q column, and their biochemical and immunological properties were examined. One of the kinases activated by osmotic stress is the 48-kD SIPK (Zhang and Klessig, 1997), identified by using immunoblot analysis and an immunocomplex kinase assay with anti-SIPK antibodies. The SIPK was activated within 5 min after osmotic stress, reaching maximum activity after another 5 min and declining to the basal level within 30 min. SIPK eluted from the Mono Q column at 150 mM NaCl (peak 1), whereas the second stress-activated kinase (42-kD enzyme) eluted at 300 mM NaCl (peak 2). The 42-kD kinase exhibited a very rapid (within 1 min) and prolonged (>2 hr) activation in response to osmotic stress.
In mammalian systems, several MAPK cascades are activated by various stresses. These MAPKs activate specific transcription factors through the phosphorylation of their transcriptional activation domains, which then leads to the expression of stress response genes (reviewed in Treisman, 1996). For example, SAPK/JNK activates transcription factors c-Jun, ATF-2, ELK-1, and ATFa; p38 kinase activates ATF-2, MEF2C, and CHOP-1; and BMK1 kinase activates MEF2C (Kyriakis and Avruch, 1996; Treisman, 1996; Whitmarsh and Davis, 1996; Kato et al., 1997). Similar pathways of transcription factor activation are found in yeast cells (Wilkinson and Millar, 1998). Assuming that plants, like yeast, possess orthologs of animal stress-activated MAPKs, we used recombinant glutathione S-transferase (GST)–ATF-2 and GST–c-Jun as in vitro substrates to identify plant stress- responsive protein kinases. The 42-kD protein kinase, which was strongly and rapidly activated by osmotic stress, phosphorylated ATF-2 and c-Jun. In contrast to animal stress-activated MAPKs, however, the plant enzyme also effectively phosphorylated casein as well as ATF-2 and c-Jun and the preferred in vitro substrate of MAPKs, MBP. Inactivation of the 42-kD kinase activity by phosphatase PP2A suggests that phosphorylation of serine and/or threonine plays a crucial role in the activation of this enzyme. In contrast to MAPKs, the activity of the tobacco 42-kD kinase was not dependent on tyrosine phosphorylation, as shown by its lack of response to tyrosine phosphatase treatment. These two features—requirement of serine/threonine but not tyrosine phosphorylation for enzymatic activity and unusually broad substrate specificity—suggest that the 42-kD osmotic stress–responsive kinase does not belong to the MAPK family. Consistent with this conclusion, we found that a variety of antibodies specific to different MAPKs failed to cross-react with the 42-kD kinase.
Besides MAPKs, other stress-activated plant kinases have been described. Their molecular masses are between ∼40 and 50 kD and they are activated by phosphorylation of serine or threonine but not tyrosine residues. For example, a 45-kD kinase that phosphorylates casein but not MBP was activated in maize primary roots in response to water deficiency (Conley et al., 1997). In green algae, transfer to a high-osmoticum medium activated a 40-kD kinase that phosphorylates casein but not MBP, whereas in low osmoticum medium, another 40-kD kinase was activated that phosphorylates MBP but not casein (Yuasa and Muto, 1996). Both of these kinase activities were abolished by alkaline phosphatase treatment. The authors of this study suggested that phosphorylation of serine and/or threonine but not tyrosine was responsible for the activation of these enzymes because the active kinases were not recognized by anti-phosphotyrosine antibodies. Another plant kinase with properties resembling those of the tobacco 42-kD kinase described in this article is the kinase activated by abscisic acid in a Ca2+-independent manner in guard cells of fava bean (Li and Assmann, 1996). This kinase also differs from MAPKs by not requiring tyrosine phosphorylation for activation (only serine was phosphorylated in the active enzyme), and it effectively phosphorylates histone H1, which is a poor MAPK substrate. However, none of these kinases has a substrate specificity as broad as that of the tobacco 42-kD enzyme.
