Abstract
A key goal of the Human Genome Project was to understand the complete set of human proteins, the proteome. Since the genome sequence by itself is not sufficient for predicting new genes and alternative splicing events that lead to new proteins, expressed sequence tags (ESTs) are used as the primary tool for these purposes. The high prevalence of artifacts in dbEST, however, often leads to invalid predictions. Here we describe a novel method for recognizing genomic DNA contamination and other artifacts that cannot be identified using current EST cleaning techniques. Our method uses the alignment of the entire set of ESTs to the human genome to identify highly contaminated EST libraries. We discovered 53 highly contaminated libraries and a subset of 24 766 ESTs from these libraries that probably represent contamination with genomic DNA, pre-mRNA, and ESTs that span non-canonical introns. Although this is only a small fraction of the entire EST dataset, each contaminating sequence could create a spurious transcript prediction. Indeed, in the clustering and assembly tool that we used, these sequences would have caused incorrect inference of 9575 new splice variants and 6370 new genes. Conclusions based on EST analysis, including prediction of alternative splicing, should be re-evaluated in light of these results. Our method, along with the identified set of contaminated sequences, will be essential for applications that depend on large EST datasets.
INTRODUCTION
An expressed sequence tag (EST) is part of a gene that results from sequencing a portion of a cDNA clone that was generated from an mRNA (1). Although expressed sequence contains the most interesting information, it constitutes only slightly >1% of the human genome (2). Extracting exonic sequences directly from genomic sequence is difficult, but ESTs have provided a convenient means of accessing them. The largest public collection of ESTs is dbEST (3), a division of GenBank that currently contains more than 11 million sequences, including more than 4.2 million human sequences.
ESTs are important tools with many applications (4,5). They have been essential to rapid gene discovery (6–10) and have been used to build a physical map (11) and a gene map (12,13) of the human genome, to annotate genomic sequence (14,15), locate exons (16,17), compare and contrast genomes of different organisms (18), find new members of gene families (19), study gene expression on a large scale (20–22) and reconstruct metabolic pathways (23). Analysis of large EST datasets led to identification of putative single nucleotide polymorphisms (24,25) and to recognition of the prevalence of alternative splicing in the human genome (26,27).
A key component of gene identification and alternative splicing prediction is the correct analysis of gene structure and splice variants. This includes precise detection of the exons in each gene; indeed, the success rate of locating exons is an important measure of the performance of gene identification algorithms (28).
A typical collection of ESTs is highly redundant, so an early step in gene structure analysis is generally to group, or cluster, the ESTs based on sequence overlaps. The ESTs in each cluster are putatively from the same gene. Some systems assemble each cluster to produce a multiple alignment that approximates the sequence of the original cDNA that generated the ESTs in the cluster. The cluster’s consensus sequence is a longer representation of the underlying gene than are any of the individual ESTs. Genomic DNA, when available, can be used to both guide the clustering and identify exon boundaries. Databases of EST clusters and/or assemblies include UniGene (29), the TIGR Human Gene Index (30), STACK (31) and GeneNest (32).
A major obstacle to correct gene identification is the high error rate in EST datasets (33). EST sequences are unedited, single pass reads, typically several hundred bases long and have a base calling error rate as high as 3% (34). Steps can be taken to mitigate the effects of sequence errors; per-base quality scores are sometimes available (35), and when they are not, GenBank entries are occasionally annotated to indicate which portion of the sequence is of high quality. Sequences can also be aligned to the genome and errors can be corrected based on the genomic sequence.
In addition to sequencing errors, ESTs suffer from various kinds of contamination, depending in part on which of many available protocols was used to construct the cDNA libraries from which the ESTs were generated (36–40). Typical problems include sequences from portions of the sequencing vector or the linker at the ends of the ESTs. Vector contamination can also occur from DNA rearrangement events within the bacterium, causing the insertion of bacterial sequence into the middle of the EST. These contaminating sequences can generally be identified by computationally comparing all EST sequences to a database of vector sequences and removed before the EST sequences are used. This process is referred to as ‘cleaning’ (10,41). Generally, when a rearrangement is recognized, the EST is discarded.
Sequences from other organisms, such as viruses, can also occur as contaminants because of laboratory contamination or infection of the human whose tissue sample was used to construct the EST library (42). These sequences can be removed by computational screening, as is done for sequencing vector contamination.
Chimeric sequences pose a more subtle problem. A chimera is a concatenation of two or more expressed sequences from different areas. It can be an artifact of cDNA cloning or sequencing (43,44). If a chimera is used as is for clustering, it will likely result in the combination of two genes into a single incorrect gene prediction. Identifying chimeras is more difficult than screening out other contamination, since the entire sequence is from the correct organism. Because the portions of a chimeric EST are joined at random, they are generally from different chromosomes or from distant regions of the same chromosome; comparison with the full genome sequence can help identify chimeric ESTs. Sophisticated algorithms that model EST properties are also used to identify chimeras (45,46).
An EST dataset can also be contaminated with genomic DNA from the organism itself. Protocol problems with DNase are one source of DNA contamination; when this happens, the oligo-dT primer used for first strand synthesis can mis-prime off genomic poly-A stretches and exacerbate the level of contamination. Such contamination may make intronic regions appear as if they are expressed, resulting in the prediction of non-existent splice variants. DNA contamination from an intergenic region can result in a false gene prediction. Existing methods are unable to detect and remove such contamination, and techniques such as eliminating all unspliced sequences that create new putative splice variants often result in true splice variants being discarded (47). In particular, single-exon genes may be missed entirely because of such filtering (48). The method we describe here avoids these pitfalls by addressing genomic DNA contamination directly.
