Abstract
Small nucleolar RNAs (designated as snoRNAs in Eukarya or sRNAs in Archaea) can be grouped into H/ACA or C/D box snoRNA (sRNA) subclasses. In Eukarya, H/ACA snoRNAs assemble into a ribonucleoprotein (RNP) complex comprising four proteins: Cbf5p, Gar1p, Nop10p and Nhp2p. A homolog for the Nhp2p protein has not been identified within archaeal H/ACA RNPs thus far, while potential orthologs have been identified for the other three proteins. Nhp2p is related, particularly in the middle portion of the protein sequence, to the archaeal ribosomal protein and C/D box protein L7Ae. This finding suggests that L7Ae may be able to substitute for the Nhp2p protein in archaeal H/ACA sRNAs. By band shift assays, we have analyzed in vitro the interaction between H/ACA box sRNAs and protein L7Ae from the archaeon Archaeoglobus fulgidus. We present evidence that L7Ae forms specific complexes with three different H/ACA sRNAs, designated as Afu-4, Afu-46 and Afu-190 with an apparent Kd ranging from 28 to 100 nM. By chemical and enzymatic probing we show that distinct bases located within bulges or loops of H/ACA sRNAs interact with the L7Ae protein. These findings are corroborated by mutational analysis of the L7Ae binding site. Thereby, the RNA motif required for L7Ae binding exhibits a structure, designated as the K-turn, which is present in all C/D box sRNAs. We also identified four H/ACA RNAs from the archaeal species Pyrococcus which exhibit the K-turn motif at a similar position in their structure. These findings suggest a triple role for L7Ae protein in Archaea, e.g. in ribosomes as well as H/ACA and C/D box sRNP biogenesis and function by binding to the K-turn motif.
INTRODUCTION
In Eukarya and Archaea, ribonucleoprotein (RNP) complexes guide two types of site-specific modifications within ribosomal RNA (rRNA) or snRNA targets: the 2′-O-methylation of riboses and the pseudouridylation of nucleotides of an RNA target (1–4). These RNP complexes contain small RNAs designated as snoRNAs (in Eukarya) or sRNAs (in Archaea) (5). The snoRNAs/sRNAs exhibit complementarity to specific sites within rRNA or snRNA sequences thereby determining the site of modification (5–7). Based on conserved sequence and structure motifs within snoRNAs (sRNAs), they can be grouped into two subclasses, the C/D box snoRNAs (sRNAs) and the H/ACA box snoRNAs (sRNAs), respectively. C/D box snoRNAs target the 2′-O-methylation of riboses, while H/ACA box snoRNAs target the conversion of uridine into pseudouridine within RNAs (7,8).
The family of C/D box snoRNA (sRNAs) contains two conserved sequence motifs: box C (RUGAUGA) and box D (CUGA) which are located close to the 5′ and 3′ ends of the snoRNA, respectively, while degenerate C and D boxes, designated as boxes C′ and D′, are located in the center of a molecule (7,9,10). The complementarity to sites within rRNAs or snRNAs is found immediately 5′ to D or D′ boxes and usually exhibits a length of 10–22 nt (1,8,9). Methylation of an RNA nucleotide occurs 5 nt upstream from D or D′ boxes, respectively (1,9).
Recently, it has been shown that C and D boxes form a secondary structure motif, designated as the K-turn (10–12). The K-turn motif is highly conserved within C/D box snoRNAs and is formed between C and D boxes via interaction of short inverted repeats present at the 5′ and 3′ ends of snoRNAs (10). Characteristic for this motif are two helical stems separated by an internal three base loop. The internal loop is always asymmetrical and purine-rich and contains three unpaired nucleotides at one side with a terminal U base. The second helix exhibits two sheared G-A base pairs usually followed by a U-U base pair and a regular Watson–Crick base pair (see Fig. 3) (12). In addition to C/D box snoRNAs, K-turn motifs have been shown to be present in various eukaryal and prokaryal RNAs including ribosomal RNAs (12,13) and U3 and U8, C/D box snoRNAs that are not functioning as methylation guides (10).
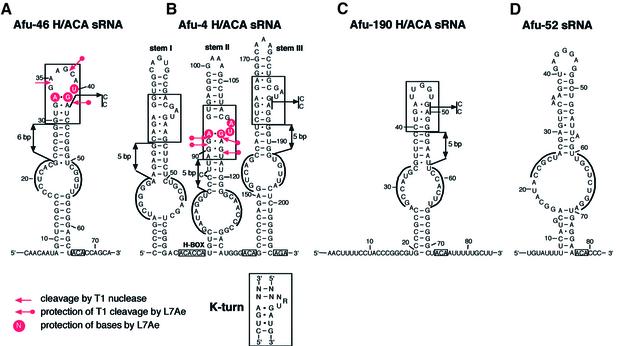
Figure 3.
Secondary structures of Afu-46 (A), Afu-4 (B), Afu-190 (C) and Afu-52 (D) sRNAs. The H or ACA/AGA motifs within sRNAs are boxed and in each pseudouridylation pocket the two tracts of complementarity to the rRNA target are overlined. Protection of bases and phosphodiester bonds within Afu-46 (A) or Afu-4 (B) sRNAs/L7Ae complexes from modification or cleavage by chemical and enzymatic probes is indicated by circles or arrows, respectively. K-turn motifs in Afu-46, Afu-4 and Afu-190 sRNAs are boxed, the consensus K-turn motif for canonical C/D box sRNAs is shown at the bottom. The mutations introduced within the K-turn motif in archaeal Afu-46, Afu-4 or Afu-190 H/ACA sRNAs are indicated by arrows. The distances between the base of stems and the K-turn motifs are indicated by double arrows.
