Abstract
Death-associated protein (DAP)-like kinase (Dlk), also known as Zipper interacting protein (ZIP) kinase, is a nuclear serine/threonine-specific kinase that phosphorylates core histones H3 and H4, and myosine light chain in vitro. It interacts with transcription and splicing factors as well as with pro-apoptotic protein Par-4 suggesting that it participates in multiple cellular processes. To explore the significance of histone phosphorylation by Dlk, we determined the phosphorylation site in H3 and generated phosphospecific antibodies for in vivo analyses. Interestingly, Dlk/ZIP kinase phosphorylated histone H3 at a novel site, Thr11, rather than Ser10, which is characteristic of mitotic chromosomes. Immunoblotting and confocal immunofluorescence analyses demonstrated that phosphoryl ation of H3 at Thr11 occurred in vivo and was restricted to mitosis as well. It was discernable from prophase to early anaphase and particularly enriched at centromeres. Strikingly, during this time interval, Dlk was associated with centromeres too, as revealed by stable expression of a green fluorescent protein (GFP)-Dlk fusion protein. These findings strongly suggest that Dlk is a centromere-specific histone kinase that might play a role in labeling centromere-specific chromatin for subsequent mitotic processes.
INTRODUCTION
Death-associated protein (DAP)-like kinase (Dlk) (1) or Zipper interacting protein (ZIP) kinase (2) is a novel serine/threonine-specific kinase that belongs to the subfamily of DAP kinases. Presently, this kinase subfamily has five members: DAP kinase itself, DAP-related protein kinase, Dlk/ZIP kinase and DAPK-related apoptosis-inducing kinases (DRAK 1 and 2). The first three of these share a high degree of sequence identity (>80%) in their catalytic domains while the latter are more distantly related (∼50% sequence identity to the first group). They all differ greatly in their non-catalytic domains (reviewed in 3,4). DAP kinase itself plays an important role in interferon γ- and TNFα/fas-induced cell death (5–7). Likewise, the DAP-related kinases have been implicated in apoptotic processes. Additionally, DAP kinase might have a tumorsuppressor function as it seems to interfere with tumor progression, particularly metastatic capacity (8).
Dlk/ZIP kinase (Dlk, for simplicity) is a nuclear kinase that is ubiquitously expressed and highly conserved (99.7 or 86% identity between rat and mouse or rat and human homologs, respectively) suggesting that it fulfills an important function. Dlk does not induce apoptosis per se. Rather, induction of cell death by Dlk requires its cytoplasmic retention and association with actin filaments. This can be achieved either by C-terminal truncation (9) or by coexpression and interaction with pro-apoptotic protein Par-4 (10). A physiological substrate in this process might be myosin light chain (MLC), since Dlk phosphorylates MLC in vitro (1) and phosphorylation of MLC has been implicated in apoptotic membrane blebbing (11). The function(s) of Dlk in the nucleus remain to be determined. Dlk contains a leucine zipper which mediates homodimerization and interaction with transcription factors ATF-4 (2,10) and AATF (12) and with transcription and splicing factor CDC5 (13). Additionally, Dlk phosphorylates core histones H3, H4 and, to lesser extent, H2A in vitro (1). Together, these data suggest a role in transcription and splicing and, perhaps, in chromatin remodeling.
Histones undergo extensive post-translational modifications, particularly acetylation, methylation, phosphorylation, ubiquitination and ADP-ribosylation (for reviews, see 14,15). While the significance of the latter two modifications is less well understood, a growing body of information about the former three allows us to deduce some general principles. Thus, acetylation of the N-terminal tails of core histones H3 and H4 has been implicated in transcriptional activation by facilitating access for the transcriptional machinery to chromatin; methylation has been implicated in long-term transcriptional silencing, and phosphorylation has been connected to chromatin condensation during mitosis. Additionally, a role of H3 phosphorylation in activation of early response genes has been demonstrated. In these latter cases, H3 phosphorylation appears to facilitate recruitment of histone acetyl transferases and subsequent acetylation of H3 at adjacent sites (Lys14) (16). Perhaps both these modifications cooperate in chromatin remodeling in promoter regions of a subset of genes and thereby contribute to full transcriptional activation. In summary, the various modifications of histones serve different functions in the dynamic changes of chromatin during transcription, replication and recombination, and during mitosis.
Phosphorylation of H3, particularly at Ser10 (and perhaps Ser28) seems to be crucial for progression through mitosis. In mammalian cells, phosphorylation of H3 at Ser10 initiates in late G2, in the pericentromeric region and spreads over the whole chromosome arms thereafter (17). It assumes its highest level during prometaphase/metaphase when chromosomes are highly condensed, and lasts until anaphase upon which it disappears. The strong correlation of H3 phosphorylation and chromosome condensation suggested a causal relationship. However, in other organisms, particularly plants, H3 phosphorylation occurs on condensed chromosomes. Thus, H3 phosphorylation may serve as a label for mitotic chromosomes as suggested recently by the ‘ready production hypothesis’ (reviewed in 15).