Premature mRNA (pre-mRNA)—mRNA that did not undergo the splicing process—constitutes another common form of EST contamination (47). It can be a result of mis-priming on intronic sequences. Although ESTs representing pre-mRNA contamination may appear to be instances of intron retention (47,49,50), they are artifacts rather than real exonic sequences.
Current approaches to cleaning EST datasets process one EST at a time. This is appropriate for sequence artifacts such as sequencing errors, vector and virus contamination and chimeric sequences, but cannot remedy problems such as contamination with genomic DNA and pre-mRNA.
We propose an approach that does address these problems. Contamination with genomic DNA and pre-mRNA often depends on the protocol for EST library construction; it therefore typically affects entire libraries. Our unit of analysis is the EST library rather than the individual EST. Although recognizing contaminated sequences on an individual basis is difficult, since they align perfectly to the genome, a contaminated library has characteristics that can be recognized by examining the library’s sequences as a group. We use information from clustering and assembly of the entire EST dataset to analyze each EST library. After identifying a library as being contaminated, we discard from it specific sequences that are likely to represent the contamination. We used this library-level analysis to screen large EST datasets for contamination with genomic DNA, pre-mRNA, and non-canonical introns.
MATERIALS AND METHODS
Our analyses were based on examining gene structure and splice variants, as predicted from alignment of human ESTs to the Human genome. ESTs were from dbEST, GenBank version 126 (17 October 2001) (www.ncbi.nlm.nih.gov/dbEST), which contained almost 3.86 million human EST sequences. The draft human genome, build 26 (17 October 2001) (www.ncbi.nlm.nih.gov/genome/guide/human), was used to guide the EST clustering and gene structure prediction.
ESTs were clustered and assembled using Compugen’s LEADS system (51). At a high level, the clustering process uses overlaps between ESTs to define EST clusters, each corresponding to a complete or partial gene. The assembly process then uses the overlap patterns to predict gene structure and splice variants for each cluster (52,53). ESTs that do not share overlaps with other expressed sequences, and so are in clusters by themselves, are called singletons.
We provide a brief description of the operation of the LEADS system; further details are available (51–53). The EST dataset was first cleaned by computationally aligning each EST to a database of typical confounding sequences, such as vectors, linkers and sequences from other organisms. Immunoglobulin and T-cell receptor sequences, whose complicated rearrangement patterns unduly complicate the clustering and assembly process, were also removed.
Repetitive elements and low complexity regions were then masked. Repetitive elements were found using a heuristic alignment model. Seeds of hits to repeats were filtered and extended using Smith–Waterman (54) alignment with the parameters match = 1, mismatch = –3, gap_open = –5, gap_extension = –5 and min_score = 22.
The remaining 3.53 million EST sequences were aligned to the genome using a splicing model that allows long gaps. Alignment at better than 94% identity was required for an EST to be included for further processing; sequences with no good genomic alignment were not considered further. ESTs with multiple good hits were analyzed in more detail, taking into account percent identity and intron content (to differentiate between genes and processed pseudogenes), to choose the correct location. Sequences with different segments aligned to multiple chromosomes, or whose alignments to a single chromosome included inferred introns of more than 400 000 bases, were suspected of being chimeras and discarded. Low quality regions from the ends of the ESTs, based on comparison to the genomic DNA, were trimmed.
The preceding steps left 3.05 million EST sequences for clustering and assembly. Multiple alignments were created from the genomic sequence and the ESTs aligned to it. Positions in which at least one EST opened or closed a long genomic gap were treated as splice sites, with preference given to gaps that began with the bases GT or GC and ended with AG.
Assembly of 14 clusters failed because of various data problems. In addition, computer processing limitations precluded analysis of 110 clusters containing more than 1000 sequences. Although other clustering and assembly tools may be able to accommodate these large clusters, omitting them would not change the nature of our results. The final input to our analysis included 2.72 million sequences from 6649 EST libraries.
RESULTS
The analyses were based on assembled clusters of ESTs created from aligning all human sequences from dbEST to the draft human genome, as described in Materials and Methods. Using the gene structures described by the assemblies, we calculated the percent of unspliced singleton sequences, the percent of sequences that overlap introns, and the percent of sequences that span non-canonical introns for each EST library. For each characteristic we calculated the mean and standard deviation for the entire set of EST libraries, and libraries with percentages more than three standard deviations above the mean were flagged as possibly being contaminated with human genomic DNA, contamination with pre-mRNA, and prevalence of non-canonical introns, respectively, as described below in detail. In each case, our results were supported by additional evidence.
For the statistics to be meaningful, we considered only EST libraries containing at least 100 ESTs that appeared in at least 50 clusters. The 1906 libraries that met these criteria included 2.52 million sequences after the processing described in Materials and Methods. The complete set of characteristics calculated for all libraries is available in the online Supplementary Material.
Human genomic DNA contamination
When a library is contaminated with human genomic DNA, the contaminating sequences will all be unspliced relative to the genome, since each contaminating sequence is contiguous genomic sequence. In addition, many contaminating sequences will be from intergenic regions, because between 64 and 75% of the genome consists of intergenic regions (2), and contaminating sequences originate randomly from different parts of the genome. The probability that a randomly chosen contaminating sequence from an intergenic region will overlap another such sequence is small, so many sequences contaminated with genomic DNA will be singletons (clusters containing only a single EST) after clustering. Libraries contaminated with genomic DNA will therefore contain an overabundance of unspliced singletons. As singleton clusters may represent rarely expressed genes, discarding all unspliced singletons would be wasteful.