The biochemical properties of the 42-kD kinase did not resemble those of any of the numerous protein kinases identified in animals or in plants; therefore, we purified the enzyme for further characterization. The sequences of the five internal tryptic peptides show that it is closely related to the following cloned kinases: CPPK1 from the resurrection plant C. plantagineum (P. Heino, M. Nylander, T. Palva, and D. Bartels, unpublished results; GenBank accession number CAA06503), the Arabidopsis ASK1 kinase (Park et al., 1993), and the Brassica napus kinases BSK1 and BSK2 (Park et al., 1995). The functions of these kinases have not been defined, although expression of the ASK1 gene is known to be controlled by light, suggesting its involvement in photoregulation of plant cells. The soybean protein kinases SPK-3 and SPK-4 are among the kinases that are related to the tobacco 42-kD kinase (Yoon et al., 1997). Interestingly, SPK-4 expression was found to be induced by dehydration and high salinity but not by exogenous abscisic acid. These results correlate with ours and suggest that this family of kinases may be involved in stress signal transduction in plants.
All the kinases mentioned in the preceding paragraph are members of the SnRK2b subfamily of SNF1-related kinases (SnRKs) (reviewed in Halford and Hardie, 1998). In animals and yeast, SNF1 family members are involved in protecting cells against nutritional and environmental stresses (Halford and Hardie, 1998). Plants have at least three subfamilies of SnRKs. The first, SnRK1, is the best characterized, whereas relatively little is known about the function of the SnRK2 and SnRK3 subfamilies. Most members of the SnRK1 subfamily are involved in carbon metabolism (reviewed in Halford and Hardie, 1998), although one member also appears to participate in the plant response to sulfur limitation (Davies et al., 1999). The best-known member of the plant SnRK2a subfamily is the protein kinase induced by abscisic acid (PKABA1). This enzyme and SPK-3, which belong to the SnRK2b subfamily, are induced by dehydration, high salt concentration, and exogenous abscisic acid (Anderberg and Walker-Simmons, 1992; Yoon et al., 1997). Together, these results suggest that plant enzymes belonging to the SNF1-related family of protein kinases function similarly to their homologs in yeast and animals and that the very poorly characterized SnRK2 subfamily may be involved in environmental stress signaling in plants.
In summary, SIPK and the 42-kD kinase, which is a member of the SnRK2b subfamily, are both activated under osmotic stress conditions in plants. In this study, we report a role for SIPK in this process as well as the biochemical features and function of the 42-kD ASK1 homolog in the immediate response of plants to osmotic stress. Because SIPK is activated by pathogen infection, pathogen-derived elicitors, and wounding (Zhang and Klessig, 1998a, 1998b; Zhang et al., 1998) as well as osmotic stress, this MAPK apparently is involved in the response to a variety of stresses. The rapid osmotic stress–induced activation of these two kinases by protein phosphorylation suggests their involvement in protein kinase cascades that probably involve two parallel signaling pathways to transduce the osmotic stress signal in plants. Construction of transgenic plants with altered expression and/or activities of SIPK and the 42-kD kinase should help to define the osmotic stress signaling pathway(s) in plants.
METHODS
Cell Culture and Treatment
BY-2 tobacco cells, kindly provided by W. Filipowicz (Friedrich-Miescher Institute, Basel, Switzerland), were cultured under conditions described elsewhere (Nagata et al., 1992); that is, cells were grown in Murashige and Skoog medium (Sigma-Aldrich Co. Ltd., Irvine, UK) supplemented with 100 mg/L myoinositol, 1 mg/L thiamine HCl, 255 mg/L KH2PO4, 0.2 mg/L 2,4-D, and 3% sucrose. They were subcultured every 7 days.
The cells were treated with NaCl (250 mM) or sorbitol (900 mM) for various periods of time (0, 1, 5, 10, 20, 30, 40, 60, and 120 min), harvested by centrifugation, quickly frozen in liquid nitrogen, and stored at −80°C until analyzed.