Four proteins represent core components of C/D box snoRNPs in Eukarya: fibrillarin, NOP56, NOP58 and the 15.5 kDa protein (7,10,14). Based on conserved sequence motifs, it had been assumed that fibrillarin serves as the methylase component in this complex (15–17). This was subsequently experimentally confirmed by in vitro analysis of the site-specific methylation guide activity of purified eukaryal C/D snoRNAs (18). Interestingly, the 15.5 kDa protein was also shown to be an integral component of the U4/U6*U5 tri-snRNP complex (10,19). It belongs to a large family of RNA binding proteins, including human ribosomal proteins L7a and S12, as well as yeast ribosomal protein L30 (11,20). The common denominator of this family of proteins is that it shares the capacity to recognize and bind to the K-turn.
Archaeal C/D box sRNAs contain all structural and sequence elements (C, C′, D, D′ boxes) characteristic for eukaryal C/D box snoRNAs. Recently, an in vitro reconstituted archaeal C/D sRNP complex was shown to be able to direct rRNA methylation in vitro (4). The protein components of this complex include aFIB, aNOP56/NOP58 proteins and ribosomal protein L7Ae which are homologous to human nucleolar proteins fibrillarin, NOP56, NOP58 and the 15.5 kDa protein, respectively. Thereby, the archaeal ribosomal protein L7Ae, exhibiting 33% identity and 60% similarity to the 15.5 kDa protein, represents a core component for C/D box sRNPs in Archaea (4,21). Recently, it was demonstrated that the archaeal L7Ae protein can serve as the functional homolog of the human 15.5 kDa protein in vitro (4,21).
The secondary structure of H/ACA box snoRNAs generally consists of two large helices connected by a single-stranded region. Conserved sequence elements: box H (ANANNA, where N represents any nucleotide) and box ACA, are located in between helices or 3 nt upstream from the 3′ end of the snoRNA, respectively (22,23). The bipartite antisense elements, which target the conversion of uridine into pseudouridine within rRNAs, are located in the internal loop within each helix. Pseudouridylation occurs at positions located 14–16 nt upstream from boxes H or ACA (2,8,24).
Eukaryal H/ACA box snoRNPs contain the evolutionary conserved proteins Cbf5p (dyskerin), Gar1p, Nhp2p and Nop10p, which are strictly required for pseudouridylation (6,25–27). Based on conserved sequence motifs, Cbf5p is thought to represent the pseudouridylase enzyme in this complex. Ribosomal RNAs of Archaea contain a few numbers of pseudouridines (24). In our recent search for small, non-messenger RNAs from the archaeon Archaeoglobus fulgidus, we have identified, for the first time, four H/ACA sRNAs guiding a total of six pseudouridylations within 16S and 23S rRNAs (3). Unlike canonical H/ACA snoRNAs, however, they exhibit a three-stem structure or a single-stem structure, as observed in trypanosomes (28).
Analysis of several archaeal genomes showed the presence of archaeal homologs to eukaryal H/ACA proteins Cbf5p (dyskerin), Gar1p and Nop10p, designated as Cbf5, Gar1 and Nop10, respectively (26,29). However, a homolog to the eukaryal Nhp2p has not been identified as of now (29). Interestingly, Nhp2p is related, particularly in the middle portion of the protein, to the C/D box protein Snu13p from yeast and its homologs, the 15.5 kDa in human and the L7Ae protein in Archaea (19). This finding suggests that L7Ae may substitute for the Nhp2p protein in archaeal H/ACA sRNAs. Interestingly, in addition to being a core component of archaeal C/D box sRNAs, the L7Ae protein serves also as an integral component of the large ribosomal subunit in Archaea as a 23S rRNA binding protein (12).
By gel retardation experiments as well as chemical and enzymatic probing we show that archaeal H/ACA box sRNAs bind to the C/D box protein L7Ae in vitro. The binding sites exhibit a common RNA structure motif typically found in all C/D box snoRNAs (sRNAs) as well as in ribosomal RNAs, designated as the K-turn.
MATERIALS AND METHODS
Materials
Dimethylsulfate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene-sulfonate (CMCT) were obtained from Fluka AG (Switzerland). The kethoxal (KE) reagent was purchased from Upjohn (UK); all other reagents and enzymes were purchased from Roche (Germany).
Synthesis of DNA templates for in vitro transcription of RNAs
DNA constructs featuring a T7 promoter followed by the coding sequence for Afu-4, Afu-46, Afu-52 and Afu-190 H/ACA sRNAs were generated by PCR using 0.5 µg of total DNA from A.fulgidus and oligonucleotides: Afu 4-T7+FORV and Afu 4-REV (for Afu-4 sRNA), Afu 46-T7+FORV and Afu 46-REV (for Afu-46 sRNA), Afu 52-T7+FORV and Afu 52-REV (for Afu-52 sRNA) and Afu 190-T7+FORV and Afu 190-REV (for Afu-190 sRNA), respectively. The obtained cDNAs were cloned into vector pCR 2.1 (TA Cloning Kit, Invitrogen) and clones were confirmed by sequence analysis. DNA templates encoding sRNAs mutant genes were generated by PCR (see above) from the same vectors using oligonucleotides: Afu 4-T7+FORV and Af4rev-mSt3(cc) for mutant Afu-4 sRNA, Afu 46-T7+FORV and Afu 46rev-mut(cc) for mutant Afu-46 sRNA and Afu 190-T7+FORV and Af190rev-m(cc) for mutant Afu-190 sRNA.
T7 in vitro transcription of RNAs
RNAs were in vitro transcribed from DNA templates (see above) containing a T7 promoter. Transcription, using T7 RNA polymerase, was performed directly from PCR-amplified DNA fragments (5 µg) in a reaction volume of 400 µl (30). RNAs were separated on denaturing 8% polyacrylamide, 7 M urea gels and passively eluted from the gels in 0.3 M NaOAc buffer (pH 5.2), 1 mM EDTA, 0.2% phenol at 4°C overnight. Subsequently, RNAs were EtOH precipitated and dissolved in 30 µl of H2O.