We used the fraction of unspliced singletons in a library to recognize libraries with human genomic DNA contamination. In order to determine what value of this fraction indicates a problem, we compared the values of these fractions from all the libraries. Libraries whose values were far from the mean were likely to be contaminated.
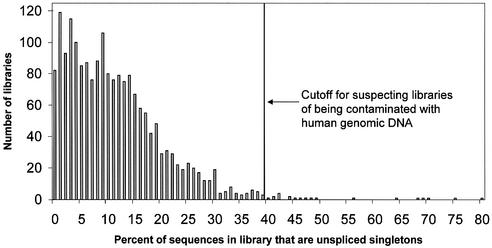
Figure 1 shows a histogram of the number of EST libraries as a function of the percentage of unspliced singletons in the library. The mean percentage of unspliced singletons among a library’s sequences was 11.3%, with a standard deviation of 9.3%. Libraries whose percentage of unspliced singletons was three or more standard deviations above the mean (39.3%) were flagged as possibly being contaminated with human genomic DNA. Although the distribution of percentages may be non-normal, this is a conservative approach to choosing data to omit from the dataset. Of the 1906 libraries considered, 21 libraries (1.1%) were so labeled; they are listed in Table 1.
Figure 1.
Number of EST libraries as a function of the percentage of unspliced singletons in the library. The percentage of unspliced singletons in each of the 1906 libraries was computed using the LEADS clustering and assembly tool. The mean percentage was 11.3%, with a standard deviation of 9.3%. The cut-off indicated by the arrow is three standard deviations above the mean, or 39.3%. The 21 libraries with percentages above this limit are probably highly contaminated with human genomic DNA.
Table 1. Libraries contaminated with human genomic DNA.
| Library | Sequences | Clusters | % Singletons | % Unspliced sequences | % Unspliced singletons |
|---|---|---|---|---|---|
| Liver III | 134 | 134 | 80.6 | 99.3 | 79.9 |
| Liver, hepatocellular carcinoma | 107 | 103 | 76.6 | 98.1 | 74.8 |
| NCI_CGAP_PHE1 | 980 | 965 | 72.3 | 95.7 | 70.2 |
| NCI_CGAP_GAS1 | 616 | 597 | 70.3 | 95.8 | 69.2 |
| Fetal brain, Stratagene | 341 | 326 | 69.2 | 93.5 | 68.3 |
| NCI_CGAP_THY4 | 152 | 140 | 70.4 | 82.2 | 64.5 |
| HTE | 233 | 230 | 64.8 | 75.5 | 56.2 |
| Fetal brain library | 138 | 107 | 52.9 | 86.2 | 48.6 |
| ET0111 | 142 | 115 | 49.3 | 86.6 | 47.9 |
| Stratagene fetal retina 937202 | 4,494 | 3,729 | 48.2 | 82.9 | 47.4 |
| Chromosome 22 exon | 445 | 376 | 48.3 | 85.4 | 45.6 |
| NN0032 | 100 | 97 | 50.0 | 75.0 | 45.0 |
| GN0139 | 123 | 110 | 47.2 | 82.1 | 43.9 |
| WATM1 | 323 | 308 | 43.7 | 78.6 | 43.7 |
| NT0217 | 128 | 116 | 42.2 | 79.7 | 42.2 |
| HTF | 2,240 | 1,766 | 50.0 | 79.2 | 41.9 |
| Selected chromosome 21 cDNA library | 439 | 328 | 43.7 | 85.4 | 41.7 |
| NCI_CGAP_SS1 | 448 | 424 | 42.4 | 73.0 | 41.5 |
| BT0677 | 118 | 109 | 43.2 | 81.4 | 40.7 |
| Outward Alu-Primed HncDNA library | 214 | 167 | 41.6 | 82.7 | 40.7 |
| ET0119 | 206 | 198 | 42.2 | 71.4 | 39.8 |
EST libraries with percentages of unspliced singletons above the cut-off of 39.3%, sorted by that percentage. These libraries are likely highly contaminated with human genomic DNA. The columns show the number of ESTs from a library, the number of clusters in which the sequences appear, and the percentages of sequences in the library that are singletons, unspliced and unspliced singletons in the clustering and assembly output.
Although most sequences from genomic DNA contamination in these libraries are singletons, some do find their way into clusters that contain other ESTs. Figure 2 illustrates the problem caused by including such a contaminating sequence in a cluster; in this example, an extra transcript is predicted, but it is likely to be spurious and will confound any conclusions drawn from this cluster. Although this sequence could be the result of alternative splicing, its source being a library that contains 70% unspliced singletons (NCI_ CGAP_PHE1) makes the alternative splicing explanation unlikely.
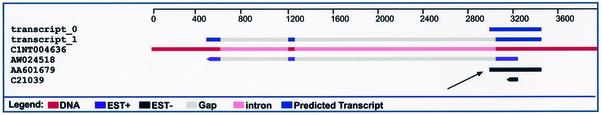
Figure 2.
A cluster that contains a sequence suspected of being human genomic DNA contamination. Although most ESTs that represent genomic DNA come from intergenic regions, some appear in clusters that contain other sequences. This figure shows an example of such a case. The two dark blue lines at the top represent predicted transcripts, the red line below them represents the genome, and the three following black and purple lines represent ESTs. The second EST (black bar) represents sequence AA601679 from EST library NCI_CGAP_PHE1, which was identified as having a high rate of genomic DNA contamination. The extension of the black bar to the left of the purple bar above it (denoted by the thin black arrow) is probably a portion of an intron that was included because of genomic DNA contamination. This resulted in the prediction of an additional transcript (transcript_0) that is likely to be spurious.