Preparation of Protein Extracts
The cells were sonicated three times, each time for 20 sec, in extraction buffer (20 mM Tris, pH 7.5, 2 mM EDTA, 2 mM EGTA, 50 mM β-glycerophosphate, 100 μM Na3VO4, 2 mM DTT, 500 μM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 1 μM leupeptin, 1 μM aprotinin, and 250 mM sucrose) with a Sonifier 250 Sonicator (Branson Ultrasonics Corp., Danbury, CT), using ∼1 mL of the extraction buffer for each 2 mL of cells. After sonication, the extracts were centrifuged at 100,000g for 60 min at 4°C.
Chromatography on the Mono Q Column
Protein extracts (i.e., supernatant after centrifugation at 100,000g) from tobacco cells treated with 250 mM NaCl for 10 min and from untreated controls containing 50 mg of protein were loaded separately onto a Mono Q HR 5/5 column attached to a fast-performance liquid chromatography system (Pharmacia). The column was equilibrated with buffer A (the extraction buffer minus sucrose). After washing with 30 mL of buffer A, the column was developed with a 100-mL linear gradient of NaCl (0 to 0.4 M) in the same buffer. The flow rate was 1 mL/min, and 1-mL fractions were collected. The chromatography was performed at 4°C.
In-Gel Kinase Activity Assay
Portions of the protein extracts (30 μg) or the Mono Q fractions (20 μL) were electrophoresed according to Laemmli (1970) in 10% SDS– polyacrylamide gels that contained one of the following copolymerized protein substrates: 0.5 mg/mL myelin basic protein (MBP), 0.5 mg/mL glutathione S-transferase (GST)–ATF-2, or 0.5 mg/mL casein. After electrophoresis, SDS was removed by washing the gels twice with the washing buffer (50 mM Tris, pH 8.0, and 20% 2-propanol) for 30 min each at room temperature; the gels were then equilibrated for 1 hr in buffer B (50 mM Tris and 5 mM 2-mercaptoethanol). Proteins were denatured with 6 M guanidine HCl in buffer A (two 30-min changes), followed by an overnight renaturation at 4°C in buffer B containing 0.04% Tween 40 (five changes). Subsequently, the gels were incubated for 60 min at room temperature in 10 mL of the reaction buffer (10 mM Tris, pH 7.5, 2 mM DTT, 0.1 mM EGTA, 15 mM MgCl2, and 20 μM ATP) supplemented with 50 μCi of γ-32P-ATP. The reaction was terminated by transferring the gels into a bath of 5% trichloroacetic acid and 1% sodium phosphate. The unincorporated γ-32P-ATP was removed by washing the gels in the bath solution until the washing solution was determined to contain no more radioactivity. Finally, the gels were stained with Coomassie Brilliant Blue R 250, dried, and exposed to Hyperfilm MP (Amersham).
Protein Kinase Assay in Solution
Phosphorylation of MBP
Twelve-microliter portions of fractions eluted from the Mono Q column were incubated with MBP as the substrate (0.3 mg/mL) and with 20 μM ATP supplemented with 1 μCi γ-32P-ATP in kinase buffer (25 mM β-glycerophosphate, pH 7.5, 1.25 mM EGTA, 1 mM DTT, 10 mM MgCl2, and 150 μM Na3VO4); the final volume of each incubation mixture was 24 μL. After 20 min at 30°C, 20 μL of the mixture was spotted onto a P81 phosphocellulose filter strip, and the reaction was terminated by washing the filter with 0.85% H3PO4. The radioactivity incorporated into the MBP was measured by scintillation counting.
Phosphorylation of ATF-2
The kinase assay was performed as described above, except that ATF-2 (0.3 mg/mL) was used as the substrate. The reaction was terminated by adding 10 μL of 3 × Laemmli sample buffer (Laemmli, 1970). Subsequently, the phosphorylated proteins were resolved on a 10% SDS–polyacrylamide gel according to Laemmli (1970) and identified by autoradiography.