Purification of L7Ae protein from A.fulgidus
A DNA fragment encoding the gene for ribosomal protein L7Ae from A.fulgidus was generated by PCR amplification of genomic DNA. The reaction was performed using a standard protocol (31) and oligonucleotides For_L7Ae_Afu and Rev_L7Ae_Afu. To facilitate cloning, NdeI and XhoI restriction sites were introduced into the oligonucleotide sequence. For cloning and expression of ribosomal protein L7Ae, vector pET 28-b+ was used. The protein was cloned by adding an additional six histidine residues and a thrombin tag at the N-terminus. The expected clones were confirmed by sequence analysis. The recombinant 6 His-Thrombin-tag-L7Ae protein was purified from Escherichia coli BL21 cells using Ni-NTA Agarose as instructed by the manufacturer (Qiagen, Hilden, Germany). The protein was dialyzed against buffer containing 20 mM HEPES–KOH (pH 7.4), 150 mM KCl, 1.5 mM MgCl2 and 5% glycerol. The His-tag of recombinant L7Ae protein was removed by incubation with 3 U of thrombin protease at 4°C overnight. After incubating the reaction for 15 min at 65°C the protease was inactivated. Protein L7Ae was concentrated by using the Ultrafree-15 Centrifugal Filter Device Biomax-5 (5 kDa concentrator) from Millipore (USA).
Gel shift assay
For binding assays to the archaeal L7Ae protein, H/ACA sRNAs were in vitro transcribed from PCR products containing a T7 promoter in the presence of [α-32P]UTP using 1000 U of T7 RNA polymerase followed by gel purification (see above). Before incubation with recombinant L7Ae protein, RNAs were heat denatured (2 min at 85°C) and subsequently quick-cooled in liquid nitrogen for 20 s, followed by renaturation of sRNAs in binding buffer (see below) for 10 min at 4°C. Complex formation of recombinant protein L7Ae and sRNAs was performed in a reaction volume of 20 µl, containing 20 mM HEPES–KOH (pH 7.4), 150 mM KCl, 1.5 mM MgCl2, 2 mM DTT, 10% glycerol, 5 µg of tRNA and 20 U of ribonuclease inhibitor (RNasin). Aliquots (0.05 pmol) of each sRNA were incubated with increasing concentrations of ribosomal protein L7Ae as indicated in the Figure legends. Reactions were incubated for 1 h at 4°C, followed by 30 min at 37°C. Subsequently, 1 µl (3 µg) of heparin was added, and complexes were incubated for an additional 5 min at 37°C. RNA and RNA–protein complexes were separated on native 8% (or 12%) (w/v) polyacrylamide gels, containing 1× TBE (90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA, pH 8.3). Electrophoresis was performed at 20°C in 1× TBE running buffer. Before exposition, gels were fixed in 10% acetic acid, 25% 2-propanol for 30 min at room temperature and dried by vacuum blotting.
In vitro mobility shift assays were performed in the presence of a large excess of protein as compared with sRNAs. Under these conditions, the apparent Kd of the RNA–protein complex corresponds to the protein concentration for which 50% of the input RNA is shifted to an RNP complex.
Chemical and enzymatic probing
Chemical and enzymatic probing of free H/ACA sRNAs and L7Ae/sRNAs complexes from A.fulgidus was performed as described previously (32) with the following modifications: reactions contained 1 pmol of sRNA and 30 or 100 pmol of L7Ae protein for Afu-4 or Afu-46 sRNAs, respectively, in a reaction volume of 50 µl. Protein–sRNA complex formation was performed as described above. Enzymatic probing by T1 RNase as well as chemical probing by DMS and kethoxal was performed in buffer A [buffer A: 20 mM HEPES–KOH (pH 7.4), 150 mM KCl, 1.5 mM MgCl2, 2 mM DTT, 5 µg of tRNA and 20 U of ribonuclease inhibitor (RNasin)]. The following concentrations of enzymatic or chemical reagents were used: 4 U of T1 RNase (1:25 dilution from 105 U/ml stock solution in probing buffer A), 1.5 µl of DMS (1:10 dilution in 95% EtOH), 5 µl of KE (1:5 dilution of 37 mg/ml stock solution in buffer A). The chemical probing by CMCT was performed in reaction buffer B [20 mM potassium borate (pH 8.0), 150 mM KCl, 1.5 mM MgCl2, 2 mM DTT, 5 µg of tRNA and 20 U of ribonuclease inhibitor (RNasin)]. CMCT was diluted to a final concentration of 37 mg/ml in buffer B and 10 µl was used for the following step. Chemical and enzymatic probing was carried out at 37°C for 5 or 20 min, respectively. The reactions were stopped by extraction with phenol and chloroform. For KE, CMCT and T1 RNase, reaction mixtures were adjusted to 20 mM potassium borate (pH 7.0), 0.3 M NaOAc and precipitated with ethanol. For DMS-modified samples, reactions were precipitated with 0.3 M NaOAc (pH 5.2) and 6 mM β-mercapto ethanol. After precipitation, RNAs were washed twice with 80% ethanol and dissolved in 10 µl of H2O. For detection of chemical modifications of T1 nuclease cleavage, primer extension reactions were performed using 5′ 32P-end-labeled primers as described previously (32). Samples were loaded onto 8% polyacrylamide, 7 M urea gels. Electrophoresis was performed at 1600 V, 25 mA for 2 h. All probing experiments were repeated at least three times with high reproducibility.
