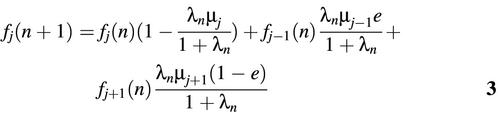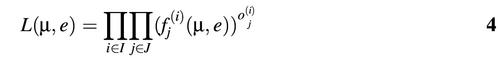Abstract
During microsatellite polymerase chain reaction (PCR), insertion–deletion mutations produce stutter products differing from the original template by multiples of the repeat unit length. We analyzed the PCR slippage products of (CA)n and (A)n tracts cloned in a pUC18 vector. Repeat numbers varied from two to 14 (CA)n and four to 12 (A)n. Data was generated on approximately 10 single molecules for each clone type using two rounds of nested PCR. The size and peak areas of the products were obtained by capillary electrophoresis. A quasi- likelihood approach to the analysis of the data estimated the mutation rate/repeat/PCR cycle. The rate for (CA)n tracts was 3.6 × 10–3 with contractions 14 times greater than expansions. For (A)n tracts the rate was 1.5 × 10–2 and contractions outnumbered expansions by 5-fold. The threshold for detecting ‘stutter’ products was computed to be four repeats for (CA)n and eight repeats for (A)n or ∼8 bp in both cases. A comparison was made between the computationally and experimentally derived threshold values. The threshold and expansion to contraction ratios are explained on the basis of the active site structure of Taq DNA polymerase and models of the energetics of slippage events, respectively.
INTRODUCTION
Microsatellite DNA sequences are short, tandem repeating DNA sequences comprised of 1–6 bp per repeating unit. Microsatellites are interspersed throughout eukaryotic genomes and are highly polymorphic in populations (1–7) due to their propensity for insertion–deletion (in–del) mutation (8) of multiples of the repeating unit during replication. Their polymorphic nature has made microsatellites useful for genetic mapping, studying genomic instability in cancer, population genetics, forensics and conservation biology. A number of factors that might contribute to repeat tract instability during DNA replication have been proposed (9).
Variation in the number of repeat units at a genetic locus is detected by amplifying the alleles by the polymerase chain reaction (PCR) using unique primers flanking the repeating sequence and resolving the PCR products by denaturing electrophoresis (3–6). Instead of detecting a single band representing the size of an allele, ‘stutter’ or ‘shadow’ bands that arise during PCR are found as well. A number of different explanations for ‘stutter’ bands and ways to limit their production during PCR have been proposed (10–17). Direct sequencing of PCR products has shown that the primary cause of ‘stutter’ bands under normal PCR conditions is a change in the number of repeating units due to slipped strand extension by Taq DNA polymerase. By slippage we mean the extension of a primer–template complex containing a loop in either the primer or template strand by a DNA polymerase. Slippage might occur either in the active site of the enzyme or before the substrate binds to the enzyme (9).
We measured the microsatellite in–del mutation frequency using the results of single-molecule PCR. A mathematical model for the microsatellite mutation mechanism during PCR was developed and a quasi-likelihood approach was devised to calculate, per PCR cycle, the mutation rate/template and mutation rate/repeat for both (A)n and (CA)n repeats. The threshold-repeat number for in–del formation was also calculated and was correlated with the number of bases that make contact with the Taq polymerase.
MATERIALS AND METHODS
Cloning of poly(CA)- and poly(A)-containing inserts
A primer {5′-TGAGGGAAGCTTCTG (CA)n [or(A)n] GCTAGTACTGCAGG-3′} and overlapping primer (5′-CCA CAGGAATTCCTGCAGTACTAGC-3′) were obtained from Operon Technologies Inc., CA. The underlined nucleotides are complementary to one another. (CA)n inserts ranged in size up to 14 repeats and (A)n inserts up to 12 repeats. A control plasmid with the same insert lacking a microsatellite sequence was also constructed.
Single-stranded templates were converted into double-stranded inserts by 20 µl of primer extension reactions containing 5 pmol each of template and primer, 7.8 U of T7 Sequenase enzyme (USB), 1× T7 Sequenase buffer, 0.2 mg/ml BSA and 200 µM each dNTP. The reaction was carried out at 37°C for 60 min after which the enzyme was inactivated at 65°C for 30 min. EcoRI and HindIII sites were engineered into the primer extension product for ease of directional cloning into the vector pUC18, such that all the clones had identical orientation of the inserts. This also had the advantage of replacing the palindrome-rich poly-cloning site of the vector.
Two microliters of the primer extension reaction and 17 ng of vector pUC18 (Invitrogen Top 10™ Cloning Kit) were then added to a 20 µl restriction digestion reaction contain ing 1× Promega MULTI-CORE™ buffer, 6 U of EcoRI, 5 U of HindIII, 5 U of BamHI restriction enzymes (the BamHI is added to digest the liberated cloning site) and 0.1 mg/ml BSA. The reaction was incubated at 37°C for 60 min and the restriction enzymes were inactivated for 30 min at 80°C.