Phosphorylation of Casein
Twelve-microliter portions of the fractions eluted from the Mono Q column were incubated with casein (0.3 mg/mL) and with 50 μM ATP supplemented with 1 μCi γ-32P-ATP in buffer containing 20 mM Tris, pH 7.5, and 20 mM MgCl2. The final incubation volume was 50 μL. After 20 min of incubation at 30°C, 40 μL of the mixture was spotted onto a 3MM filter strip (Whatman International Ltd., Maidstone, UK) and then washed with 5% trichloroacetic acid. The radioactivity incorporated into casein was determined by scintillation counting.
Phosphorylation of RRRDDDSDDD Peptide
The kinase assay was performed as described for the MBP phosphorylation, except that the concentration of the peptide was 1 mM.
Immunoblot Analysis
Proteins from crude extracts or from fractions obtained after Mono Q chromatography were separated on 10% SDS–polyacrylamide gels and transferred to nitrocellulose by tank electroblotting, according to the procedure recommended in the Bio Rad Laboratories manual. The membrane was blocked for 2 hr at room temperature in TBST buffer (10 mM Tris, pH 7.5, 100 mM NaCl, and 0.1% Tween 20) containing 5% milk, and then incubated for 2 hr in the same buffer with appropriate primary antibodies: anti–ERK1/ERK2, anti-JNK, or anti-p38, purchased from Santa Cruz Biotechnology (Santa Cruz, CA); anti-CK2α, raised against peptide C (amino acids 66 to 86 from human CK2α), which is present in all currently known CK2 proteins (kindly provided by L.A. Pinna [University of Padova, Italy]), and either anti-SIPK or anti-WIPK. The anti-SIPK and anti-WIPK antibodies were raised against the N-terminal peptides unique for these kinases: MDGSGQQTDTMMSDAGAEQPPTAP and MADANMGAGGGQFPDFPS, respectively (Zhang and Klessig, 1998a; Zhang et al., 1998).
After incubation with an alkaline phosphatase–conjugated secondary antibody, the results were visualized by staining with p-nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate p-toluidine salt.
Immunocomplex Kinase Activity Assay
Protein from crude extracts (50 μg) was incubated with anti-SIPK or anti-WIPK antibody (5 μg) in immunoprecipitation buffer (20 mM Tris, pH 7.5, 2 mM EDTA, 2 mM EGTA, 50 mM β-glycerophosphate, 100 μM Na3VO4, 2 mM DTT, 500 μM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 1 μM leupeptin, 1 μM aprotinin, 1% Triton X-100, and 150 mM NaCl) at 4°C for 4 hr on a rocker. Approximately 25 μL of packed volume of protein A–agarose was added, and the incubation was continued for another 2 hr. Agarose bead–protein complexes were pelleted by brief centrifugation and washed three times with immunoprecipitation buffer.
Ten microliters of 3 × Laemmli sample buffer was added to the pelleted agarose bead–protein complex, and the sample was heated at 95°C for 3 min. After brief centrifugation, the supernatant was assayed by using the MBP in-gel kinase, as described before.
Treatment with Protein Phosphatases
The phosphatase inhibitors β-glycerophosphate and vanadate that had been included in the buffer used to elute the fractions from the Mono Q column were removed by filtration on a PD-10 column (Pharmacia) equilibrated with 20 mM Tris, pH 7.5, 2 mM EGTA, and 2 mM EDTA. Fifty-microliter portions were supplemented with 50 ng of maize serine/threonine protein phosphatase type 2A (PP2A) purified to homogeneity from maize seedlings, according to the procedure previously described by Tung et al. (1985) or with 2 microunits of protein tyrosine phosphatase (PTP; Boehringer Mannheim, Germany). In control experiments, okadaic acid purchased from Alexis Biochemicals (San Diego, CA) was added with PP2A, and Na3VO4 was added with PTP. The reaction mixture was preincubated for 20 min at 30°C, after which okadaic acid was added to a final concentration of 4.8 μM to inhibit PP2A or Na3VO4 was added to a final concentration of 1 mM to inhibit PTP. Twelve-microliter portions were withdrawn, and the activities of the protein kinases were assayed by using MBP, ATF-2, or casein as substrate.