Oligonucleotides
All oligonucleotides were synthesized by MWG Biotech (Ebersberg, Germany). Oligonucleotides used for amplification of the L7Ae gene from A.fulgidus genomic DNA are: For_L7Ae_Afu, CTGACATATGTACGTGAGATTTGAGG TTC and Rev_L7Ae_Afu, CTGACTCGAGTTACTTCTGA AGGCCTTTAATC. Oligonucleotides used for generation of DNA templates for T7 transcription, analysis of chemical and enzymatic probing by primer extension as well as mutagenesis, are: Afu 4-T7+FORV, TAATACGACTCACTATAG GCCCCGATCGGGGAAGAGGCAGAGG; Afu 4-REV, CTGCCCCCAGAGTAACACCAC; Afu 46-T7+FORV, TAATACGACTCACTATAGGCAACAATAGCTCCGCC CCCTCA; Afu 46-REV, TGCTGGTGTAGCTCCGC CAAC; Afu 52-T7+FORV, TAATACGACTCACTATA GGTGTATTTTTCATGCAGGCATACCG; AFu52-REV, GGGTGTTTCCTCATCCAGAG; Afu 190-T7+FORV, TAATACGACTCACTATAGGAACTTTTCCTACCCGGC GTGCC; Afu 190-REV, AAGCAAAAATTGTAGCCC GCCAAG; Af4rev-St2, CTGTCCCATGTCCGGGTTGC; Af4rev-mSt3(cc), TCTGCCCCCAGAGTAACACCACCCC GGATCGCAGG; Afu 46rev-mut(cc), TGCTGGTGTAG CTCCGCCAACCGACCCGGGAGGATGCTTCTC and Af190rev-m(cc), AATTGTAGCCCGCCAAGTGGAATT CCCCGGACCAATCAG.
RESULTS AND DISCUSSION
Binding of archaeal H/ACA sRNAs to the C/D box protein L7Ae
Thus far, three of the four expected homologs for eukaryal H/ACA snoRNA binding proteins have been identified in Archaea, designated as Cbf5p, Gar1p and Nop10p (29). For the fourth eukaryal H/ACA snoRNA protein, designated as Nhp2p, no homolog has been reported so far. However, significant sequence similarity over a region of 53 amino acids between eukaryal Nhp2p and the archaeal C/D box protein L7Ae has been observed (25). Therefore, we investigated whether C/D box protein L7Ae could substitute for Nhp2p in archaeal H/ACA sRNAs.
We expressed and purified L7Ae protein from A.fulgidus by affinity chromatography (see Materials and Methods). Previously, four archaeal H/ACA sRNAs had been identified by our group in an approach designated as ‘experimental RNomics’, and were shown to represent the first members of H/ACA snoRNA homologs (Afu-4, Afu-46, Afu-52 and Afu-190 H/ACA sRNAs) in an archaeal species (3). By in vitro binding assays, we analyzed whether the four archaeal H/ACA sRNAs were able to specifically interact and form stable complexes with the archaeal C/D box protein L7Ae.
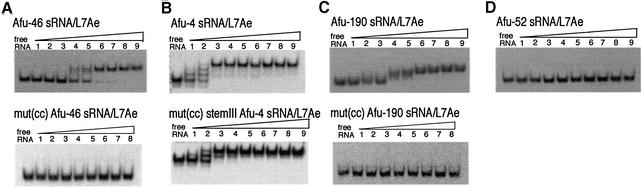
The in vitro synthesized sRNAs were radioactively labeled and a gel retardation assay was performed using increasing concentrations of L7Ae protein (see Materials and Methods). Even in the presence of a vast excess of non-specific competitor RNA (total yeast tRNA added in a molar ratio of about 3700:1) Afu-46 and Afu-190 formed specific sRNP complexes with the L7Ae protein with apparent Kd values of 100 and 75 nM, respectively (Fig. 1A and C). When Afu-4 sRNA and increasing concentrations of L7Ae protein were assayed under the same conditions, we detected three distinct mobility shifts of the sRNP complex (Fig. 1B). Unlike Afu-46 and Afu-190, which exhibit a single-stem structure, the Afu-4 sRNA exhibits a three-stem structure (see Fig. 3). Thus, the three distinct complexes observed in the gel retardation assay might correlate with three potential binding sites for the L7Ae protein present within each of the three stems of the sRNA (see below). The Afu-4/L7Ae sRNP complex was formed with an apparent Kd of 28 nM. Thereby, binding constants between H/ACA sRNAs to the L7Ae protein are in the same range as those observed for its interaction with C/D box sRNAs (10,21,33).
Figure 1.
Interaction of L7Ae protein with archaeal H/ACA sRNAs and sRNA mutants as assessed by gel retardation analysis. (A) Afu-46 H/ACA sRNA (top) and mutant Afu-46 H/ACA sRNA (bottom); (B) Afu-4 H/ACA sRNA (top) and mutant Afu-4 H/ACA sRNA (bottom); (C) Afu-190 H/ACA sRNA (top) and mutant Afu-190 sRNA (bottom); (D) Afu-52 sRNA. 32P-labeled sRNAs were incubated with recombinant protein L7Ae from A.fulgidus as follows: free RNA, no protein added; (A, C and D) lanes 1–9, 2.5, 12.5, 25, 50, 100, 150, 200, 250 and 375 nM L7Ae protein added; (B) lanes1–9, 20, 25, 30, 35, 40, 45, 50, 55 and 60 nM L7Ae protein added.
In contrast to Afu-4, Afu-46 and Afu-190 sRNAs, we failed to detect specific complex formation between Afu-52 and the L7Ae protein by our binding assay (Fig. 1D). Previously, Afu-52 sRNA has been predicted to guide pseudouridylation of U 2878 from 23S rRNA of A.fulgidus (3). However, a more detailed study could not experimentally verify the presence of the predicted pseudouridine in rRNA (data not shown). Moreover, the thermodynamic stability of the predicted guide duplex involving Afu-52 was significantly lower compared with the other three sRNAs. In contrast, the pseudouridylation targets of Afu-46, Afu-190 and Afu-4 sRNAs could be experimentally confirmed in ribosomal RNAs (3 and data not shown). Based on these findings, we propose a functional interaction between the C/D box protein L7Ae and the three archaeal H/ACA sRNAs Afu-4, Afu-46 and Afu-190, but not Afu-52 sRNA.