Ten microliters of the digested sample was added to a 15 µl ligation reaction containing 1× T4 ligase buffer (Promega) and 3 U of T4 DNA ligase. The reaction was held at 15°C overnight after which 3 µl was used to transform competent Escherichia coli cells (Invitrogen Top 10™ Cloning Kit) according to the manufacturer’s protocol.
The transformants were plated on LB-agar plates containing 100 µg/ml ampicillin and 30 µg/ml X-Gal. The plates were incubated at 37°C for 12–16 h and white colonies were picked and grown overnight in liquid LB containing ampicillin. The inserts from individual plasmid clones were sequenced.
PCR
For single-molecule PCR, the plasmid clones were serially diluted to an average of approximately 0.5 molecules per microliter. Only 9% of the samples with a PCR product would have been expected to contain more than one initial starting template. One microliter of DNA dilution was amplified using two rounds of nested PCR. Fifty microliter PCRs consisted of 1× PCR buffer (10 mM Tris–HCl pH 8.3, 50 mM KCl, 0.01 mg/ml gelatin), 2.5 mM MgCl2, 100 µM each dNTP and 4 pmol each of forward primer (5′-CGGCATCAGAGCAGATTGTA-3′) and reverse primer (5′-GCGTTGGCCGATTCATTAA-3′). The primers are complementary to pUC18 sequences. The 5′ end of one primer is 31 bp away from the beginning of the HindIII site of the insert, while the 5′ end of the other is 70 bp away from the end of the EcoRI site of the insert. First-round PCR conditions were 95°C for 3 min, followed by 10 cycles of 95°C for 30 s and 63°C for 3 min, and 20 cycles of 95°C for 30 s and 63°C for 2 min. Final extension was at 72°C for 5 min. One microliter of the first-round product was further amplified in the second round using 8 pmol each of forward primer (5′-GTCACGACGTTGTAAAACGA-3′) and reverse primer (5′-GGCTCGTATGTTGTGTGGAA-3′). The reverse primer used in the second round was labeled at its 5′ end with the Beckman CEQ WellRED Dye D4 (blue). The second-round amplification conditions were 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 62°C for 30 s and 72°C for 30 s. The usual final extension step at 72°C for 5–10 min was omitted in order to minimize non-templated nucleotide addition by Taq polymerase. Amplification of the control plasmid gave rise to a 132 bp PCR product. The size of PCR products from plasmids with microsatellite markers was equal to 132 bp + the number of (A)n repeats or, in the case of (CA)n tracts, 132 bp + two times the number of repeats. The PCR products were resolved on a Beckman CEQ2000 denaturing microcapillary electrophoresis system. Molecular sizes (in nucleotides) and peak areas of all bands were collected for each of the clones. Clones without any repeat units in the insert were used as controls.
In a second set of experiments, a single round of PCR was performed starting with 100 or 1000 molecules of each clone for 40, 50 and 60 cycles using the second-round PCR conditions described above.
Kinetic PCR
Kinetic PCR (kt-PCR) (18) was used to calculate PCR efficiency (Perkin-Elmer 5700) in the second round using 1 µl of first-round product (initiated with a single target molecule). 5(6)Carboxy–X-rhodamine (2 µM) and 0.2× Sybr Green I were added to 50 µl of a second-round PCR mix. Real-time relative dye fluorescence intensities were used to estimate the efficiency (λ) of the PCR between PCR cycles n1 and n2 (n2 > n1) according to the equation:
Rfn2 = Rfn1 × (1 + λ)n2–n1
that is,
λ = 1 – (Rfn2 / Rfn1)1 / (n2–n1)
where Rfn2 is the relative dye fluorescence intensity value at the PCR cycle n2, and Rfn1 is the relative fluorescence intensity value at the PCR cycle n1. In reactions with a known number of starting templates, the efficiency values were obtained for various intervals along the PCR curve. These efficiency values were then used in the mathematical model for calculating mutation rates/cycle for templates with different number of repeats.
A mathematical model and quasi-maximum likelihood estimation method
We consider a mathematical model for PCR similar to that described by Sun (19). During the nth PCR cycle, each template generates a new copy with probability λn. λn is referred to as the efficiency of PCR at the nth PCR cycle. During the copying process the newly synthesized copy, but not the template, can undergo a mutation.
Let S(n) be the expected total number of templates after ‘n’ PCR cycles, we have:
S(n) = (1 + λn)S(n – 1)1
Next, we consider the mutation process. It is observed from the experiment that the mutation rate of a template increases with the number of repeat units. We do not assume any specific relationship between the mutation rate and the number of repeat units. Let µj be the mutation rate for a template with ‘j’ number of repeat units. We assume that when a mutation occurs, the probability that it is an expansion is e and the probability that it is a contraction is 1 – e.