Purification of the 42-kD Kinase
All purification steps were performed at 4°C.
Protein extract from ∼500 g of BY-2 cells treated for 5 min with 250 mM NaCl was loaded onto a 15-mL SOURCE 15Q (Pharmacia) column previously equilibrated with buffer A, the same buffer used for the Mono Q column described earlier. After washing with 150 mL of buffer A, the column was developed with a 300-mL linear gradient (0 to 0.4 M NaCl) in the same buffer. The 42-kD kinase was eluted at ∼300 mM NaCl. The fractions containing the highest kinase activity were pooled and, after the conductivity was adjusted to 500 mM NaCl, were loaded onto a 10-mL phenyl-Sepharose CL-6B (Pharmacia) column equilibrated with buffer A supplemented with 500 mM NaCl. The column was washed with 50 mL of the above buffer and then with 20 mL of buffer A plus 20% ethylene glycol. The enzyme was finally eluted with buffer A containing 60% ethylene glycol. Two-milliliter fractions were collected.
Active fractions were pooled and directly applied onto a 2-mL heparin-Sepharose CL-6B (Pharmacia) column equilibrated with buffer A. The column was washed with 20 mL of buffer A, and for enzyme elution, the following NaCl step gradient was applied: 5 mL of buffer A plus 200 mM NaCl followed by 5 mL of buffer A plus 500 mM NaCl. The kinase eluted with the 200 mM NaCl. The peak fractions were pooled, desalted with a PD10 column (Pharmacia), and applied to a Mono Q H5/5 column. The chromatography was performed as described earlier (see Chromatography on the Mono Q Column).
Microsequencing of Internal Tryptic Peptides
Active fractions from the final Mono Q column were pooled and concentrated by using a Centricon 30 concentrator (Amicon Inc., Beverly, MA), and the proteins were separated on preparative 12% SDS–polyacrylamide gels. After Coomassie blue staining, the gel was washed in water for 1.5 hr, and the 42-kD protein band was excised. The gel containing the protein of interest was lyophilized and sent to D. Winant at the Protein and Nucleic Acid Facility (Beckman Center, Stanford University Medical Center, Palo Alto, CA) for microsequencing. The 42-kD protein was subjected to digestion with trypsin, and the resulting peptides were separated by reversed-phase HPLC. After analysis by mass spectrometry, selected peptides were sequenced.
NOTE ADDED IN PROOF
While this manuscript was being accepted for publication, the sequence of a plant homolog of the p38/HOG1 MAPK was published (Dixon, K.P., Xu, J.-R., Smirnoff, N., and Talbot, N.J. [1999]. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11, 2045–2058).
Acknowledgments
We thank Professor Edwin Krebs and Drs. Jonathan Graves and Dongxia Li for providing the GST–ATF-2— and GST–c-Jun—expressing clones, antibodies against p38, and ERK1/ERK2 kinases, and for advice and helpful discussions. We are grateful to Professor Lorenzo A. Pinna for antibodies against the CK2α catalytic subunit and Dr. Witold Filipowicz for BY-2 tobacco cells, to Professor Alina Kacperska for critical reading of the manuscript and comments during realization of this project, and to Dr. Dick Winant for help in sequence analysis. We are also very grateful to all members of our laboratory for stimulating discussions and especially to Ireneusz Jurkowski, Katarzyna Onyszczuk, and Marcin Górski for help in preparation of the manuscript. This work was supported by grants from the State Committee for Scientific Research (KBN, Poland, Grant No. 6 PO4A 028 12) to G.D., the European Community (INCO-Copernicus, Grant No. IC 15-CT96-0901) to G.M., and Grant Nos. 9802200 from the U.S. Department of Agriculture and MCB 97-23952 from the National Science Foundation to D.F.K.