Structure analysis of L7Ae-H/ACA sRNA complexes
Secondary structure prediction of archaeal H/ACA sRNAs. Based on thermodynamic features for RNA structure prediction (34), the secondary structures for Afu-46, Afu-52 and Afu-190 H/ACA sRNAs consist of a single, stable stem with the ACA motif positioned downstream from the stem structure (3). The same single-hairpin structure has been shown for eukaryal H/ACA box snoRNAs in the trypanosome Leptomonas collosoma (28). In contrast, Afu-4 H/ACA sRNA was predicted to fold into a highly stable secondary structure exhibiting three long stems connected by two single-stranded regions (3). The large internal loop of each long stem contains a bipartite antisense element thereby targeting three sites in ribosomal RNA for modification (3). The single-stranded region between stems 1 and 2 represents a typical H box (sequence: AcAccA) motif also found between stems I and II of eukaryal H/ACA snoRNAs. An ACA box motif (sequence: 5′-ACA-3′) is placed in the single-stranded region in between stems 2 and 3. Downstream of stem 3 of the Afu-4 sRNA, a degenerate ACA motif, AGA, can be observed; an AGA sequence at the 3′ end of eukaryal H/ACA snoRNAs has also been observed in the yeast Saccaromyces cerevisiae and trypanosomes (2,28).
Analysis of the L7Ae/Afu-46 sRNA complex by chemical and enzymatic probing. In order to refine the computer folding predictions of archaeal H/ACA sRNAs as well as to identify the precise binding sites for ribosomal protein L7Ae, we performed chemical and enzymatic probing analysis on free or L7Ae-bound Afu-46 H/ACA sRNA. To that end, we chose chemical probes DMS, CMCT, KE, as well as the enzymatic probe, T1 nuclease. Chemical probes modify bases of an RNA not involved in Watson–Crick interactions (32). RNase T1 cleaves the phosphodiester backbone of an RNA following a G residue preferentially in single-stranded regions of the molecule (35). In the unbound state, most modifications of bases and phosphodiester bond cleavages within Afu-46 sRNA correlated well with the simple stem–loop secondary structure predicted by the m-fold program (3).
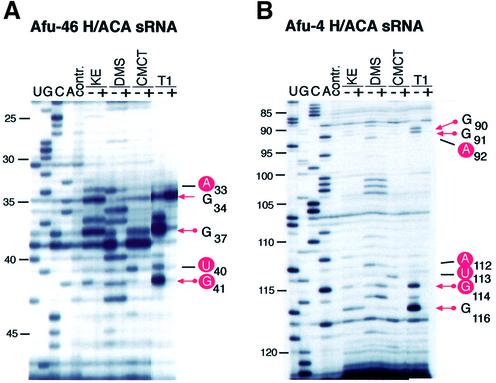
Comparing the probing results of free Afu-46 sRNA with the L7Ae/Afu-46 RNP complex, the most obvious differences were observed within the terminal loop region of the RNA hairpin. Upon L7Ae binding, base U40 is strongly, and base U43 is weakly, protected from modification with CMCT (Figs 2A and 3A). In addition, A33 and G41 become protected from modification with DMS and KE, respectively. Accordingly, in the absence of the L7Ae protein enzymatic probing of free Afu-46 sRNA with T1 nuclease shows strong cleavage at positions G34, G37 and G41 in the terminal loop. In the L7Ae/Afu-46 complex, however, the phosphodiester bonds following G37 and G41, but not G34 showed strongly reduced accessibility to T1 nuclease (Figs 2A and 3A). These results are consistent with protein L7Ae binding to the terminal loop of Afu-46 sRNA.
Figure 2.
Chemical and enzymatic probing of free Afu-46 sRNA and the Afu-46/L7Ae complex (A) and free Afu-4 H/ACA sRNA and the Afu-4/L7Ae complex (B). Chemical and enzymatic probing of free sRNA (–) and the sRNA/L7Ae protein complex (+); U, G, C and A, sequencing lanes; contr., control lane, no chemical or enzymatic probes added. KE, DMS, CMCT and T1, addition of chemical probes KE, DMS and CMCT or enzymatic probe T1 nuclease.
Analysis of the L7Ae/Afu-4 sRNA complex by chemical and enzymatic probing. We next investigated the interaction of an H/ACA sRNA exhibiting a three-stem structure with the L7Ae protein. Free Afu-4 sRNA and the Afu-4 sRNA/L7Ae RNP complex were subjected to chemical and enzymatic probing. Due to the high G/C content of stems I and III which impeded annealing of oligonucleotide primers, primer extension analysis of modified nucleotides could be performed for stem II of Afu-4 sRNA only (see Fig. 3B).
When we compared the probing patterns of free Afu-4 sRNA with the L7Ae/Afu-4 RNP complex, differences in accessibility to chemical and enzymatic probes were observed exclusively within a small internal bulge of stem II (Figs 2B and 3B). Nucleotides A92, G111, A112, U113 and G114 were protected by the L7Ae protein from chemical modification with DMS, KE or CMCT, respectively. Enzymatic probing experiments showed that L7Ae protein protected the phosphodiester backbone of the RNA following nucleotides G90 and G91 and G114 and G116 from T1 RNase cleavage. These results are consistent with L7Ae protein binding to the central bulge within the upper portion of stem II of the Afu-4 sRNA (Fig. 3B).
L7Ae binding sites within Afu-46 and Afu-4 sRNAs represent K-turn motifs
The L7Ae protein has been shown to bind within C/D box sRNAs as well as 23S ribosomal RNA to a motif designated as the K-turn (12,21 and see above). In addition, the selenocysteine inserting RNA motif (SECIS-element) exhibits a K-turn motif which is recognized by the human SBP2 protein (36). SPB2 shares sequence similarity to the human homolog of the L7Ae protein, the 15.5 kDa protein. Binding to the K-turn motif was shown to involve the same conserved amino acids between 15.5 kDa and SBP2 proteins (36).