Interestingly, H3 or at least Ser10-phosphorylated H3 appeared to be excluded from centromeres, where it is replaced by its relative centromere protein (CENP-A) (18–20). CENP-A, a constitutive centromere protein is incorporated into chromatin in late S-phase and seems to provide a specific, epigenetic label for centromeric chromatin serving as platform for assembly of centromere/kinetochore complexes (21,22). However, a recent study by Blower et al. (23) using a novel chromatin-stretching technique showed that centromeres contain both CENP-A and H3, perhaps in alternating arrays. Interestingly, both H3 and its relative CENP-A seem to be phosphorylated by aurora B kinase (24–26). However, it is not understood why this kinase phosphorylates H3 in chromosomal arms but not in centromeres where it prefers CENP-A. Obviously, other factors are necessary to mediate the distinction between different substrates in their respective contexts.
In the present study, we have investigated the significance of Dlk as a histone kinase. We found that Dlk phosphorylates H3 at a novel site, Thr11, which was specifically phosphorylated during mitosis. H3 bearing this modification seemed to be enriched in centromeric chromatin while H3 phosphorylated at Ser10 was found in the peripheral regions. Strikingly, Dlk was also associated with centromeres during mitosis and this association coincided precisely with phosphorylation of histone H3 at Thr11 and of CENP-A. We hypothesize that Dlk might be a centromere-specific histone kinase which plays a role in labeling centromeric chromatin for subsequent mitotic processes.
MATERIALS AND METHODS
Cell culture
Rat 1 fibroblasts and rat embryo fibroblast 52.2 were maintained in DMEM (Life Technologies, Inc., Karlsruhe, Germany) supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin. Human breast carcinoma (MCF-7) cells were grown in RPMI medium (Life Technologies, Inc.) containing 5% fetal calf serum and antibiotics.
Expression plasmids and transfection
Plasmids encoding the full-length green fluorescent protein (GFP)-Dlk fusion protein have been described (1). For the generation of stable Dlk-expressing cell lines, MCF-7 cells were transfected with GFP-Dlk expression plasmid using Lipofectamine™ (Life Technologies, Inc.) as specified by the manufacturer, and selected in the presence of 1 mg/ml Geneticin (G418) (Life Technologies, Inc.). After 3 weeks, surviving colonies were isolated and cells expressing GFP-Dlk were screened by fluorescence microscopy and further subjected to single-cell cloning.
Generation of antibodies
Phosphospecific antibodies (Pineda Antibody Service, Berlin, Germany) were generated in rabbits against a synthetic phosphopeptide of the sequence SARKST(P)GGK corresponding to residues 6–14 of histone H3. The threonine residue, which is normally at position 6, was replaced by serine to avoid the second threonine. Antibodies were affinity purified on a peptide column. Antibodies against Dlk were generated by immunizing rabbits with a synthetic peptide GLKRR LCRLENRYDA corresponding to residues 403–417 of Dlk (Genosphere, Paris, France).
Immunofluorescence microscopy
For immunofluorescence microscopy, MCF-7 or Rat 1 cells grown on coverslips were fixed with methanol at –20°C for 5 min. After washing with PBS, cells were treated with 5% non-fat dry milk for 1 h and incubated with the respective primary antibodies at the following dilutions: CREST autoimmune serum specific to centromere proteins CENP-A, B, C (27) (DPC Biermann, Bad Nauheim, Germany) at 1:200; rabbit polyclonal antibody specific for phospho-Thr11 of histone H3 (Pineda, Berlin) at 1:2000; rabbit polyclonal antibody specific for phospho-Ser10 of histone H3 (Upstate, Lake Placid, USA) and rabbit polyclonal antibody specific for CENP-F (clone D10, Novus Biologicals Inc., Littleton, CO, USA) at 1:5000. All primary antibodies were incubated for 1 h at room temperature. As secondary antibodies, Cy3-conjugated goat serum (Dianova, Hamburg, Germany) was used; goat anti-mouse and goat anti-rabbit were used at 1:500, or goat anti-human IgG at 1:200 dilutions and incubated for 30 min at room temperature. Nuclei were stained with 4,6-diamidino-2-phenylindole or propidium iodide for 15 min and subsequent washing as described above.
Images were captured with an Axiophot fluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a CCD-camera using filters optimized for double-label experiments and a 63× oil immersion objective. Confocal microscopy was performed either with a Leica DM-RBE fluorescence microscope coupled with a TCS-4D Laser Scanning Confocal Apparatus (Leica, Bensheim, Germany) or with a Zeiss Axioplan fluorescence microscope coupled with a ZEISS LSM 510. Images were treated with Adobe Photoshop™.