In order to remove as many contaminating ESTs as possible without losing too many informative sequences, we suggest discarding all singleton sequences from contaminated libraries, as well as all unspliced sequences from contaminated libraries that appear in clusters that contain more than one EST. In our dataset, this corresponded to discarding 11 667 sequences. Of these, 6370 were singletons; discarding them removed an equal number of falsely predicted genes. The remaining 5297 sequences were contained in 3481 clusters; discarding them eliminated 1175 possibly spurious transcript predictions.
The first five entries in Table 1, the libraries with the largest percentages of unspliced singletons, are of particular interest. Over 90% of the sequences in each library are unspliced, and 68–80% of their sequences are unspliced singletons. These libraries are probably almost entirely contamination; that is, almost all their sequences represent genomic DNA rather than portions of mRNAs.
Seeking confirmation of these results, we compared the repeat content of the discarded sequences to that of a randomly chosen set of EST sequences. Nearly 45% of the genome consists of interspersed repeats, but these appear mainly in introns and intergenic regions (48). Although expressed sequences contain repeats, the repeat rate is much higher in regions that are not expressed. Sequences contaminated with genomic DNA will therefore tend to contain more repetitive regions than will ESTs.
Of the 11 667 sequences that were discarded because of suspected contamination with human genomic DNA, 3524 (30.2%) contained significant hits to human repetitive elements (see Materials and Methods for details). In contrast, in a sample of 10 000 randomly chosen ESTs, only 806 (8.1%) contained such hits. This indicates that the eliminated sequences were indeed genomic contamination.
Pre-mRNA contamination
Unlike the sequences that contain DNA contamination, pre-mRNA sequences should appear mostly within genes, since by definition they come from expressed regions. Because genes consist mainly of introns (48), most ESTs derived from pre-mRNA sequences will be partly or completely contained within introns. As a result, they will cause clustering and assembly tools to incorrectly treat the intronic portions of the pre-mRNA sequences as exons and will lead to the prediction of spurious transcripts.
In order to identify contamination from pre-mRNA, we first identified introns on the genome. For this purpose, an intron was defined as a gap of at least 15 bases in the alignment of an expressed sequence to the genome that begins with the bases ‘GT’ or ‘GC’ and ends with ‘AG’ (see Non-canonical introns, below, for further details about common sequences that start and end introns). We then looked for putative unspliced EST sequences that overlap (but are not completely contained in) introns and measured the percentage of such sequences in each EST library.
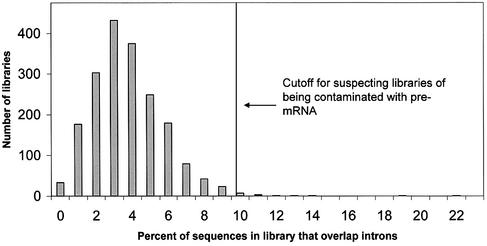
A histogram of the number of EST libraries as a function of the percentage of sequences in the library that overlap introns appears in Figure 3. The mean percentage of library sequences that overlap introns was 3.7%, with a standard deviation of 2.0%. Using the criterion of three standard deviations, the cut-off for labeling a library as being contaminated with pre-mRNA was 9.8%. Table 2 lists the 14 libraries (0.7% of the libraries analyzed) with percentages above the cut-off. Interestingly, all libraries listed in Table 2 were prepared with a protocol that uses random primers for cDNA amplification (40,55). The different protocol may explain the high contamination in these libraries, since most other protocols use poly-T primers for cDNA amplification, and poly-T primers prefer mature mRNAs that contain poly-A tails.
Figure 3.
Number of EST libraries as a function of the percentage of sequences in the library that overlap introns. The percentage of sequences that overlap introns in each of the 1906 libraries was computed using the LEADS clustering and assembly tool. The mean percentage was 3.7%, with a standard deviation of 2.0%. The cut-off indicated by the arrow is three standard deviations above the mean, or 9.8%. The 14 libraries with percentages above this limit are probably highly contaminated with pre-mRNA sequences.
Table 2. Libraries contaminated with pre-mRNA.
| Library | Sequences | Clusters | % Sequences that partially overlap introns | % Unspliced sequences |
|---|---|---|---|---|
| KT0031 | 169 | 94 | 22.5 | 69.8 |
| ST0256 | 149 | 61 | 18.8 | 69.8 |
| ST0186 | 540 | 266 | 14.1 | 57.0 |
| CT0352 | 170 | 75 | 12.9 | 43.5 |
| ST0173 | 134 | 73 | 11.9 | 60.4 |
| HT0397 | 174 | 59 | 11.5 | 57.5 |
| MT0226 | 101 | 74 | 10.9 | 59.4 |
| ST0118 | 245 | 69 | 10.6 | 51.4 |
| UT0047 | 134 | 94 | 10.4 | 71.6 |
| HT1146 | 106 | 88 | 10.4 | 44.3 |
| ET0193 | 164 | 130 | 10.4 | 51.8 |
| UT0116 | 281 | 167 | 10.3 | 52.7 |
| SN0010 | 146 | 124 | 10.3 | 69.2 |
| PT0001 | 142 | 67 | 9.9 | 51.4 |
EST libraries with percentages of pre-mRNA contamination above the cut-off of 9.8%, sorted by that percentage. The columns show the number of ESTs from a library, the number of clusters in which the sequences appear, the percentage of sequences in the library that partially overlap introns, and the percentage of sequences that are unspliced in the clustering and assembly output.
As a conservative approach for excluding sequences that may be problematic, we suggest omitting all sequences from these libraries that overlap an intron (as defined above), or that are unspliced relative to the genome. In our test set, 2128 sequences were deleted from 1104 clusters, resulting in the elimination of 538 transcript predictions that were probably spurious.