With the above model, the expected number of template molecules with j repeat units after n – 1 PCR cycles, Sj(n), satisfies the following recursive equation:
Sj(n + 1) = Sj(n) + Sj(n)λn(1 – µj) + Sj – 1(n)λnµj–1e + Sj+ 1(n)λnµj+1(1 – e)2
The above equation can be explained as follows. The template molecules with j repeat units after n PCR cycles are composed of four sets of molecules: (i) those with j repeat units after the (n – 1)-st PCR cycle [Sj(n)], (ii) newly generated templates from parent molecules of j repeat units with no mutations [Sj(n)λn(1 – µj)], (iii) newly generated templates from parent molecules of j – 1 repeat units with one repeat unit added [Sj – 1(n)λnµj– 1e], and (iv) newly generated templates from parent molecules of j + 1 repeat units with one repeat unit deleted [Sj+ 1(n)λnµj– 1(1 – e)]. Let fj(n) be the fraction of molecules with j repeats after n PCR cycles. Then fj(n) can be approximated by Sj(n) / E[S(n)]. From equations 1 and 2, fj(n) satisfies the following recursive equation:
For given efficiencies at different PCR cycles, we can calculate the values of fj(n) at any values of (µα, µα + 1, …, µβ, e) = (µ, e), where α and β are the lower and upper range of the number of repeat units for PCR products of any given template and µ is the vector (µα, µα + 1, …, µβ).
Let oj(i) be the observed fraction of molecules with j repeats in the ith PCR experiment, i ∈ I, and j ∈ J, where I is the set of all the experiments and J is the set of repeat units of interest. Let fj(i) be the predicted fraction of molecules using equation 3 such that i ∈ I, and j ∈ J. The quasi-likelihood L(µ, e) is then defined by:
µ and e were estimated by maximizing L(µ, e). Note that L(µ, e) is not the true but rather the quasi-likelihood of the observed data because the branching process of mutations during PCR creates a dependency among the different sized PCR products. There are β – α + 1 mutation rates (µα, µα + 1, …, µβ) and one expansion rate e with a total of β – α + 2 parameters to be estimated. Due to the high dimension of the parameter space, we use the Kiefer-Wolfowitz (20) algorithm to locate the maximum point of L(µ, e). Theoretical studies have shown that the above approach can accurately estimate the mutation rates as well as the expansion probabilities if the number of PCR cycles is greater than 40 (Y. Lai, D. Shinde, N. Arnheim and F. Sun, unpublished results).
For a simple example of how equation 4 (above) is evaluated, suppose we have experimental data only on (CA)6 and (CA)8. For (CA)6, the fraction of PCR products with five and six repeats is 17 and 83%. For (CA)8, the fraction of products with six, seven and eight repeats is 3, 20 and 77%, respectively. The goal will be to use the stutter pattern frequencies to estimate the mutation rate/template/cycle for all possible templates from (CA)5 to (CA)8. We use the quasi-likelihood function:
L(µ5,µ6,µ7,µ8,e)=(f5(1))0.17(f6(1))0.83(f6(2))0.03(f7(2))0.20(f8(2))0.77
where f5(1) and f6(1) are given by the recursive formula 3 with initial value f6(1) = 1and f6(2), f7(2), and f8(2) are given by the recursive formula 3 with initial value f8(2) = 1.
There are a total of five parameters to be estimated and we can use the Kiefer-Wolfowitz algorithm (20) to find the maximum point of L(µ5, µ6, µ7, µ8, e).
We first analyzed the experimental data assuming that all the mutation rates (for instance µ5, µ6, µ7, µ8 in the above example) are independent. When the whole data set was examined we found that this analysis yielded an obvious linear relationship between the mutation rates and the number of repeat units. We therefore assumed µj = aj + b and estimated a and b using the quasi-likelihood approach (above). The theoretical basis of the computational method and simulation studies to test the validity of this approach will be published elsewhere (Y. Lai, D. Shinde, N. Arnheim and F. Sun, unpublished results).
RESULTS
Single-molecule PCR experiments
We examined the role of Taq DNA polymerase in the formation of in–del mutations during PCR of templates containing dinucleotide and mononucleotide repeats. A possible confounding factor is that microsatellite sequences in the cloned PCR templates can vary in size due to mutations that arose during propagation in E.coli (21–23). To eliminate any influence of these in vivo generated in–dels, we amplified single DNA molecules of each clone.