Preparation of cell extracts, SDS–PAGE and western blotting
Randomly growing or mitotic cells were sequentially extracted to remove soluble cytoplasmic and nucleoplasmic proteins and to obtain nuclear extracts. Mitotic cells were obtained by treatment with nocodazole (Sigma-Aldrich, Taufkirchen, Germany) (0.1 µg/ml) for 16 h, prior to harvest and shake off. Cells were washed with ice-cold PBS and lysed in isotonic lysis buffer (10 mM NaPO4 pH 8.0, 140 mM NaCl, 3 mM MgCl2, 1 mM dithiothreitol, 0.5% Nonidet-P40, 50 µM leupeptin) for 10 min on ice. Where appropriate, 10 mM NaMoO4 was added as additional phosphatase inhibitor. The lysate and nuclei were separated by low speed centrifugation, nuclei were resuspended in lysis buffer and briefly treated with benzonase (La Roche Applied Science, Mannheim, Germany) to reduce the viscosity of DNA after denaturation of chromatin in SDS sample buffer. For western blot analyses, nuclear extracts were separated on SDS–polyacrylamide gels and electrophoretically transferred onto nitrocellulose membranes according to standard protocols. Membranes were blocked with 5% non-fat dry milk in TBS following incubation with the polyclonal anti (P)-Ser10 or anti (P)-Thr11 antibody (1:5000 and 1:2000, respectively); anti-CENP-A (Upstate) was used at 1:1000. Bound antibodies were detected with a peroxidase-conjugated secondary goat anti-rabbit antibody (Dianova, Hamburg, Germany) using an enhanced chemoluminescence detection kit (Pierce/Perbio, Bonn, Germany). As loading control, aliquots identical to those used for immunoblotting were run on the same gel and stained with Coomassie brilliant blue to visualize the proteins.
In vitro kinase assay
In vitro kinase assays were carried out with baculovirus-expressed purified His-tagged Dlk essentially as described previously (1). As substrates a mixure of purified histones from calf thymus (Roche) were employed. To achieve quantitative phosphorylation, as required for immunoblotting with phospho-specific antibodies, the kinase reaction was carried out with 100 µM unlabeled ATP.
Phosphoamino acid analysis and two-dimensional phosphopeptide mapping
In vitro phosphorylated histones H3 and H4 were subjected to two-dimensional tryptic phosphopeptide mapping and phosphoamino acid analysis on thin-layer cellulose plates essentially as described previously (28).
RESULTS
Dlk phosphorylates core histones H3 and H4 at threonine residues in vitro
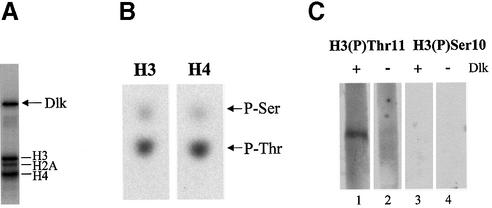
Dlk was employed in an in vitro kinase reaction with a mixture of purified histones from calf thymus as substrates. Dlk efficiently phosphorylated core histones H3 and H4 and, to a lesser extent, H2A, as published (1) (Fig. 1A). The phosphorylated proteins were isolated and subjected to phosphoamino acid analysis (see Materials and Methods). Interestingly, all three histones were phosphorylated at threonine, rather than serine residues (shown for H3 and H4 in Fig. 1B). Since this modification has not been described so far, we asked whether it also occurs in vivo. Emphasis was put on histone H3, since modification of H3 appears to play an extraordinary role in processes involving chromatin remodeling. We determined the phosphorylation site in H3, generated phosphospecific antibodies and employed these in western blotting and immunofluorescence analyses.
Figure 1.
Dlk phosphorylates core histones H3 and H4 at threonine residues. (A) A mixture of purified histones from calf thymus was phosphorylated by Dlk in vitro and analyzed by SDS–PAGE and autoradiography. The phosphorylated histones and autophosphorylated Dlk are indicated. (B) The phosphoproteins were blotted onto nitrocellulose membrane, isolated and subjected to partial acid hydrolysis. Phosphoamino acids were separated by electrophoresis on thin layer cellulose plates and visualized by autoradiography. (C) Histones were left untreated (–) or quantitatively phosphorylated by Dlk (+) in the presence of unlabeled ATP and subjected to immunoblot analysis. Each of the samples was probed with antibodies specific for (P)-Thr11 or (P)-Ser10 of histone H3 as described in Materials and Methods.
Inspection of the amino acid sequence of H3 revealed only one potential threonine phosphorylation site, which fulfils the requirements for a Dlk phosphorylation site, an Arg residue at the –3 position (K.H.Scheidtmann, unpublished results). This site, Thr11 in the sequence Ala-Arg-Thr-Lys-Gln-Thr-Ala-Arg-Lys-Ser-Thr11-Gly-Gly-Lys was verified by two-dimensional peptide analysis (data not shown). Interestingly, this site is adjacent to the well established Ser10 site. Antibodies against a synthetic phosphopeptide encompassing residues 6–14 of H3 and including (P)-Thr11 were generated and assayed for their specificity. Histones from calf thymus were quantitatively phosphorylated by purified Dlk in vitro and tested for their reactivity with the antibodies. As shown in Figure 1C (lanes 1 and 2), the antibodies reacted strongly with phosphorylated H3 but not with the unphosphorylated form or any of the other histones. Moreover, this modification was not recognized by (P)-Ser10-specific antibodies (lanes 3 and 4).
Histone H3 phosphorylation at Thr11 occurs in vivo and is restricted to mitosis
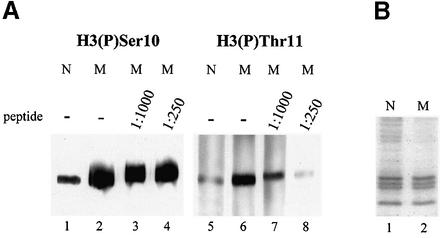
We next asked whether phosphorylation of H3 at Thr11 occurs in vivo. Nuclear extracts from Rat 1 cells were separated by SDS–PAGE and analyzed by western blotting using antibodies specific for (P)-Thr11 or, as control, (P)-Ser10 of H3. Indeed, both antibodies reacted specifically with cellular H3, but not with other proteins (Fig. 2A, lanes 1 and 5). Similar results were obtained with nuclear extracts from other cell lines (data not shown). The signal obtained with the (P)-Thr11-specific antibodies was much weaker than that observed with the (P)-Ser10-specific antibodies. This difference could be due to different affinities of the antibodies or reflect the relative abundance of the respective modifications. In the latter case phosphorylation at Thr11 might occur only in a subfraction of H3, either in a particular phase of the cell cycle or within specific chromosomal regions.