To verify our results, we compared the repeat content of the discarded sequences to that of a random set of ESTs. Of the 2128 deleted sequences, 346 (16.3%) contained significant hits to human interspersed repeats, compared with 8.1% of the control set. The difference, though significant, is smaller than the difference for genomic DNA contamination. This is probably because pre-mRNA sequences contain some expressed sequence, but genomic contamination is rarely from expressed regions.
Non-canonical introns
Introns that begin with the bases GT or GC and end with the bases AG are referred to as ‘canonical introns’. The overwhelming majority (98.12%) of introns are of the GT/AG kind, and 0.76% are of the GC/AG kind (48). We have found, though, that some libraries have a much larger than normal fraction of sequences that span non-canonical introns. Although this could, in principle, happen in nature, it is probably a symptom of other problems that adversely affect use of these libraries in generating consensus sequences from EST clusters.
In order to evaluate this kind of contamination in EST libraries, we identified intron ends by examining the alignment of the ESTs to the genome, and flagged ESTs that span at least one non-canonical intron. In virtually all libraries analyzed (1878 of 1906), no more than 5% of the sequences in the library were flagged; the mean percentage was 1% and the standard deviation was 2.28%. The mean percentage of non-canonical introns is much lower than 1%, as most sequences contain multiple introns.
Using a cut-off of three standard deviations above the mean, libraries in which more than 7.8% of the sequences were flagged were identified as contaminated with sequences spanning non-canonical introns. The 19 libraries (1%) so identified are listed in Table 3. One library, HTE, is also listed in Table 1 as probably contaminated with human genomic DNA.
Table 3. Libraries exhibiting non-canonical introns.
| Library | Sequences | Clusters | % Sequences with non-canonical introns |
|---|---|---|---|
| FHTA | 137 | 120 | 39.4 |
| FHTB | 127 | 116 | 31.5 |
| GLC | 12 714 | 4617 | 27.0 |
| ADC | 1431 | 1122 | 26.6 |
| DCB | 2831 | 1480 | 26.3 |
| HTC | 3206 | 2091 | 25.5 |
| GKC | 11 420 | 3607 | 24.8 |
| CDA | 1498 | 1163 | 23.4 |
| ADA | 297 | 160 | 22.9 |
| ADB | 4608 | 2780 | 22.3 |
| PLACE4 | 367 | 130 | 16.6 |
| GKB | 981 | 662 | 16.3 |
| DCA | 1104 | 708 | 15.4 |
| CU | 883 | 575 | 15.2 |
| MDS | 2785 | 1717 | 10.5 |
| NPA | 794 | 217 | 9.6 |
| NPC | 3143 | 2177 | 8.5 |
| NPD | 1304 | 1040 | 8.4 |
| HTE | 233 | 230 | 8.2 |
EST libraries with percentages of non-canonical introns above the cut-off of 7.84%, sorted by that percentage. Library PLACE4 is not considered problematic, as discussed in the text. The columns show the number of ESTs from a library, the number of clusters in which the sequences appear, and the percentage of sequences in the library that span a non-canonical intron when aligned to the genome.
Although we do not understand why some libraries have such a large fraction of sequences that span non-canonical introns, several factors indicate that these non-canonical introns are experimental artifacts rather than biological reality. First, all the libraries but one (PLACE4 is the exception) were generated in the same institute. Secondly, these putative introns differ dramatically in size from typical introns. In particular, 64% of the non-canonical introns from these libraries (excluding those from library PLACE4) are 51–59 bases long, with 46% being exactly 54 bases long. Over the entire human genome, the average intron size is more than 3000 bases (48). This points to some kind of error in the creation of these ESTs or in the reporting of their sequences.
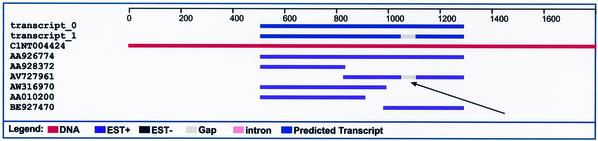
Figure 4 illustrates a cluster containing a sequence from a library that exhibits non-canonical introns. The gap denoted by the black arrow is a non-canonical intron (it begins with AA and ends with TT) and is 54 bases long. It appears only in the one sequence in that cluster that comes from a library detected as possibly contaminated. This gap may represent sequence that has somehow been incorrectly deleted from the EST.
Figure 4.
A cluster that exhibits a non-canonical intron. The two dark blue lines at the top represent predicted transcripts, the red line below them represents the genome, and the six following purple lines represent ESTs. The gap indicated by the black arrow begins with AA and ends with TT (i.e., it is a non-canonical intron). The gap is 54 bases long, which is similar to many of the observed non-canonical introns found in problematic libraries. The gap occurred only in the sequence that came from a library detected as possibly contaminated; it may correspond to sequence that was incorrectly deleted from some ESTs. This gap resulted in the prediction of a splice variant (transcript_1) that is likely spurious.
The library PLACE4 merits special attention. All sequences from this library that contain non-canonical introns share a single non-canonical intron in a single cluster. This is quite different from the other libraries. The fact that all other libraries listed in Table 3 are from a single source, and PLACE4 is from a different source, leads us to conjecture that this cluster exhibits a rare but real splicing pattern. We therefore did not discard data from PLACE4, even though it has a large percentage of sequences with non-canonical introns.
As with the problems discussed above, we suggest a conservative approach to eliminating sequences to retain as much useful information as possible. We recommend discarding all sequences from these libraries that have non-canonical introns. This resulted in the omission of 10 971 sequences and 7862 possibly spurious transcript predictions contained in 5074 clusters.