Upon closer inspection of the predicted secondary structures for Afu-4 and Afu-46 sRNAs we were able to fold the L7Ae binding sites manually into K-turn motifs: we observed the two canonical sheared G-A base pairs at both binding sites within Afu-4 and Afu-46 sRNAs, as well as the 3 nt bulge at one side of the helix, including the terminal U base (Fig. 3A and B). Interestingly, in Afu-4 sRNA we observed K-turn motifs within stem I and stem III as well, in addition to stem II (Fig. 3B). Thus, the three-stem–loop structure of Afu-4 sRNA might contain three bindings sites for the L7Ae protein. Consistent with this model, in a gel retardation experiment (Fig. 2B, see above) we observed three distinct shifts in mobility of Afu-4 sRNA upon addition of increasing concentrations of L7Ae protein. The three shifts in mobility might be indicative of successive L7Ae protein binding to the K-turn motifs within the three stems (see below).
The L7Ae binding site within Afu-46 sRNA reflects a different form of the K-turn motif located within the apical stem and loop of the RNA hairpin (Fig. 3A). It corresponds to one of the two L7Ae binding sites present in archaeal C/D RNAs (4). Thereby, the two sheared G-A pairs followed by a U-U pair are present as observed in canonical K-turn motifs. Also, the terminal U base juxtaposed to the first G-A base pair can be observed. However, the 3 nt bulge found in canonical K-turns is substituted by a 7 nt loop. Similar to Afu-46 sRNA, in Afu-190 sRNA the K-turn motif is located in the apical stem and loop region of the RNA hairpin, with a loop size of 4 nt (Fig. 3C). Interestingly, in all three archaeal H/ACA sRNAs the K-turn motif in the upper stem is invariably positioned 5–6 bp away from the basis of this stem.
Consistent with the lack of binding to L7Ae protein (see above), we could not find any evidence for the presence of a K-turn motif within Afu-52 H/ACA sRNA (Fig. 3D). Since we were also unable to confirm the predicted pseuduridylation site within archaeal ribosomal 23S RNA targeted by the H/ACA candidate, Afu-52 sRNA might exert another function, unrelated to pseudouridylation, in the cell.
Mutation of bases within the K-turn motif of archaeal H/ACA sRNAs eliminate L7Ae binding
To provide further evidence that the K-turn motif found in archaeal H/ACA sRNAs is involved in binding to the L7Ae protein, we mutated canonical bases from this motif in Afu-46, Afu-190 and Afu-4 sRNAs. Subsequently, we assessed the ability of the mutant RNAs to bind L7Ae protein by gel retardation experiments (see above). By in vitro mutagenesis, bases G41 and A42 within the K-turn motif of Afu-46 sRNA were replaced by bases C41 and C42, respectively (Fig. 3A). These mutations were generated in order to impede formation of the two sheared non-Watson–Crick G-A base pairs found within all canonical K-turn motifs. Previously, it has been shown that these two base pairs are required for binding of L7Ae to the K-turn in eukaryal and archaeal C/D box snoRNAs and sRNAs, respectively (10,21). As expected, we did not observe a shift in mobility of mutant Afu-46 sRNA when incubated with the L7Ae protein, reflecting a lack of specific RNP complex formation (Fig. 1A, bottom).
For Afu-190 sRNA, corresponding mutations were introduced into the K-turn motif, disrupting the two sheared G-A base pairs (Fig. 3C). As observed for Afu-46 sRNA, we failed to detect specific binding to the L7Ae protein (Fig. 1C, bottom) as assessed by gel retardation analysis. These findings provide additional evidence for the presence of the predicted K-turn motif located within the apical stem–loop of Afu-190 sRNA.
Accordingly, the Afu-4 sRNA mutant was generated by replacement of the two non-Watson–Crick G-A pairs located in stem III (Fig. 3B). Stem III was chosen for mutagenesis in order to provide further experimental evidence for the presence of the predicted K-turn motif in this stem which could not be analyzed by chemical probing (see above). In line with our model, only two shifts in mobility could be observed when mutant Afu-4 sRNA was analyzed for binding to protein L7Ae (Fig. 1B, bottom). In contrast, three distinct shifts in mobility were shown for the wild-type Afu-4 sRNA (Fig. 1B, top). This is consistent with the formation of a complex involving binding of two molecules of L7Ae protein to the two wild-type K-turn motifs in stems I and II, while binding of the L7Ae protein to stem III is impeded by the introduced mutations (Fig. 3B). These findings are consistent with our model that each stem within Afu-4 sRNA serves as a specific binding site for L7Ae (see above).
Novel H/ACA sRNAs detected in distantly related Archaea also exhibit the K-turn motif at a similar location
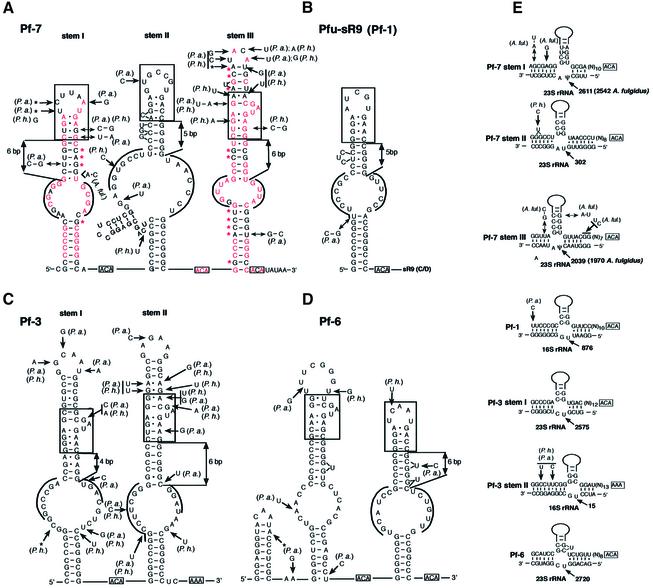
Recently, several non-messenger RNAs with a high degree of secondary structure were identified in the genomes of AT-rich hyperthermophilic Archaea by computational search of GC-rich regions associated with a screen based on comparative analysis in phylogenetically related species (37). Interestingly, one of the small RNAs detected in three Pyrococcus species, three-hairpin Pf7, is unambiguously related to Afu-4 sRNA, both structurally and by its potential to guide pseudouridylation (Figs 3B and 4A, E). Sequence similarity is most apparent over hairpin domain III, with conservation of the large internal loop sequence and accumulation of several compensatory base changes over the lower and upper stems. Each of the three hairpin domains of Pf7 exhibits typical features of pseudouridylation guide domains (Fig. 4A).