Figure 2.
Phosphorylation of H3 at Thr11 occurs in vivo during mitosis. Rat 1 cells were left untreated or arrested with nocodazol to collect mitotic cells. Cells were lysed in phosphate lysis buffer, soluble cyto-/nucleoplasmic proteins were discarded, residual nuclear proteins were dissolved in SDS sample buffer and subjected to immunoblot analysis using antibodies specific for H3 (P)-Ser10 (lanes 1–4) or H3 (P)-Thr11 (lanes 5–8); incubation with primary antibodies was without (–) or with (shown are peptide dilutions) competing peptide phosphorylated at Thr11, dilutions were made from a 1 mM solution; N, nontreated normal cells, M, nocodazol-treated mitotic cells; after SDS–PAGE, aliquots of N and M extracts equal to the ones loaded on the gels shown in (A) and (B) have been stained with Coomassie blue to assure that identical amounts of proteins were loaded on each lane.
To investigate this issue, and to further explore the specificity of the antibodies, we performed immunoblot analyses with nuclear extracts prepared from asynchronously growing, normal (N) or nocodazol-arrested mitotic (M) Rat 1 cells. Since phosphorylation of H3 at Ser10 is a hallmark of mitotic chromatin, we used (P)-Ser10-specific antibodies as reference. (P)-Ser10-specific antibodies reacted much more strongly with H3 from mitotic cells than with H3 from normal cells, as expected (Fig. 2A, lanes 2 and 1, respectively). Interestingly, the (P)-Thr11-specific antibodies exhibited the same preference (compare lanes 5 and 6 in Fig. 2A). Figure 2B shows that equal amounts of proteins from normal or mitotic cells were loaded. To exclude the possibility that the (P)-Thr11-specific antibodies simply cross-reacted with H3 phosphorylated at Ser10, we performed the immune reactions in the presence of competing peptide encompassing the phosphorylated Thr11. In this case the reaction was blocked in a concentration-dependent manner for the (P)-Thr11- (lanes 7 and 8) but not for the (P)-Ser10-specific antibodies (lanes 3 and 4). Thus, both these antibodies specifically recognized the respective modification. Together, these data suggest that, during mitosis, histone H3 is phosphorylated at both Ser10 and, to lesser extent, Thr11.
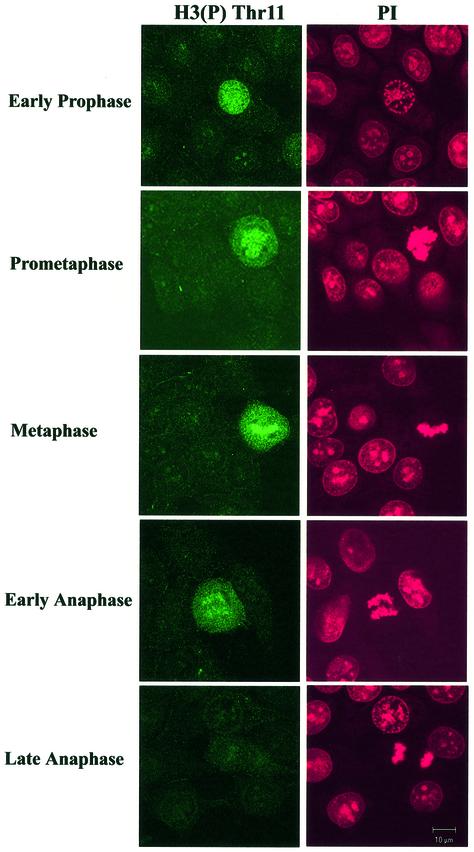
To further confirm these results and to investigate the possibility that phosphorylation at Thr11 was restricted to a short time interval within mitosis or to particular chromosomal regions, we performed immunofluorescence analyses. Cells were counterstained with propidium iodide to visualize chromatin and to evaluate mitotic stages. Again, Thr11 phosphorylation was restricted to mitotic cells, particularly between prophase and early anaphase after which the signal faded (Fig. 3). In contrast, phosphorylation of Ser10 appeared earlier in late G2/early prophase and was discernable until anaphase (data not shown), in agreement with published observations (17,29).
Figure 3.
Phosphorylation of H3 at Thr11 is restricted from prophase to early anaphase. Immunofluorescence analyses were performed on MCF-7 cells using (P)-Thr11-specific antibodies. Cells were counterstained with propidium iodide (PI) to visualize the chromatin and to evaluate mitotic stages. Please note that interphase cells are not stained by the antibody.