DISCUSSION
We have demonstrated a novel method for recognizing EST contamination that cannot be identified using previously known approaches. Rather than analyzing ESTs one at a time, entire EST libraries are examined en masse. Characteristics of the libraries, rather than those of individual ESTs, are used to find contamination. The result is EST datasets that give more accurate predictions of gene structure and alternative splicing, and so are more effective in the myriad applications for which ESTs are used.
Applying these methods to dbEST, version 126, eliminated 24 766 ESTs (0.9% of the sequences contained in the output from clustering and assembly) from 16 029 clusters. Removing these sequences caused 6370 singleton clusters to disappear, and 9575 probably spurious transcript predictions were dropped. The numbers of ESTs removed as a result of each problem described earlier, and the effect of such removal on the numbers of gene and transcript predictions, are summarized in Table 4. Any study that uses ESTs for purposes of prediction must consider ignoring these contaminating sequences.
Table 4. The numbers of ESTs removed as a result of each kind of problem, and the effect of removing the sequences on the numbers of predicted genes and transcripts.
| Problem | Sequences removed | Singletons eliminated | Sequences removed from clusters | Clusters affected | Transcripts eliminated |
|---|---|---|---|---|---|
| Human genomic DNA contamination | 11 667 | 6370 | 5297 | 3481 | 1175 |
| Pre-mRNA contamination | 2128 | 0 | 2128 | 1104 | 538 |
| Non-canonical introns | 10 971 | 0 | 10 971 | 5074 | 7862 |
| Total | 24 766 | 6370 | 18 396 | 9659 | 9575 |
Each removed singleton corresponds to eliminating a spurious gene prediction.
We note that because our approach is based on statistical characteristics of a library, it does not work well for small libraries or libraries whose sequences appear in only a small number of clusters. Many libraries are smaller than the cut-offs used for this analysis and may contain contamination that we cannot detect.
This research can be extended in several directions. Some EST libraries are derived by pooling cDNA clones from multiple tissues. Problems in sequences from one tissue might be reduced to apparently reasonable levels when several tissues are combined. The source tissue of each clone can, however, be determined by using specially inserted tags (56). One could separate pooled libraries into tissue-specific subsets and apply our method to the sequences from each tissue separately.
Since the sequence of the human genome is still at a draft stage, applying our method using other versions of the human genome may alter the numbers slightly. Doing so should not affect the overall per-library results, though, because the analysis used only EST libraries that appeared in at least 50 different EST clusters, corresponding to at least 50 different genomic loci.
We point out in addition that our method does not depend on use of a specific clustering and assembly tool. It could therefore be implemented using other EST clustering and assembly tools, such as STACK (31) or TAP (17).
Our approach can also be used to improve existing methods for cleaning EST datasets. For example, one could identify ESTs that align well to the genome, except at the end. A library with a large percentage of such sequences should be investigated to see if it was prepared with a vector or linker that does not appear in the screening databases. Similarly, protocol errors may have occurred in generating a library with a large fraction of chimeric sequences. In general, this methodology can be used to implement a suite of tests for cleaning EST data in a more thorough manner than is currently done.
Our method and the possibly contaminated libraries that it identified have significant implications for projects that use ESTs to predict gene structure and alternative splicing patterns. The improved predictions will translate into more accurate results for the wide variety of applications based on EST data.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Alex Golubev for computing support; Zurit Levine, Doug Smith and Amir Toporik for helpful conversations; Gila Naveh for literature assistance; and Pini Akiva, Dvir Dahary, Raveh Gill-More, Dan Graur, Erez Levanon, Galit Rotman, Amir Toporik, Alon Wasserman, Frances Zetland and the anonymous referees for commenting on earlier versions of the manuscript. This work was performed in partial fulfilment of the requirements for a Ph.D. degree of Rotem Sorek, Sackler Faculty of Medicine, Tel Aviv University, Israel.
REFERENCES
- 1.Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A., et al. (2001)The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Boguski M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) dbEST–database for ‘expressed sequence tags’. Nature Genet., 4, 332–333. [DOI] [PubMed] [Google Scholar]
- 4.Boguski M.S. (1995) The turning point in genome research. Trends Biochem. Sci., 20, 295–296. [DOI] [PubMed] [Google Scholar]
- 5.Marra M.A., Hillier,L. and Waterston,R.H. (1998) Expressed sequence tags—ESTablishing bridges between genomes. Trends Genet., 14, 4–7. [DOI] [PubMed] [Google Scholar]
- 6.Adams M.D., Dubnick,M., Kerlavage,A.R., Moreno,R., Kelley,J.M., Utterback,T.R., Nagle,J.W., Fields,C. and Venter,J.C. (1992) Sequence identification of 2,375 human brain genes. Nature, 355, 632–634. [DOI] [PubMed] [Google Scholar]
- 7.Adams M.D., Kerlavage,A.R., Fields,C. and Venter,J.C. (1993) 3,400 new expressed sequence tags identify diversity of transcripts in human brain. Nature Genet., 4, 256–267. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov R., Preston-Hurlburt,P. and Janeway,C.A.,Jr (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature, 388, 394–397. [DOI] [PubMed] [Google Scholar]
- 10.Liang F., Holt,I., Pertea,G., Karamycheva,S., Salzberg,S.L. and Quackenbush,J. (2000) Gene index analysis of the human genome estimates approximately 120,000 genes. Nature Genet., 25, 239–240. [DOI] [PubMed] [Google Scholar]