References
- Adam, A.L., Pike, S., Hoyos, E., Stone, J.M., Walker, J.C., and Novacky, A. (1997). Rapid and transient activation of a myelin basic protein kinase in tobacco leaves treated with harpin from Erwinia amylovora. Plant Physiol. 115 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderberg, R.J., and Walker-Simmons, M.K. (1992). Isolation of a wheat cDNA clone for an abscisic acid–inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA 89 10183–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre, L., Ligterink, W., Heberle-Bors, E., and Hirt, H. (1996). Mechanosensors in plants. Nature 383 489–490. [DOI] [PubMed] [Google Scholar]
- Bögre, L., Ligterink, W., Meskiene, I., Barker, P.J., Heberle-Bors, E., Huskinsson, N.S., and Hirt, H. (1997). Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert, H.J., Nelson, D.E., and Jensen, R.G. (1995). Adaptation to environmental stresses. Plant Cell 7 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2 48–54. [Google Scholar]
- Canman, C.E., and Kastan, M.B. (1996). Signal transduction. Three paths to stress relief. Nature 384 213–214. [DOI] [PubMed] [Google Scholar]
- Cano, E., and Mahadevan, L.C. (1995). Parallel signal processing among mammalian MAPKs. Trends Biochem. Sci. 20 117–122. [DOI] [PubMed] [Google Scholar]
- Cheeseman, J.M. (1988). Mechanisms of salinity tolerance in plants. Plant Physiol. 87 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley, T.R., Sharp, R.E., and Walker, J.C. (1997). Water deficit rapidly stimulates the activity of a protein kinase in the elongation zone of the maize primary root. Plant Physiol. 113 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J.P., Yildiz, F.H., and Grossman, A.R. (1999). Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, A., Mayer, J.E., and Hahlbrock, K. (1990). Fungal elicitor triggers rapid, transient, and specific protein phosphorylation in parsley cell suspension cultures. J. Biol. Chem. 265 6360–6368. [PubMed] [Google Scholar]
- Errede, B., and Levin, D.E. (1993). A conserved kinase cascade for MAP kinase activation in yeast. Curr. Opin. Cell Biol. 5 254–260. [DOI] [PubMed] [Google Scholar]
- Fallon, K.M., and Trewavas, A.J. (1994). Phosphorylation of a renatured protein from etiolated wheat leaf protoplasts is modulated by blue and red light. Plant Physiol. 105 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G., Grosskopf, D.G., Regenass, M., and Boller, T. (1991). Rapid changes of protein phosphorylation are involved in transduction of the elicitor signal in plant cells. Proc. Natl. Acad. Sci. USA 88 8831–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford, N.G., and Hardie, D.G. (1998). SNF1-related protein kinases: Global regulators of carbon metabolism in plants? Plant Mol. Biol. 37 735–748. [DOI] [PubMed] [Google Scholar]
- Harper, J.F., Sussman, M.R., Schaller, G.E., Putnam-Evans, C.L., Charbonneau, H., and Harmon, A.C. (1991). A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science 252 951–954. [DOI] [PubMed] [Google Scholar]
- Herskowitz, I. (1995). MAP kinase pathways in yeast: For mating and more. Cell 80 187–197. [DOI] [PubMed] [Google Scholar]
- Hirt, H. (1997). Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci. 2 11–15. [Google Scholar]
- Hotta, H., Aoki, N., Matsuda, T., and Adachi, T. (1998). Molecular analysis of a novel protein kinase in maturing rice seed. Gene 213 47–54. [DOI] [PubMed] [Google Scholar]
- Jonak, C., Heberle-Bors, E., and Hirt, H. (1994). MAP kinases: Universal multi-purpose signaling tools. Plant Mol. Biol. 24 407–416. [DOI] [PubMed] [Google Scholar]