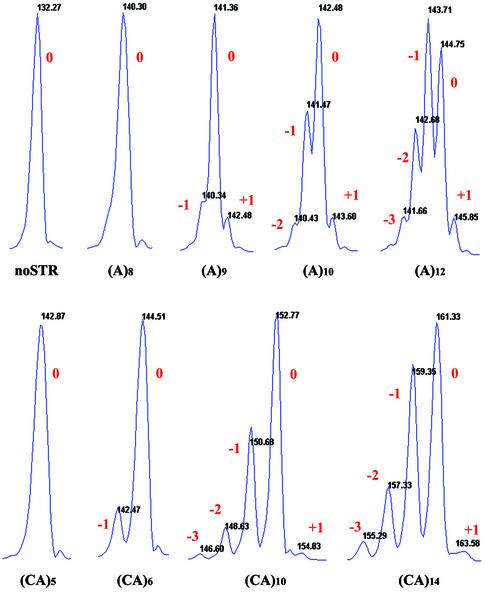
The products of single-molecule amplification from the clone without a microsatellite insert (noSTR) show a single peak with a molecular size of 132 nt (Fig. 1). Under the conditions of our assay, little if any product coincides with a +1 peak due to 3′ end addition of an A (10). The absolute length of each PCR product was determined by comparison with the length of the noSTR control. Due to the stochastic nature of both PCR and the mutation process, the stutter patterns may vary when starting from different single molecules with the same number of repeat units. For example, starting with single (CA)14 molecules, the fraction of mutated PCR products [(CA)13 + (CA)15, etc.] varied from 64 to 49% among the 12 replicates. For (CA)12 the fraction of mutated PCR products [(CA)11 + (CA)13, etc.] varied from 55 to 34%. The complete data set for all templates can be found at http://www-hto.usc.edu/∼fsun.
Figure 1.
‘Stutter’ profiles of products after single-molecule PCR. The integral numbers refer to the change in repeat number from the starting template size. ‘Zero’ refers to the original number of repeats, and ‘plus’ and ‘minus’ refer to additions or deletions of repeats. ‘noSTR’ is the PCR product from a template without repeats. Note that the main peak in the (A)12 profile after 60 cycles does not correspond to the initial size of the single molecule but rather to the one repeat deletion product. This would be expected if the chance of a contraction mutation was greater than that for an expansion given the large number of PCR cycles. The apparent +1 peak in (A)8 was not called as a mutation product based on the software that considers both the peak height and slope threshold.
In clones with a microsatellite insert, the stutter profiles become more spread out as the number of repeat units in the template increases (Figs 1 and 2). This is consistent with an increase in the template mutation rate as the number of repeat units in the template increases. Note that as more repeats are added to the template, a greater effect is seen on the dispersion of the stutter profile of an (A) tract than a (CA) tract (Figs 1 and 2). This indicates that the average mutation rate/repeat/cycle is higher for (A)n than for (CA)n tracts. Finally, even after 60 PCR cycles, we did not observe any mutations for (A)n tracts with eight or less repeat units or (CA)n tracts with five or less repeat units (data not shown).
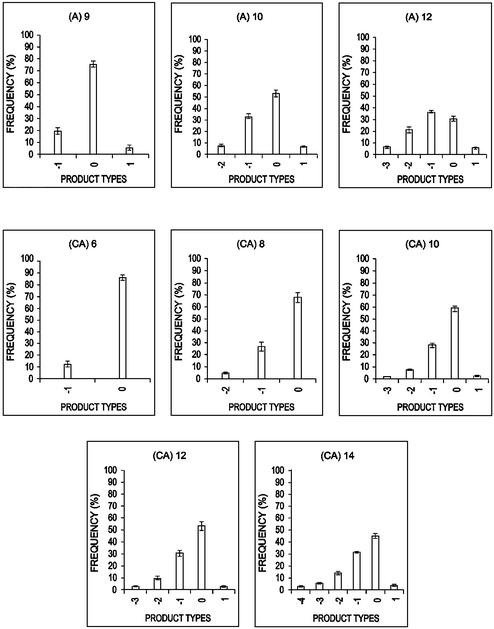
Figure 2.
The distribution of the PCR products after single-molecule amplification using (A)n and (CA)n templates. The x-axis shows the product size where ‘zero’ refers to the original number of repeats, and ‘plus’ or ‘minus’ refer to additions or deletions of repeats. Each bar represents the fraction of PCR products with that size averaged over the 10 single molecules. Error bars (±95% CI) representing the variation between amplification of individual molecules are also shown.
PCR efficiency
In order to study the relationship between mutation rate and number of repeat units, we need to know the efficiency of PCR over different cycle numbers. kt-PCR experiments using the first-round PCR product as template were amplified to estimate the PCR efficiency in the second round (note that beginning with a single molecule, the amount of product generated in the first round of 30 PCR cycles is too small to be detected by real-time PCR). The kt-PCR results (data not shown) provide us with the concentration of template molecules at different PCR cycles. We calculated the efficiency at the nth cycle (λn) as λn = 0.85, 1 ≤ n ≤ 50; λn = 0.85 – 0.05(n – 50), 51 ≤ n ≤ 58; and λn = 0.15, n = 59, 60 to use in our computational analysis.
Estimation of the mutation rate
We collected data on 10 or more single molecules of template with a defined number of repeats. In each case we could calculate what fraction of the PCR product was in each mutation class (+1 or +2, … or –1 or –2, …). Using these fractions and the above efficiencies, we applied the quasi-likelihood method using the experimental data on (CA)6, (CA)8, (CA)10, (CA)12 and (CA)14. Note that all of the stutter profiles for all of the single molecules for each of the five (CA)n template sizes listed above were used together to estimate the mutation rate/template/cycle in the same manner given in the example (see Materials and Methods) where data from only two different template sizes were used.