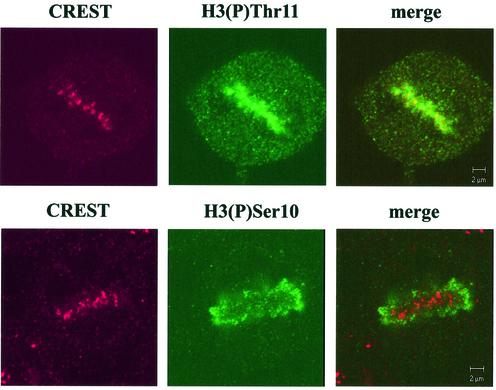
Detailed inspection of mitotic cells by confocal microscopy revealed that Thr11-phosphorylated H3 was not distributed over the whole chromosomes but occurred preferentially at centromere-like dots, as shown on the metaphase picture of Figure 4, upper panel. Centromeres can be visualized with autoimmune sera containing antibodies against various centromere constituents, particularly the so-called constitutive centromere proteins CENP-A, B, C, etc. (27). Co-staining with centromere-specific CREST serum and subsequent confocal immunofluorescence analyses revealed that both antisera revealed very similar staining patterns; the dots stained by anti-H3-(P)Thr11 antibodies largely co-localized with centromere-specific dots (Fig. 4, upper panel). In contrast, the Ser10-specific antibodies stained whole chromosomes, preferentially in distal regions, and did not colocalize with centromeres stained by CREST antibodies (Fig. 4, lower panel). These results demonstrate that phosphorylation of H3 at Thr11 is largely restricted to centromeric regions and suggest that Ser10 or Thr11 phosphorylation are mutually exclusive.
Figure 4.
H3 phosphorylation at Thr11 occurs predominantly in centromeres. MCF-7 cells were stained with centromere-specific human CREST autoimmune serum and either H3 (P)-Thr11- (upper panel) or H3 (P)-Ser10-specific antibodies (lower panel) and analyzed by confocal immunofluorescence microscopy. The yellow signals in the merged images indicate co-localization. Note that (P)-Ser10-specific antibodies do not react with centromeres at all.
Dlk associates with centromeres during mitosis
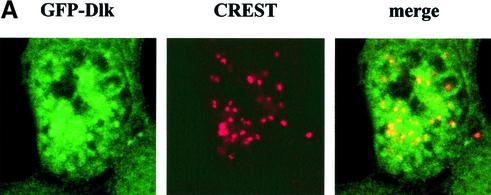
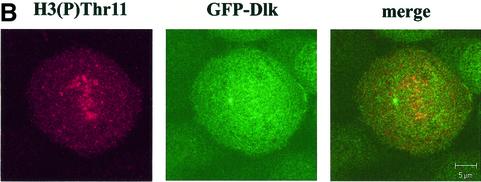
If Dlk was responsible for phosphorylating H3 and, perhaps, CENP-A in vivo, then one would expect fluctuations of Dlk in its level of expression, activity or its subcellular distribution. To address this issue, we established cell clones of MCF7 cells that stably expressed Dlk as GFP-fusion protein. One of these clones, denoted MCF-7-C2, which expressed GFP-Dlk at a low level (compared with transient overexpression) was used for further studies. Ectopic expression of GFP-Dlk revealed no phenotypic effects on these cells, i.e. the growth properties (doubling time, saturation density, serum requirement) of MCF-7-C2 cells were indistinguishable from the parental cell line. In most cells, GFP-Dlk exhibited a diffuse nuclear distribution with a fraction of cells (∼30%) showing a punctuate staining in a diffuse background (data not shown). This picture changed completely during mitosis. Strikingly, Dlk was found associated with different mitotic structures, particularly centromeres (Fig. 5A) but also with centrosomes and the contractile ring, but not with the mitotic spindle (data not shown; U.Preuss, P.Buchenau and K.H.Scheidtmann, submitted). Since association with centromeres might be related to the centromere-specific histone modifications reported above, we studied this association in more detail. Centromeres were visualized by co-staining with CREST serum as shown above. Confocal immunofluorescence microscopy revealed a clear co-localization of GFP-Dlk and centromere proteins in most mitotic cells (Fig. 5A), but not in interphase cells. If both H3 phosphorylated at Thr11 and Dlk individually co-localized with centromeres, then they should both co-localize directly. This assumption was confirmed by the pictures shown in Figure 5B. (Note that in this image one of the centrosomes is in focus showing association of GFP-Dlk with these structures.)
Figure 5.