Figure 4.
Secondary structures of novel H/ACA sRNAs identified among previously reported Pyrococcus non-coding RNAs and their presumptive rRNA target sites for pseudouridylation. (A) Pf7 RNA, the Pyrococcus homolog of Afu-4. Nucleotides conserved in A.fulgidus (in red) and base-pairings exhibiting compensatory changes between A.fulgidus and the Pyrococcus (red stars) are denoted. (B) Pf1 RNA: this single-hairpin H/ACA domain is linked to a C/D sRNA, sR19, present in both a shorter form, similar to other archaeal C/D RNAs, and a longer form including the upstream H/ACA domain (37). (C) Pf3 RNA. (D) Pf6 RNA. (A–D) All sequences shown are for Pyrococcus furiosus. In the large internal loop nucleotides able to base-pair with Pyrococcus 16S or 23S rRNA (as depicted in E) are overlined. The K-turn motifs are boxed. (B–D) Differences in Pyrococcus abyssi (Pa) and Pyrococcus horikoshii (Ph) are denoted for Pf1, Pf3 and Pf6 for which no reported homologs in A.fulgidus are known. (E) Potential base-pairings directing a rRNA pseudouridylation. Each predicted site of pseudouridylation is denoted by an arrow with indication of its position in the P.furiosus rRNA sequence. All guide duplexes shown here for P.furiosus are conserved in P.abyssi and P.horikoshii. The H/ACA sRNA sequence in a 5′ to 3′ orientation is shown (top strand) with its long upper stem schematized by a solid line and the K-turn motif depicted by a box with indication of its distance from the basis of the stem. Proven target uridines are indicated by ψ, predicted by U. Guide duplexes involving stems I and III of Pf7 are supported by compensatory base changes (base-pairing denoted by double arrows) in A.fulgidus Afu-4. Note that Afu-4 hairpin domains I and III have each an additional, experimentally verified rRNA target uridine (3).
Nucleotide sequences in each interior loop can form a canonical bipartite guide duplex of 11–13 bp around a Pyrococcus rRNA uridine target positioned 14–16 nt away from a downstream single-stranded ACA motif (Fig. 4E). Hairpin domains I of both Afu-4 and Pf7 target the same 23S rRNA uridine corresponding to a pseudouridine conserved in eukaryal 28S rRNA (38). Hairpin domains III of both RNAs have also the same target, a 23S rRNA uridine which is pseudouridylated not only in Eukarya but also in E.coli (27). In contrast, hairpin domains II have different potential targets in A.fulgidus and the Pyrococcus 23S rRNA, respectively, due to nucleotide changes in the corresponding antisense elements of Afu-4 and Pf7 (Fig. 4E). Remarkably, while the upper stem of each hairpin domain substantially diverges in sequence between Pf7 and Afu-4, a K-turn motif is conserved in all cases, at an identical location relative to the basis of each upper stem in A.fulgidus and the Pyrococcus (Figs 3B and 4A).
Finally, through a detailed examination of all other small RNAs identified by Klein et al. (37) we found that four of these highly structured RNAs exhibit the H/ACA hallmarks. The rRNA uridines predicted to be targeted by three of them, Pyrococcus Pf1, Pf3 and Pf6, were readily identified (Fig. 4B–E), while the RNA target of Methanococcus jannaschii Mj2 (data not shown) remains unknown. Remarkably, each long hairpin domain of the four additional archaeal H/ACA candidates displays a K-turn motif in its upper stem, invariably positioned 5–6 bp away from the basis of this stem (with one exception, Pf3, stem I, Fig. 4C). This arrangement is identical to A.fulgidus Afu-46, Afu-4 and Afu-190 RNAs (Fig. 3A–C) and its Pyrococcus homolog, Pf7. The systematic presence and conserved positioning of the motif in a wide range of H/ACA sRNA specimens exhibiting one, two or three hairpin domains and originating from distantly related archaeal species suggests L7Ae plays a fundamental role in H/ACA sRNP assembly and pseudouridylation guide function.
CONCLUSIONS
In this study, we show that a core protein for C/D box sRNAs, the L7Ae protein, also specifically interacts with archaeal H/ACA sRNAs. The RNA structure in H/ACA sRNAs from A.fulgidus required for binding was identified as the K-turn motif. RNP complexes were formed with an apparent Kd between 28 and 100 nM, similar to those observed for binding of L7Ae protein to C/D box sRNAs. We also observed the K-turn motif in H/ACA sRNAs from three distantly related archaeal Pyrococcus species. In general, K-turn motifs are located five to six bases above the basis of the respective stem structure. These data are consistent with a model that archaeal H/ACA box sRNAs share the K-turn as a common RNA binding motif with C/D box sRNAs. These findings point, apart from its function as a ribosomal protein, to a dual role for the L7Ae protein in snoRNA biogenesis and function for both sub-classes of sRNAs in Archaea. In contrast, in Eukarya, a typical K-turn motif is not readily found in H/ACA snoRNAs. In some cases, a K-turn motif can be folded manually which is, however, energetically less stable than alternate structures not exhibiting the K-turn. In addition, the eukaryal homolog Nhp2p does not specifically bind to RNA (24 and Nicolas Watkins, personal communication).