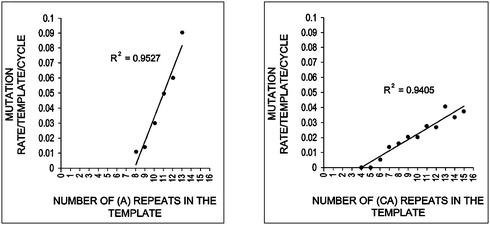
The estimated mutation rate/template/cycle of clones with different numbers of (CA)n or (A)n repeat units are given in Figure 3 and the mutation rate/repeat/cycle shown in Table 1. Extensive simulation studies have shown that the quasi-likelihood approach can recover the relationship between template mutation rates and number of repeat units (Y. Lai, D. Shinde, N. Arnheim and F. Sun, unpublished results). Variation in the efficiency parameter has also been shown to have an insignificant effect on the mutation rate estimates (Y. Lai, D. Shinde, N. Arnheim and F. Sun, unpublished results).
Figure 3.
Relationship between the estimated mutation rate/template/cycle and the number of repeats in the template.
Table 1. Mutation rates/repeat/cycle estimated by the quasi-likelihood method.
| Total µ | Expansion µe | Contraction µ(1 – e) | Calculated threshold j0 (repeats) | Bases | |
|---|---|---|---|---|---|
| (A)n | 1.5 × 10–2 | 2.3 × 10–3 | 1.3 × 10–2 | 8 | 8 |
| (CA)n | 3.6 × 10–3 | 2.4 × 10–4 | 3.3 × 10–3 | 4 | 8 |
The same calculation and considerations were used to estimate the mutation rate for An based on the single-molecule data from templates with A9, A10 and A12 tracts.
The estimated mutation rates increase approximately linearly with an increase in repeat number for both (CA)n and (A)n tracts (Fig. 3). Note that the calculated linear relationship between mutation rate and repeat number includes templates that we have not analyzed but that are predicted to lie on the line based solely on the data we did collect. Using the linear assumption, the same quasi-likelihood approach estimated the template mutation rate µj for (CA)j as: µj = aj + b = a(j – 4) – c, a = 3.6 × 10–3, b = 1.5 × 10–2, c = 4 × 10–4. The term a can be regarded as the mutation rate/repeat/cycle. The template mutation rate depends upon a as well as the number of repeats, j, after the latter is corrected for the threshold of four repeat units estimated computationally. The value of parameter c is ∼10-fold lower than the value of a (see also below) and, in practice, may be taken as equal to zero. The 95% confidence interval for a based on 1000 bootstrap simulations is quite narrow. The probability of (CA)n expansion per repeat per PCR cycle e is estimated at 0.06755 with 95% confidence interval (0.06750, 0.06759). The contraction rate is ∼14-fold higher than the expansion rate.
The template mutation rate for the (A)n tract is µj = a(j – 8) – c, a = 1.5 × 10–2, c = 2.3 × 10–3. The 95% confidence interval (based on 1000 bootstrap simulations) for a is (1.517, 1.522) × 10–2. The probability of (A)n expansion (e) is estimated at 0.15788 with 95% confidence interval (0.15782, 0.15795). The contraction rate is ∼5-fold higher than the expansion rate.
Amplification of a population of molecules
We examined the effect of the number of initial templates (100 or 1000 molecules) and PCR cycles (a single round of 40, 50 or 60) on the mutation rates per cycle of templates with (A)n and (CA)n repeats. In the case of templates with five or fewer (CA) repeats or eight or fewer (A) repeats, no stutter products are detected even after 60 cycles at both DNA amounts (data not shown) agreeing with the single-molecule data. For most of the templates the distribution of PCR mutation products obtained after 40 or more cycles, starting with 100 or 1000 molecules, also does not differ significantly from that obtained after single-molecule PCR. This suggests that the mutation rate/template/cycle should not be affected significantly by the change in the number of starting template molecules from 1 to 1000 when the number of PCR cycles is 40 or more.
DISCUSSION
Mutation assays
The mutation rate of rare in–del events in microsatellite sequences has been measured in several different ways in vitro. One method detects rare mutations that arise during a single gap-filling reaction by a DNA polymerase (24–26). The mutants are identified by an in vivo selection step. In this method, only one of the many different possible kinds (+1 or –1, etc.) of frameshifts can be studied in any one assay.
A strictly in vitro method to measure in–del mutation rates uses a single round of extension with a primer annealed to a template containing a microsatellite sequence (27). In this case only expansion mutations have been studied since incomplete extensions of the primer mimic contractions. The approach described in our paper has an in vitro experimental and a computational component. In a single PCR assay, the mutation rates (/template/cycle and /repeat/cycle) are computed as is the ratio of expansion to contraction mutations. The model takes into consideration that the mutation rate of the template may change as repeats are added or deleted during PCR. In a sense, the mutation rate/template/cycle is analogous to in vivo assays that estimate a rate/template/cell division (21,28,29).