Dlk associates with centromeres. MCF-7-C2 cells stably expressing GFP-Dlk fusion protein were grown on cover slips and subjected to confocal immunofluorescence analyses. (A) Co-localization of GFP-Dlk (green) and centromere proteins stained with CREST serum (red), as in Figure 4. The yellow signals in the merged images indicate co-localization. (B) Co-localization of GFP-Dlk with H3 phosphorylated at Thr11. Please note that GFP-Dlk is also associated with centrosomes, one of which is in the focal plane
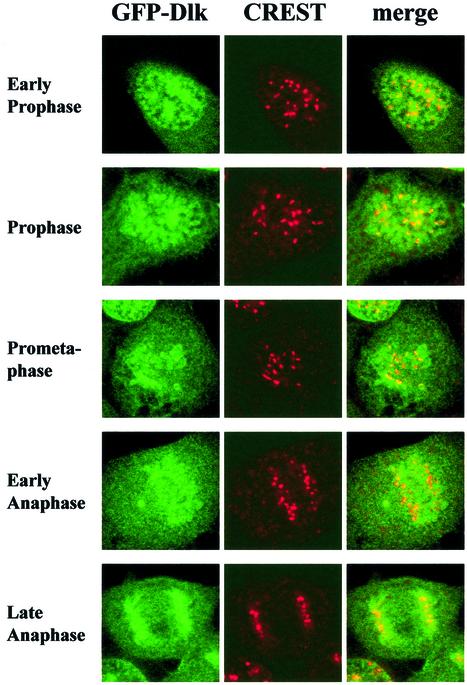
We next determined the time period during which Dlk was associated with centromeres. Inspection of different mitotic phases revealed co-localization of GFP-Dlk with centromeres from prophase, when chromatin condensation was advanced, until anaphase. Co-localization was lost at telophase (Fig. 6). This time course was confirmed by co-staining for CENP-F, a mitotic centromere protein that associates with centromeres from early prophase to anaphase (30) (data not shown). Together, our data demonstrate that during mitosis GFP-Dlk associates with centromeres and that this association coincided precisely with phosphorylation of histone H3 at a novel site, Thr11.
Figure 6.
Dlk associates with centromeres from prophase to early anaphase. Detailed analyses of different mitotic phases show that GFP-Dlk associates with centromeres from early prophase (upper panel) until early anaphase (lower panel). In late anaphase, the GFP signal is no longer restricted to centromeres.
DISCUSSION
In this study we investigated the possible significance of Dlk as a histone kinase. In vitro, Dlk phosphorylated H3 at a novel site, Thr11, rather than Ser10. This modification occurred in vivo as well, specifically during mitosis from prophase to early anaphase. Furthermore, Thr11-phosphorylated H3 was concentrated in centromeric regions, while Ser10-phosphorylated H3 was distributed over the entire chromosomes, particularly in peripheral regions. Thus, this novel modification of H3 appears to be specific for centromeric chromatin. This might explain why it has not been detected earlier. Interestingly, the kinetics of phosphorylation of H3 at Thr11 paralleled that of the H3 variant CENP-A at Ser7, as published recently (29). This coincidence suggested that both proteins are phosphorylated by the same kinase. Indeed, Dlk is also capable of phosphorylating an N-terminal fragment of CENP-A, in vitro (U.Preuss, P.Buchenau and K.H.Scheidtmann, submitted).
Does Dlk phosphorylate H3, and perhaps CENP-A, in vivo? It was striking that recombinant Dlk was associated with centromeres too, and that this association coincided precisely with H3 phosphorylation at Thr11. A critical question is whether the same is true for endogenous Dlk. So far, we have not been able to verify these results for endogenous Dlk because none of the available antibodies against Dlk proved to be suitable for immunofluorescence studies. Additionally, the level of endogenous Dlk might be too low for detection by immunofluorescence. Similar cases are reported in the literature. For example, the association of aurora B kinase and Polo-like kinase 1 with kinetochores was revealed only by using GFP tagging (31,32). To obtain information about the subnuclear localization of endogenous Dlk we performed cell fractionation experiments. Cellular proteins were separated into soluble, nuclease released chromatin and high salt fractions and the distributions of endogenous Dlk and GFP-Dlk were analyzed by immunoprecipitation and SDS–PAGE. These experiments revealed that ectopically expressed GFP-Dlk was 5- to 10-fold more abundant than endogenous Dlk. However, the vast majority of the overexpressed GFP-fusion protein was extracted with the soluble protein while the nuclease-released chromatin and high salt fractions showed a similar distribution of GFP-tagged Dlk and endogenous Dlk (data not shown; U.Preuss, P.Buchenau and K.H.Scheidtmann, submitted). It should be pointed out that the overexpression of Dlk in MCF7-C2 cells is quite moderate compared with transient transfection experiments. Cells are rather sensitive to changes in the composition of critical structures like centromere/kinetochore complexes and usually respond by inducing apoptosis. However, the growth properties of the GFP-Dlk-expressing MCF-7-C2 clone were indistinguishable from those of the parental MCF-7 cells. Finally, phosphorylation of H3 at Thr11 was not restricted to GFP-Dlk-expressing cells but occurred in a number of different cell lines investigated (parental MCF-7 cells, Rat 1, human prostate carcinoma PC3 cells). These data argue against artifacts due to over-expression of Dlk and we are quite convinced that the observations made for GFP-Dlk apply to endogenous Dlk as well. Under these considerations, our data strongly suggest that Dlk is a centromere-specific histone H3 kinase.