Our observations suggest a common evolutionary origin of both sub-classes of sRNAs, previously proposed by the group of Tamas Kiss (23). Since L7Ae represents also an integral part of the ribosome by binding to the K-turn in 23S rRNA, this common ancestor might even be ribosome derived. In addition, it has been shown that the human homolog of the archaeal L7Ae protein, the 15.5 kDa protein, binds not only to C/D box snoRNAs but also to U4 snRNA (19). This finding further bridges the gap between snRNAs and snoRNAs. In line with that notion, it has recently been proposed that certain tissue-specific snoRNAs interact with mRNAs rather than with rRNAs and thus might also be related to snRNAs on a functional level as well (39–41).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Nick Watkins for helpful discussions and Johannes Heilmann for technical assistance. This work was supported by the German Human Genome Project through the BMBF (#01KW9966) to A.H. and J.B., an IZKF grant (Teilprojekt IKF G6, Münster) to A.H., by laboratory funds from the Centre National de la Recherche Scientifique and Université Paul Sabatier, Toulouse, and by a grant from the Ministère de l’Education Nationale, de la Recherche et de la Technologie (Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires, 2001–2002) to J.P.B.
REFERENCES
- 1.Kiss-Laszlo Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 2.Ganot P., Bortolin,M.L. and Kiss,T. (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 3.Tang T.H., Bachellerie,J.P., Rozhdestvensky,T., Bortolin,M.L., Huber,H., Drungowski,M., Elge,T., Brosius,J. and Huttenhofer,A. (2002) Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl Acad. Sci. USA, 99, 7536–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omer A.D., Ziesche,S., Ebhardt,H. and Dennis,P.P. (2002) In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl Acad. Sci. USA, 99, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filipowicz W. and Pogacic,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell. Biol., 14, 319–327. [DOI] [PubMed] [Google Scholar]
- 6.Lafontaine D.L. and Tollervey,D. (1998) Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci., 23, 383–388. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein L.B. and Steitz,J.A. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell. Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- 8.Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol., 260, 178–195. [DOI] [PubMed] [Google Scholar]
- 9.Cavaille J., Nicoloso,M. and Bachellerie,J.P. (1996) Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature, 383, 732–735. [DOI] [PubMed] [Google Scholar]
- 10.Watkins N.J., Segault,V., Charpentier,B., Nottrott,S., Fabrizio,P., Bachi,A., Wilm,M., Rosbash,M., Branlant,C. and Luhrmann,R. (2000) A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- 11.Vidovic I., Nottrott,S., Hartmuth,K., Luhrmann,R. and Ficner,R. (2000) Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell, 6, 1331–1342. [DOI] [PubMed] [Google Scholar]
- 12.Klein D.J., Schmeing,T.M., Moore,P.B. and Steitz,T.A. (2001) The kink-turn: a new RNA secondary structure motif. EMBO J., 20, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler W.C., Grundy,F.J., Murphy,B.A. and Henkin,T.M. (2001) The GA motif: an RNA element common to bacterial antitermination systems, rRNA and eukaryotic RNAs. RNA, 7, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman D.R., Kuhn,J.F., Shanab,G.M. and Maxwell,E.S. (2000) Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA, 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tollervey D., Lehtonen,H., Jansen,R., Kern,H. and Hurt,E.C. (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation and ribosome assembly. Cell, 72, 443–457. [DOI] [PubMed] [Google Scholar]
- 16.Niewmierzycka A. and Clarke,S. (1999) S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem., 274, 814–824. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Boisvert,D., Kim,K.K., Kim,R. and Kim,S.H. (2000) Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J., 19, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galardi S., Fatica,A., Bachi,A., Scaloni,A., Presutti,C. and Bozzoni,I. (2002) Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol., 22, 6663–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nottrott S., Hartmuth,K., Fabrizio,P., Urlaub,H., Vidovic,I., Ficner,R. and Luhrmann,R. (1999) Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J., 18, 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao H., White,S.A. and Williamson,J.R. (1999) A novel loop–loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nature Struct. Biol., 6, 1139–1147. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn J.F., Tran,E.J. and Maxwell,E.S. (2002) Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res., 30, 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- 23.Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- 24.Ofengand J. (2002) Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett., 514, 17–25. [DOI] [PubMed] [Google Scholar]
- 25.Henras A., Henry,Y., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Gelugne,J.P. and Caizergues-Ferrer,M. (1998) Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J., 17, 7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watkins N.J., Gottschalk,A., Neubauer,G., Kastner,B., Fabrizio,P., Mann,M. and Luhrmann,R. (1998) Cbf5p, a potential pseudouridine synthase and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA, 4, 1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogacic V., Dragon,F. and Filipowicz,W. (2000) Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol., 20, 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang X.H., Liu,L. and Michaeli,S. (2001) Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem., 276, 40313–40318. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe Y. and Gray,M.W. (2000) Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res., 28, 2342–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullis K.B. and Faloona,F.A. (1987) Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol., 155, 335–350. [DOI] [PubMed] [Google Scholar]
- 32.Stern S., Moazed,D. and Noller,H.F. (1988) Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol., 164, 481–489. [DOI] [PubMed] [Google Scholar]
- 33.Tang T.H., Rozhdestvensky,T.S., d’Orval,B.C., Bortolin,M.L., Huber,H., Charpentier,B., Branlant,C., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2002) RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res., 30, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M. (2000) Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol., 10, 303–310. [DOI] [PubMed] [Google Scholar]
- 35.Shelness G.S. and Williams,D.L. (1985) Secondary structure analysis of apolipoprotein II mRNA using enzymatic probes and reverse transcriptase. Evaluation of primer extension for high resolution structure mapping of mRNA. J. Biol. Chem., 260, 8637–8646. [PubMed] [Google Scholar]
- 36.Allmang C., Carbon,P. and Krol,A. (2002) The SBP2 and 15.5/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA, 8, 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein R.J., Misulovin,Z. and Eddy,S.R. (2002) Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc. Natl Acad. Sci. USA, 99, 7542–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ofengand J. and Bakin,A. (1997) Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol., 266, 246–268. [DOI] [PubMed] [Google Scholar]
- 39.Liang X.H., Xu,Y.X. and Michaeli,S. (2002) The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA, 8, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavaille J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]