From the perspective of the amount of labor required, our method is best suited to the analysis of thermostable polymerases, but could be applied to thermolabile enzymes if desired. When starting with cloned templates, single molecule analysis is not required to estimate mutation rates using the quasi-likelihood method unless, of course, the mutation rates in the cloning host are so high as to introduce significant size heterogeneity in the population of cloned molecules. This does not appear to have been the case with the (A)n and (CA)n tract lengths we used.
Estimating mutation rates by the quasi-likelihood method using more than a single PCR template might be a problem in certain circumstances. In our experiments, only PCR of (A)9 tracts illustrates a potential difficulty. All 10 of the single-molecule amplifications showed evidence of stutter product. However, when 100 or 1000 template molecules were amplified for 40, 50 or 60 cycles, only 25–50% of the reactions showed stutter products. Because of the stochastic nature of the mutation process, templates with repeat numbers close to the threshold may need to experience many PCR cycles before it is assured that stutter products can be detected. In essence, the number of efficient PCR cycles that can be achieved is greater starting with one template than with 100, which is greater than starting with 1000 templates. Given a constant mutation rate/template/cycle, the lower the PCR efficiency the fewer the number of molecules that are duplicated each cycle and therefore the smaller the number of new mutant molecules produced. This potential problem can be circumvented by running many aliquots with larger template numbers if single-molecule analysis is not carried out. Note that increasing the number of cycles beyond 60 would not help since the efficiency of PCR drops radically as this cycle number is approached.
Threshold for instability
Assuming the commonly accepted simple slippage model (each mutation event usually inserts or deletes only one repeat), a single, mutation rate/repeat/cycle can account for the data on (CA)n or (A)n tracts. For (A)n tracts, quasi-likelihood analysis gave the same threshold estimate [(A)8] as our experimental results.
For (CA)n tracts, a threshold size of (CA)5 had to be exceeded in order to detect mutation events in our assay. When estimated independently by the computational approach, the threshold was found to be (CA)4. Surprisingly, the mutation rate per repeat per cycle computed using the threshold value of (CA)4, suggested that that we should have been able to detect the expected, albeit low, level of mutations with (CA)5 templates considering the sensitivity of the microcapillary electrophoresis assay. This discrepancy might result from an overestimate of our ability to detect low mutation frequencies using our electrophoresis system. It is also possible that the mutation rate/repeat/cycle may not be constant (as is assumed in our calculation) over the range of repeat tract sizes we used in our experiments and may drop significantly for templates with five or fewer repeats compared with those with >(CA)5. In theory, running more PCR cycles would allow the detection of rarer mutation events. However, amplification efficiency is too low after 60 cycles for this to be possible.
Our results on (CA)n tracts suggest that templates with more than 8–10 nt [(CA)4–(CA)5] have a constant mutation rate/repeat/cycle (at least up to 14 repeats). There appear to be no in vitro data on (CA)n tracts to compare with our threshold estimates.
There is some evidence that the mutation rate/repeat/cycle is not the same for all tract sizes at least in the case of (T)n templates. A single in vitro gap-filling reaction using T7 polymerase with an M13mp2 template followed by in vivo selection of the mutants, showed that the mutation rate/template for a single round of primer extension increased significantly as the number of bases increased from five to eight (30). Similar experiments using Taq polymerase are limited to the study of –1 base frameshifts in a (T)5 tract. In this case an error rate no greater than 4.7 × 10–5 per detectable nucleotide incorporated (equivalent to our mutation rate/repeat/cycle) was observed (25). This rate is a minimum of 300-fold less than the mutation rate/repeat/cycle we calculated for >(A)8 tracts. Even though there are a number of differences between the two experimental protocols (e.g. in the PCR assay, mutations can occur on either the A or T template strand) we suggest that the gap-filling data support a transition to a higher mutation rate/repeat/cycle at a tract size somewhere between (T)5 and (A)9 and that no additional threshold is reached at least up to (A)13.
It is interesting that for both types of microsatellites, the total number of nucleotides in a template that undergoes mutation at the maximal rate/repeat/cycle is approximately the same [9 for (A)n and 10–12 for (CA)n]. X-ray data on the structure of Taq polymerase’s active site suggest that during extension of a primer–template by Taq polymerase, the active site is filled with ∼7–9 bp of DNA (31,32). This is consistent with the number of contacts demonstrated for other polymerases (33–36). Thus, it is possible that the mutation rate/repeat/cycle may be maximal when the active site can be completely filled with the repeated sequence. Perhaps some mutation intermediate is stabilized when all the possible active site amino acid contacts are made with repeated nucleotides.
Another possibility is that slipped mutation intermediates do not arise within the enzyme’s active site (24,37). Because of the low processivity of Taq polymerase, slipped intermediates may form in the repeated region of a partially extended template before the binding of the next enzyme molecule. Since the active site can accommodate 9–12 nt, templates with more than this number of repeats have the opportunity to form slipped structures in solution that, as a result of branch migration, form a loop at the 5′ end of the repeated region. A loop would then be absent at the 3′ region of the template and therefore bind more readily to the active site of the enzyme.