With regard to CENP-A, it was recently shown to be phosphorylated by aurora B, in vitro (25). Aurora B is involved in regulation of different mitotic processes (reviewed in 33) and is responsible for phosphorylating the bulk of H3 at Ser10 and Ser28 (24,26,34). Phosphorylation at these sites differs in space and time from the modifications reported here (17,26,29). In prophase, aurora B is recruited to centromeres by inner centromere protein (INCENP) and remains there until the onset of anaphase when it relocalizes in complex with INCENP to the central spindle and later on to the contractile ring (35). Thus, both Dlk and aurora B are localized to centromeres from prophase to early anaphase. However, both kinases differ clearly in their substrate specificities. Aurora B phosphorylates H3 at Ser10 and Ser28, which are both preceeded by Ala-Arg-Lys, Arg at –2 position probably being the critical residue for substrate recognition. These sites are clearly not targeted by Dlk which requires an Arg residue at the –3 position, as for Thr11 (K.H.Scheidtmann, unpublished results). On the contrary, aurora B does not phosphorylate H3 at Thr11. As for CENP-A, it contains overlapping recognition sequences for both kinases, the sequence preceeding Ser7 being Arg-Arg-Arg-Ser (29). However, it is not yet clear why aurora B phosphorylates the bulk of H3 in chromosomal arms while it ignores H3 in centromeres, where it prefers CENP-A as substrate. This issue requires further investigation.
What causes association of Dlk with centromeres? Centromeres and kinetochores are assembled in a stepwise fashion, some factors presumably serving as targeting factors. Thus, CENP-A is required for assembly of constitutive centromere proteins (20–22), INCENP is required for association of aurora B and survivin (35), CENP-F seems to be required for subsequent assembly of CENP-E and BubR (36), and Bub1 is required for association of Mad1, Mad2 and Bub3 (37). Dlk appears at centromeres after CENP-F (not shown). Whether its centromere association depends on CENP-F or INCENP, or occurs independently, needs to be investigated.
What might be the function(s) of H3 Thr11 and CENP-A phosphorylation? It is generally assumed that in higher eukaryotes centromeres are defined epigenetically by establishing or maintaining specific chromatin structures (reviewed in 19,38,39). Histone H3 appeared to be excluded from this centromere-specific chromatin, where it is replaced by CENP-A (20,40). CENP-A, together with CENP-B and CENP-C serve as platform for centromere and subsequent kinetochore assembly. However, only Ser10-phosphorylated H3 is clearly excluded from centromeres. A recent study by Blower et al. (23) using a novel chromatin-stretching technique demonstrated that centromeres do contain both CENP-A and H3 in alternating arrays. Our data suggest that this centromeric H3 is phosphorylated at Thr11, instead of Ser10. Thus, Dlk could be involved in establishing centromere-specific chromatin. However, phosphorylation of H3 (at Thr11) and CENP-A occurs well after establishment of centromere complexes after the onset of mitosis. We propose that these modifications play a role in kinetochore rather than centromere assembly. They might serve as a recognition code for kinetochore proteins. Whether Dlk targets any of the kinetochore components and whether it actively participates in regulation of kinetochore formation, attachment of spindle fibers or even the spindle checkpoint is the subject of further investigation.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the expert introduction to confocal microscopy by Dr P. Buchenau, Botanical Institute, and Carsten Euwens Institute of Genetics, University of Bonn, and the initial characterization of the (P)-Thr11-specific antibodies by V. Weinl, Bonn. This work was supported by Deutsche Forschungsgemeinschaft, grant Sche 246/12-2 and the Fund of the Chemical Industry.
REFERENCES
- 1.Kögel D., Plöttner,O., Landsberg,G., Christian,S. and Scheidtmann,K.H. (1998) Cloning and characterization of Dlk, a novel serine/threonine kinase that is tightly associated with chromatin and phosphorylates core histones. Oncogene, 17, 2645–2654. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T., Matsumoto,M., Takeda,K., Sanjo,H. and Akira,S. (1998) ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol. Cell. Biol., 18, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kögel D., Prehn,J.H. and Scheidtmann,K.H. (2001) The DAP kinase family of pro-apoptotic proteins: novel players in the apoptotic game. Bioessays, 23, 352–358. [DOI] [PubMed] [Google Scholar]
- 4.Cohen O. and Kimchi,A. (2001) DAP-kinase: from functional gene cloning to establishment of its role in apoptosis and cancer. Cell Death Differ., 8, 6–15. [DOI] [PubMed] [Google Scholar]
- 5.Deiss L.P., Feinstein,E., Berissi,H., Cohen,O. and Kimchi,A. (1995) Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev., 9, 15–30. [DOI] [PubMed] [Google Scholar]
- 6.Cohen O., Feinstein,E. and Kimchi,A. (1997) DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J., 16, 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen O., Inbal,B., Kissil,J.L., Raveh,T., Berissi,H., Spivak-Kroizaman,T., Feinstein,E. and Kimchi,A. (1999) DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J. Cell Biol., 146, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inbal B., Cohen,O., Polak-Charcon,S., Kopolovic,J., Vadai,E., Eisenbach,L. and Kimchi,A. (1997) DAP kinase links the control of apoptosis to metastasis. Nature, 390, 180–184. [DOI] [PubMed] [Google Scholar]
- 9.Kögel D., Bierbaum,H., Preuss,U. and Scheidtmann,K.H. (1999) C-terminal truncation of Dlk/ZIP kinase leads to abrogation of nuclear transport and high apoptotic activity. Oncogene, 18, 7212–7218. [DOI] [PubMed] [Google Scholar]