- Jonak, C., Kiegerl, S., Ligterink, W., Baker, P.J., Huskisson, N.S., and Hirt, H. (1996). Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc. Natl. Acad. Sci. USA 93 11274–11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin, M. (1994). Signal transduction from the cell surface to the nucleus through the phosphorylation of transcription factors. Curr. Opin. Cell Biol. 6 415–424. [DOI] [PubMed] [Google Scholar]
- Kato, Y., Kravchenko, V.V., Tapping, R.I., Han, J., Ulevitch, R.J., and Lee, J.-D. (1997). BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16 7054–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knetsch, M.L.W., Wang, M., Snaar-Jagalska, B.E., and Heimovaara-Dijkstra, S. (1996). Abscisic acid induces mitogen-activated protein kinase activation in barley aleurone protoplasts. Plant Cell 8 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritsas, V.M. (1988). Effect of ethylene and ethylene precursors on protein phosphorylation of xylogenesis in tuber explants of Helianthus tuberosus (L.). J. Exp. Bot. 39 375–386. [Google Scholar]
- Kyriakis, J.M., and Avruch, J. (1996). Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays 18 567–577. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Li, J., and Assmann, S.M. (1996). An abscisic acid–activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell 8 2359–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligterink, W., Kroj, T., zur Nieden, U., Hirt, H., and Scheel, D. (1997). Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science 276 2054–2057. [DOI] [PubMed] [Google Scholar]
- Marin, O., Meggio, F., and Pinna, L.A. (1994). Design and synthesis of two new peptide substrates for the specific and sensitive monitoring of casein kinases −1 and −2. Biochem. Biophys. Res. Commun. 198 898–905. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Gotoh, Y., Nishida, E., Yamaguchi-Shinozaki, K., Hayashida, N., Iwasaki, T., Kamada, H., and Shinozaki, K. (1994). Characterization of two cDNAs that encode MAP kinase homologues in Arabidopsis thaliana and analysis of the possible role of auxin in activating such kinase activities in cultured cells. Plant J. 5 111–122. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Irie, K., Hirayama, T., Hayashida, N., Yamaguchi-Shinozaki, K., Matsumota, K., and Shinozaki, K. (1996). A gene encoding a mitogen-activated protein kinase kinase is induced simultaneously with genes for mitogen protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Ichimura, K., and Shinozaki, K. (1997). Environmental stress response in plants: The role of mitogen-activated protein kinases. Trends Biotechnol. 15 15–19. [DOI] [PubMed] [Google Scholar]
- Monroy, A.F., Sarhan, F., and Dhindsa, R.S. (1993). Cold-induced changes in freezing tolerance, protein phosphorylation, and gene expression: Evidence for a role of calcium. Plant Physiol. 102 1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T., Nemoto, Y., and Hasezawa, S. (1992). Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int. Rev. Cytol. 132 1–30. [Google Scholar]
- Nishihama, R., Banno, H., Shibata, W., Hirano, K., Nakashima, M., Usami, S., and Machida, Y. (1995). Plant homologues of components of MAPK (mitogen-activated protein kinase) signal pathways in yeast and animal cells. Plant Cell Physiol. 36 749–757. [DOI] [PubMed] [Google Scholar]
- Park, Y.S., Hong, S.W., Oh, S.A., Kwak, J.M., Lee, H.H., and Nam, H.G. (1993). Two putative protein kinases from Arabidopsis thaliana contain highly acidic domains. Plant Mol. Biol. 22 615–624. [DOI] [PubMed] [Google Scholar]
- Park, Y.S., Song, O.K., Hong, S.W., Kwak, J.M., Cho, M.J., and Nam, H.G. (1995). Frequent in-frame length variations are found in the diverged simple repeat sequences of the protein-coding regions of two putative protein kinase genes of Brassica napus. Plant Mol. Biol. 27 829–833. [DOI] [PubMed] [Google Scholar]