It has also been suggested that stutter profiles may include molecules that arose by neither of the above mechanisms but rather by ‘out of register’ annealing of truncated extension PCR products or template switching (38). If primer extensions were terminated prematurely and specifically in the repeated region throughout PCR, and the concentration of such fragments in the last few PCR cycles was sufficiently high, annealing would be possible between complementary repeated sequence bases at their 3′ ends. Primer extension of such substrates by polymerase could occur, leading to expansion and/or deletion products depending on the rules for repeated sequence annealing of the prematurely terminated products. Our results do not support the above model. The microcapillary electrophoresis profiles used to size the PCR products did not show any of the expected prematurely terminated products. Finally, true template switching models (38) should produce no stutter profiles unless it is accompanied by polymerase slippage.
Ratio of expansions and contractions
A preference for contractions in vitro is seen with (T)n tracts in experiments using DNA polymerase β, DNA polymerase α, DNA polymerase γ and T4 DNA polymerase (24,30). Our PCR results show the contraction to expansion mutation ratio is 14 for (CA)n and 5 for (A)n tracts. This preference for contractions could be due, at least in part, to the fact that Taq polymerase lacks a 3′ to 5′ proofreading activity (39). Studies on T7 polymerase show the influence of proofreading on instability depends upon repeat number (30). Using T7 polymerase, tracts with three repeats show 160-fold more mutations using enzyme that lacks proofreading activity. However, (T)8 tracts (the longest studied) are approximately seven times more stable with the proofreading enzyme. This proofreading activity is restricted to the elongating primer. Since expansions, but not contractions, form loops on the primer strand, one would expect more efficient proofreading of lesions leading to expansions and therefore an increase in the contraction to expansion ratio in enzymes that lack this activity (reviewed in 30). The true in vivo situation of course is more complicated since it has been shown that an interaction of an accessory protein with a polymerase (thioredoxin with T7 polymerase) (37) can also influence the ratio of contractions and expansions.
As discussed in an earlier section, it is conceivable that mutation intermediates form in the absence of polymerase. If a loop in the template strand (which can be extended to form a contraction) is accommodated within the enzyme’s active site more easily than a mutation intermediate containing a loop in the nascent strand (24), this would tend to promote contraction mutations.
Finally, energetic arguments have also been made to explain why, in general, one should expect a preference for contractions over expansions (13,24,40). A substrate with a loop in the template strand will yield a contraction but a loop in the nascent strand will yield an expansion. For mononucleotide repeat contractions, one base in the nascent strand must dissociate from its complementary base in the template and anneal to the next template nucleotide. Two bases in the nascent strand must dissociate to generate an expansion. Similarly, a minimum of three bases must dissociate to produce a dinucleotide repeat expansion, whereas only two are required for a contraction. Thus, for any one type of microsatellite repeat, expansion mutations are energetically less favorable than contractions.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank David Gelfand and Nancy Schoenbrunner (Roche Molecular Systems) as well as John Petruska (USC) for helpful discussions.
REFERENCES
- 1.Miesfeld R., Krystal,M. and Arnheim,N. (1981) A member of a new repeated sequence family which is conserved throughout eucaryotic evolution is found between the human delta and beta globin genes. Nucleic Acids Res., 9, 5931–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamada H., Petrino,M.G. and Kakunaga,T. (1982) A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc. Natl Acad. Sci. USA, 79, 6465–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber J.L. and May,P.E. (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am. J. Hum. Genet., 44, 388–396. [PMC free article] [PubMed] [Google Scholar]
- 4.Litt M. and Luty,J.A. (1989) A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet., 44, 397–401. [PMC free article] [PubMed] [Google Scholar]
- 5.Tautz D. (1989) Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res., 17, 6463–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Economou E.P., Bergen,A.W., Warren,A.C. and Antonarakis,S.E. (1990) The polydeoxyadenylate tract of Alu repetitive elements is polymorphic in the human genome. Proc. Natl Acad. Sci. USA, 87, 2951–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckman J.S. and Weber,J.L. (1992) Survey of human and rat microsatellites. Genomics, 12, 627–631. [DOI] [PubMed] [Google Scholar]
- 8.Streisinger G., Okada,Y., Emrich,J., Newton,J., Tsugita,A., Terzaghi,E. and Inouye,M. (1966) Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol., 31, 77–84. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel T.A. and Bebenek,K. (2000) DNA replication fidelity. Annu. Rev. Biochem., 69, 497–529. [DOI] [PubMed] [Google Scholar]
- 10.Clark J.M. (1988) Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res., 16, 9677–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauge X.Y. and Litt,M. (1993) A study of the origin of ‘shadow bands’ seen when typing dinucleotide repeat polymorphisms by the PCR. Hum. Mol. Genet., 2, 411–415. [DOI] [PubMed] [Google Scholar]
- 12.Murray V., Monchawin,C. and England,P.R. (1993) The determination of the sequences present in the shadow bands of a dinucleotide repeat PCR. Nucleic Acids Res., 21, 2395–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hite J.M., Eckert,K.A. and Cheng,K.C. (1996) Factors affecting fidelity of DNA synthesis during PCR amplification of d(C-A)n.d(G-T)n microsatellite repeats. Nucleic Acids Res., 24, 2429–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnuson V.L., Ally,D.S., Nylund,S.J., Karanjawala,Z.E., Rayman,J.B., Knapp,J.I., Lowe,A.L., Ghosh,S. and Collins,F.S. (1996) Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. Biotechniques, 21, 700–709. [DOI] [PubMed] [Google Scholar]
- 15.Walsh P.S., Fildes,N.J. and Reynolds,R. (1996) Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic Acids Res., 24, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bovo D., Rugge,M. and Shiao,Y.H. (1999) Origin of spurious multiple bands in the amplification of microsatellite sequences. Mol. Pathol., 52, 50–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krebs S., Seichter,D. and Forster,M. (2001) Genotyping of dinucleotide tandem repeats by MALDI mass spectrometry of ribozyme-cleaved RNA transcripts. Nat. Biotechnol., 19, 877–880. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi R. and Watson,R.M. (1999) Kinetic PCR analysis using a CCD-camera and without using oligonucleotide probes. In Innis,M.A., Gelfand,D.H. and Sninsky,J.J. (eds), PCR Methods Manual. Academic Press Inc., San Diego, CA, USA, pp. 263–284.