- 10.Page G., Kögel,D., Rangnekar,V. and Scheidtmann,K.H. (1999) Interaction partners of Dlk/ZIP kinase: co-expression of Dlk/ZIP kinase and Par-4 results in cytoplasmic retention and apoptosis. Oncogene, 18, 7265–7273. [DOI] [PubMed] [Google Scholar]
- 11.Mills J.C., Stone,N.L., Erhardt,J. and Pittman,R.N. (1998) Apoptotic membrane blebbing is regulated by myosin light chain phosphorylation. J. Cell Biol., 140, 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page G., Lödige,I., Kögel,D. and Scheidtmann,K.H. (1999) AATF, a novel transcription factor that interacts with Dlk/ZIP kinase and interferes with apoptosis. FEBS Lett., 462, 187–191. [DOI] [PubMed] [Google Scholar]
- 13.Engemann H., Heinzel,V., Page,G., Preuss,U. and Scheidtmann,K.H. (2002) DAP-like kinase interacts with the rat homolog of Schizosaccharomyces pombe CDC5 protein, a factor involved in pre-mRNA splicing and required for G2/M phase transition. Nucleic Acids Res., 30, 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 15.Hans F. and Dimitrov,S. (2001) Histone H3 phosphorylation and cell division. Oncogene, 20, 3021–3027. [DOI] [PubMed] [Google Scholar]
- 16.Clayton A.L., Rose,S., Barratt,M.J. and Mahadevan,L.C. (2000) Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J., 19, 3714–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendzel M.J., Wei,Y., Mancini,M.A., Van Hooser,A., Ranalli,T., Brinkley,B.R., Bazett-Jones,D.P. and Allis,C.D. (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma, 106, 348–360. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan K.F., Hechenberger,M. and Masri,K. (1994) Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol., 127, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanHooser A.A., Mancini,M.A., Allis,C.D., Sullivan,K.F. and Brinkley,B.R. (1999) The mammalian centromere: structural domains and the attenuation of chromatin modeling. FASEB J., 13 (Suppl. 2), S216–S220. [DOI] [PubMed] [Google Scholar]
- 20.Van Hooser A.A., Ouspenski,I.I., Gregson,H.C., Starr,D.A., Yen,T.J., Goldberg,M.L., Yokomori,K., Earnshaw,W.C., Sullivan,K.F. and Brinkley,B.R. (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci., 114, 3529–3542. [DOI] [PubMed] [Google Scholar]
- 21.Howman E.V., Fowler,K.J., Newson,A.J., Redward,S., MacDonald,A.C., Kalitsis,P. and Choo,K.H. (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA, 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oegema K., Desai,A., Rybina,S., Kirkham,M. and Hyman,A.A. (2001) Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol., 153, 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blower M.D., Sullivan,B.A. and Karpen,G.H. (2002) Conserved organization of centromeric chromatin in flies and humans. Dev. Cell, 2, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giet R. and Glover,D.M. (2001) Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol., 152, 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeitlin S.G., Shelby,R.D. and Sullivan,K.F. (2001) CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol., 155, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crosio C., Fimia,G.M., Loury,R., Kimura,M., Okano,Y., Zhou,H., Sen,S., Allis,C.D. and Sassone-Corsi,P. (2002) Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol., 22, 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Earnshaw W.C. and Rothfield,N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma, 91, 313–321. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs B., Hecker,D. and Scheidtmann,K.H. (1995) Phosphorylation studies on rat p53 using the baculovirus expression system. Manipulation of the phosphorylation state with okadaic acid and influence on DNA binding. Eur. J. Biochem., 228, 625–639. [DOI] [PubMed] [Google Scholar]
- 29.Zeitlin S.G., Barber,C.M., Allis,C.D. and Sullivan,K. (2001) Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci., 114, 653–661. [DOI] [PubMed] [Google Scholar]
- 30.Liao H., Winkfein,R.J., Mack,G., Rattner,J.B. and Yen,T.J. (1995) CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol., 130, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murata-Hori M., Tatsuka,M. and Wang,Y.L. (2002) Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell, 13, 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnaud L., Pines,J. and Nigg,E.A. (1998) GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma, 107, 424–429. [DOI] [PubMed] [Google Scholar]
- 33.Nigg E.A. (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nature Rev. Mol. Cell Biol., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- 34.Goto H., Yasui,Y., Nigg,E.A. and Inagaki,M. (2002) Aurora-B phosphorylates histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells, 7, 11–17. [DOI] [PubMed] [Google Scholar]
- 35.Adams R.R., Wheatley,S.P., Gouldsworthy,A.M., Kandels-Lewis,S.E., Carmena,M., Smythe,C., Gerloff,D.L. and Earnshaw,W.C. (2000) INCENP binds the aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol., 10, 1075–1078. [DOI] [PubMed] [Google Scholar]
- 36.Chan G.K., Schaar,B.T. and Yen,T.J. (1998) Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol., 143, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp-Baker H. and Chen,R.H. (2001) Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol., 153, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choo K.H. (2000) Centromerization. Trends Cell Biol., 10, 182–188. [DOI] [PubMed] [Google Scholar]
- 39.Warburton P.E. (2001) Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet., 17, 243–247. [DOI] [PubMed] [Google Scholar]
- 40.Warburton P.E., Cooke,C.A., Bourassa,S., Vafa,O., Sullivan,B.A., Stetten,G., Gimelli,G., Warburton,D., Tyler-Smith,C., Sullivan,K.F. et al. (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol., 7, 901–904. [DOI] [PubMed] [Google Scholar]