- 11.Hudson T.J., Stein,L.D., Gerety,S.S., Ma,J., Castle,A.B., Silva,J., Slonim,D.K., Baptista,R., Kruglyak,L., Xu,S.H. et al. (1995) An STS-based map of the human genome. Science, 270, 1945–1954. [DOI] [PubMed] [Google Scholar]
- 12.Schuler G.D., Boguski,M.S., Stewart,E.A., Stein,L.D., Gyapay,G., Rice,K., White,R.E., Rodriguez-Tome,P., Aggarwal,A., Bajorek,E. et al. (1996) A gene map of the human genome. Science, 274, 540–546. [PubMed] [Google Scholar]
- 13.Deloukas P., Schuler,G.D., Gyapay,G., Beasley,E.M., Soderlund,C., Rodriguez-Tome,P., Hui,L., Matise,T.C., McKusick,K.B., Beckmann,J.S. et al. (1998) A physical map of 30,000 human genes. Science, 282, 744–746. [DOI] [PubMed] [Google Scholar]
- 14.Waterston R., Martin,C., Craxton,M., Huynh,C., Coulson,A., Hillier,L., Durbin,R., Green,P., Shownkeen,R., Halloran,N. et al. (1992) A survey of expressed genes in Caenorhabditis elegans. Nature Genet., 1, 114–123. [DOI] [PubMed] [Google Scholar]
- 15.McCombie W.R., Adams,M.D., Kelley,J.M., FitzGerald,M.G., Utterback,T.R., Khan,M., Dubnick,M., Kerlavage,A.R., Venter,J.C. and Fields,C. (1992) Caenorhabditis elegans expressed sequence tags identify gene families and potential disease gene homologues. Nature Genet., 1, 124–131. [DOI] [PubMed] [Google Scholar]
- 16.Brody L.C., Abel,K.J., Castilla,L.H., Couch,F.J., McKinley,D.R., Yin,G., Ho,P.P., Merajver,S., Chandrasekharappa,S.C., Xu,J. et al. (1995) Construction of a transcription map surrounding the BRCA1 locus of human chromosome 17. Genomics, 25, 238–247. [DOI] [PubMed] [Google Scholar]
- 17.Kan Z., Rouchka,E.C., Gish,W.R. and States,D.J. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res., 11, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tugendreich S., Bassett,D.E.,Jr, McKusick,V.A., Boguski,M.S. and Hieter,P. (1994) Genes conserved in yeast and humans. Hum. Mol. Genet., 3, 1509–1517. [DOI] [PubMed] [Google Scholar]
- 19.Papadopoulos N., Nicolaides,N.C., Wei,Y.F., Ruben,S.M., Carter,K.C., Rosen,C.A., Haseltine,W.A., Fleischmann,R.D., Fraser,C.M., Adams,M.D. et al. (1994) Mutation of a mutL homolog in hereditary colon cancer. Science, 263, 1625–1629. [DOI] [PubMed] [Google Scholar]
- 20.Adams M.D., Kerlavage,A.R., Fleischmann,R.D., Fuldner,R.A., Bult,C.J., Lee,N.H., Kirkness,E.F., Weinstock,K.G., Gocayne,J.D., White,O. et al. (1995) Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature, 377, 3–174. [PubMed] [Google Scholar]
- 21.Braren R., Firner,K., Balasubramanian,S., Bazan,F., Thiele,H.G., Haag,F. and Koch-Nolte,F. (1997) Use of the EST database resource to identify and clone novel mono(ADP-ribosyl)transferase gene family members. Adv. Exp. Med. Biol., 419, 163–168. [DOI] [PubMed] [Google Scholar]
- 22.Allikmets R., Gerrard,B., Glavac,D., Ravnik-Glavac,M., Jenkins,N.A., Gilbert,D.J., Copeland,N.G., Modi,W. and Dean,M. (1995) Characterization and mapping of three new mammalian ATP-binding transporter genes from an EST database. Mam. Genome, 6, 114–117. [DOI] [PubMed] [Google Scholar]
- 23.Nelson P.S., Han,D., Rochon,Y., Corthals,G.L., Lin,B., Monson,A., Nguyen,V., Franza,B.R., Plymate,S.R., Aebersold,R. et al. (2000) Comprehensive analyses of prostate gene expression: convergence of expressed sequence tag databases, transcript profiling and proteomics. Electrophoresis, 21, 1823–1831. [DOI] [PubMed] [Google Scholar]
- 24.Garg K., Green,P. and Nickerson,D.A. (1999) Identification of candidate coding region single nucleotide polymorphisms in 165 human genes using assembled expressed sequence tags. Genome Res., 9, 1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buetow K.H., Edmonson,M.N. and Cassidy,A.B. (1999) Reliable identification of large numbers of candidate SNPs from public EST data. Nature Genet., 21, 323–325. [DOI] [PubMed] [Google Scholar]
- 26.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 27.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reese M.G., Hartzell,G., Harris,N.L., Ohler,U., Abril,J.F. and Lewis,S.E. (2000) Genome annotation assessment in Drosophila melanogaster. Genome Res., 10, 483–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuler G.D. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 30.Quackenbush J., Cho,J., Lee,D., Liang,F., Holt,I., Karamycheva,S., Parvizi,B., Pertea,G., Sultana,R. and White,J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res., 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller R.T., Christoffels,A.G., Gopalakrishnan,C., Burke,J., Ptitsyn,A.A., Broveak,T.R. and Hide,W.A. (1999) A comprehensive approach to clustering of expressed human gene sequence: the sequence tag alignment and consensus knowledge base. Genome Res., 9, 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krause A., Haas,S.A., Coward,E. and Vingron,M. (2002) SYSTERS, GeneNest, SpliceNest: exploring sequence space from genome to protein. Nucleic Acids Res., 30, 299–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfsberg T.G. and Landsman,D. (1997) A comparison of expressed sequence tags (ESTs) to human genomic sequences. Nucleic Acids Res., 25, 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillier L.D., Lennon,G., Becker,M., Bonaldo,M.F., Chiapelli,B., Chissoe,S., Dietrich,N., DuBuque,T., Favello,A., Gish,W. et al. (1996) Generation and analysis of 280,000 human expressed sequence tags. Genome Res., 6, 807–828. [DOI] [PubMed] [Google Scholar]
- 35.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 36.Bonaldo M.F., Lennon,G. and Soares,M.B. (1996) Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res., 6, 791–806. [DOI] [PubMed] [Google Scholar]
- 37.Peterson L.A., Brown,M.R., Carlisle,A.J., Kohn,E.C., Liotta,L.A., Emmert-Buck,M.R. and Krizman,D.B. (1998) An improved method for construction of directionally cloned cDNA libraries from microdissected cells. Cancer Res, 58, 5326–5328. [PubMed] [Google Scholar]
- 38.Krizman D.B., Chuaqui,R.F., Meltzer,P.S., Trent,J.M., Duray,P.H., Linehan,W.M., Liotta,L.A. and Emmert-Buck,M.R. (1996) Construction of a representative cDNA library from prostatic intraepithelial neoplasia. Cancer Res., 56, 5380–5383. [PubMed] [Google Scholar]
- 39.Kotewicz M.L. and Gerard,G.F. (1997) United States Patent 6,063,608.