- 19.Sun F. (1995) The polymerase chain reaction and branching processes. J. Comput. Biol., 2, 63–86. [DOI] [PubMed] [Google Scholar]
- 20.Kiefer J. and Wolfowitz,J. (1952) Stochastic estimation of the maximum of a regression function. Ann. Math. Stat., 23, 462–466. [Google Scholar]
- 21.Levinson G. and Gutman,G.A. (1987) High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res., 15, 5323–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schumacher S., Fuchs,R.P. and Bichara,M. (1997) Two distinct models account for short and long deletions within sequence repeats in Escherichia coli. J. Bacteriol., 179, 6512–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bichara M., Pinet,I., Schumacher,S. and Fuchs,R.P. (2000) Mechanisms of dinucleotide repeat instability in Escherichia coli. Genetics, 154, 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel T.A. (1986) Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J. Biol. Chem., 261, 13581–13587. [PubMed] [Google Scholar]
- 25.Eckert K.A. and Kunkel,T.A. (1990) High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res., 18, 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bebenek K. and Kunkel,T.A. (1995) Analyzing fidelity of DNA polymerases. Methods Enzymol., 262, 217–232. [DOI] [PubMed] [Google Scholar]
- 27.da Silva E.F. and Reha-Krantz,L.J. (2000) Dinucleotide repeat expansion catalyzed by bacteriophage T4 DNA polymerase in vitro. J. Biol. Chem., 275, 31528–31535. [DOI] [PubMed] [Google Scholar]
- 28.Henderson S.T. and Petes,T.D. (1992) Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 2749–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farber R.A., Petes,T.D., Dominska,M., Hudgens,S.S. and Liskay,R.M. (1994) Instability of simple sequence repeats in a mammalian cell line. Hum. Mol. Genet., 3, 253–256. [DOI] [PubMed] [Google Scholar]
- 30.Kroutil L.C., Register,K., Bebenek,K. and Kunkel,T.A. (1996) Exonucleolytic proofreading during replication of repetitive DNA. Biochemistry, 35, 1046–1053. [DOI] [PubMed] [Google Scholar]
- 31.Eom S.H., Wang,J. and Steitz,T.A. (1996) Structure of Taq polymerase with DNA at the polymerase active site. Nature, 382, 278–281. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Korolev,S. and Waksman,G. (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J., 17, 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ollis D.L., Brick,P., Hamlin,R., Xuong,N.G. and Steitz,T.A. (1985) Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature, 313, 762–766. [DOI] [PubMed] [Google Scholar]
- 34.Beese L.S., Derbyshire,V. and Steitz,T.A. (1993) Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science, 260, 352–355. [DOI] [PubMed] [Google Scholar]
- 35.Doublie S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- 36.Kiefer J.R., Mao,C., Braman,J.C. and Beese,L.S. (1998) Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature, 391, 304–307. [DOI] [PubMed] [Google Scholar]
- 37.Kunkel T.A., Patel,S.S. and Johnson,K.A. (1994) Error-prone replication of repeated DNA sequences by T7 DNA polymerase in the absence of its processivity subunit. Proc. Natl Acad. Sci. USA, 91, 6830–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odelberg S.J., Weiss,R.B., Hata,A. and White,R. (1995) Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res., 23, 2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tindall K.R. and Kunkel,T.A. (1988) Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry, 27, 6008–6013. [DOI] [PubMed] [Google Scholar]
- 40.Streisinger G. and Owen,J. (1985) Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics, 109, 633–659. [DOI] [PMC free article] [PubMed] [Google Scholar]