- Pinna, L.A. (1990). Casein kinase 2: An “eminence grise” in cellular regulation? Biochim. Biophys. Acta 1054 267–284. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S., Chengappa, S., and Poovaiah, B.W. (1987). Auxin-regulated changes in protein phosphorylation in pea epicotyls. Biochem. Biophys. Res. Commun. 144 944–950. [DOI] [PubMed] [Google Scholar]
- Robinson, M.J., and Cobb, M.H. (1997). Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9 180–186. [DOI] [PubMed] [Google Scholar]
- Seger, R., and Krebs, E.G. (1995). The MAPK signaling cascade. FASEB J. 9 726–735. [PubMed] [Google Scholar]
- Seo, S., Okamoto, M., Seto, H., Ishizuka, K., Sano, H., and Ohashi, Y. (1995). Tobacco MAP kinase: A possible mediator in wound signal transduction pathways. Science 270 1988–1992. [DOI] [PubMed] [Google Scholar]
- Seo, S., Sano, H., and Ohashi, Y. (1999). Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell 11 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274 1900–1902. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol. 115 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short, T.W., and Briggs, W.R. (1990). Characterization of a rapid, blue light–mediated change in detectable phosphorylation of a plasma membrane protein from etiolated pea (Pisum sativum L.) seedlings. Plant Physiol. 92 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J.M., and Walker, J.C. (1995). Plant protein kinase families and signal transduction. Plant Physiol. 108 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., and Shinshi, H. (1995). Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with a fungal elicitor. Plant Cell 7 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman, R. (1996). Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 8 205–215. [DOI] [PubMed] [Google Scholar]
- Tung, H.Y.L., Alemany, S., and Cohen, P. (1985). The protein phosphatases involved in cellular regulation. II. Purification, subunit structure and properties of protein phosphatases-2A0, 2A1, and 2A2 from rabbit skeletal muscle. Eur. J. Biochem. 148 253–263. [DOI] [PubMed] [Google Scholar]
- Usami, S., Banno, H., Ito, Y., Nishihama, R., and Machida, Y. (1995). Cutting activates a 46-kilodalton protein kinase in plants. Proc. Natl. Acad. Sci. USA 92 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriard, M.-P., Martin, F., Pugin, A., Ricci, P., and Blein, J.-P. (1994). Protein phosphorylation is induced in tobacco cells by the elicitor cryptogein. Plant Physiol. 104 1245–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh, A.J., and Davis, R.J. (1996). Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74 589–607. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.G., and Millar, J.B.A. (1998). SAPKs and transcription factors do the nucleocytoplasmic tango. Genes Dev. 12 1391–1397. [DOI] [PubMed] [Google Scholar]
- Yoon, H.W., Kim, M.C., Shin, P.G., Kim, J.S., Lee, S.Y., Hwang, I., Bahk, J.D., Hong, J.C., Han, C., and Cho, M.J. (1997). Differential expression of two functional serine/threonine protein kinases from soybean that have an unusual acidic domain at the carboxy terminus. Mol. Gen. Genet. 255 359–371. [DOI] [PubMed] [Google Scholar]
- Yuasa, T., and Muto, S. (1996). Activation of 40-kDa protein kinases in response to hypo- and hyperosmotic shock in the halotolerant green alga Dunaliella tertiolecta. Plant Cell Physiol. 37 35–42. [Google Scholar]
- Zhang, S., and Klessig, D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1998. a). The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc. Natl. Acad. Sci. USA 95 7225–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1998. b). Resistance gene N–mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 95 7433–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Du, H., and Klessig, D.F. (1998). Activation of the tobacco SIP kinase by both a cell wall–derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]