- 40.Camargo A.A., Samaia,H.P., Dias-Neto,E., Simao,D.F., Migotto,I.A., Briones,M.R., Costa,F.F., Nagai,M.A., Verjovski-Almeida,S., Zago,M.A. et al. (2001) The contribution of 700,000 ORF sequence tags to the definition of the human transcriptome. Proc. Natl Acad. Sci. USA, 98, 12103–12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou H.H. and Holmes,M.H. (2001) DNA sequence quality trimming and vector removal. Bioinformatics, 17, 1093–1104. [DOI] [PubMed] [Google Scholar]
- 42.Weber G., Shendure,J., Tanenbaum,D.M., Church,G.M. and Meyerson,M. (2002) Identification of foreign gene sequences by transcript filtering against the human genome. Nature Genet., 30, 141–142. [DOI] [PubMed] [Google Scholar]
- 43.Burke J., Wang,H., Hide,W. and Davison,D.B. (1998) Alternative gene form discovery and candidate gene selection from gene indexing projects. Genome Res., 8, 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aaronson J.S., Eckman,B., Blevins,R.A., Borkowski,J.A., Myerson,J., Imran,S. and Elliston,K.O. (1996) Toward the development of a gene index to the human genome: an assessment of the nature of high-throughput EST sequence data. Genome Res., 6, 829–845. [DOI] [PubMed] [Google Scholar]
- 45.Ewing B. and Green,P. (2000) Analysis of expressed sequence tags indicates 35,000 human genes. Nature Genet., 25, 232–234. [DOI] [PubMed] [Google Scholar]
- 46.Huang X. and Madan,A. (1999) CAP3: a DNA sequence assembly program. Genome Res., 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Croft L., Schandorff,S., Clark,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]
- 48.International Human Genome Sequencing Consortium. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 49.Ding W.Q., Kuntz,S.M. and Miller,L.J. (2002) A misspliced form of the cholecystokinin-B/gastrin receptor in pancreatic carcinoma: role of reduced cellular U2AF35 and a suboptimal 3′-splicing site leading to retention of the fourth intron. Cancer Res., 62, 947–952. [PubMed] [Google Scholar]
- 50.Flowers J.M., Powell,J.F., Leigh,P.N., Andersen,P. and Shaw,C.E. (2001) Intron 7 retention and exon 9 skipping EAAT2 mRNA variants are not associated with amyotrophic lateral sclerosis. Ann. Neurol., 49, 643–649. [PubMed] [Google Scholar]
- 51.Shoshan A., Grebinskiy,V., Magen,A., Scolnicov,A., Fink,E., Lehavi,D. and Wasserman,A. (2001) In Bittner,M.L., Chen,Y., Dorsel,A.N. and Dougherty,E.R. (eds), Proceedings of SPIE: Microarrays: Optical Technologies and Informatics, Vol. 4266, pp. 86–95.
- 52.Sorek R., Ast,G. and Graur,D. (2002) Alu-containing exons are alternatively spliced. Genome Res., 12, 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie H., Zhu,W., Wasserman,A., Grebinskiy,V., Olson,A. and Mintz,L. (2002) Computational analysis of alternative splicing using EST tissue information. Genomics, 80, 326–330. [DOI] [PubMed] [Google Scholar]
- 54.Smith T.F. and Waterman,M.S. (1981) Comparison of biosequences. Adv. Appl. Math., 2, 482–489. [Google Scholar]
- 55.deSouza S.J., Camargo,A.A., Briones,M.R., Costa,F.F., Nagai,M.A., Verjovski-Almeida,S., Zago,M.A., Andrade,L.E., Carrer,H., El-Dorry,H.F. et al. (2000) Identification of human chromosome 22 transcribed sequences with ORF expressed sequence tags. Proc. Natl Acad. Sci. USA, 97, 12690–12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gavin A.J., Scheetz,T.E., Roberts,C.A., O’Leary,B., Braun,T.A., Sheffield,V.C., Soares,M.B., Robinson,J.P. and Casavant,T.L. (2002) Pooled library tissue tags for EST-based gene discovery. Bioinformatics, 18, 1162–1166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.