Abstract
Human centromeres remain poorly characterized regions of the human genome despite their importance for the maintenance of chromosomes. In part this is due to the difficulty of cloning of highly repetitive DNA fragments and distinguishing chromosome-specific clones in a genomic library. In this work we report the highly selective isolation of human centromeric DNA using transformation-associated recombination (TAR) cloning. A TAR vector with alphoid DNA monomers as targeting sequences was used to isolate large centromeric regions of human chromosomes 2, 5, 8, 11, 15, 19, 21 and 22 from human cells as well as monochromosomal hybrid cells. The alphoid DNA array was also isolated from the 12 Mb human mini-chromosome ΔYq74 that contained the minimum amount of alphoid DNA required for proper chromosome segregation. Preliminary results of the structural analyses of different centromeres are reported in this paper. The ability of the cloned human centromeric regions to support human artificial chromosome (HAC) formation was assessed by transfection into human HT1080 cells. Centromeric clones from ΔYq74 did not support the formation of HACs, indicating that the requirements for the existence of a functional centromere on an endogenous chromosome and those for forming a de novo centromere may be distinct. A construct with an alphoid DNA array from chromosome 22 with no detectable CENP-B motifs formed mitotically stable HACs in the absence of drug selection without detectable acquisition of host DNAs. In summary, our results demonstrated that TAR cloning is a useful tool for investigating human centromere organization and the structural requirements for formation of HAC vectors that might have a potential for therapeutic applications.
INTRODUCTION
The centromere is a chromosomal domain required for proper chromosome segregation during mitotic and meiotic cell division. A typical human centromere appears as a primary constriction in metaphase chromosomes and contains primarily alpha (or alphoid) satellite DNA. Alphoid DNA has repeating units 171 bp in length organized into higher order repeats (HORs). HOR units in a single chromosome share 95–99% sequence homology (1). More degenerate repeats are observed in the pericentromeric regions. The length of the entire centromeric alphoid array varies from 0.2 to 5 Mb. The nucleotide sequence of alphoid DNA and its organization into HORs varies in different human chromosomes. These variants are classified into 12 distinct monomer types and five families. In addition, two ancestral alphoid repeats have been identified: type B contains the binding site for centromeric protein CENP-B and type A lacks the CENP-B binding site (2). Pericentromeric regions of human chromosomes also contain classical satellites I, II and III as well as beta- and gamma-satellites (3).
The organization of the human centromere has been studied extensively, in part because of its importance in biology and also because the structure and function of the centromere must be understood in order to develop a human artificial chromosome (HAC) system, which may offer a new approach for creating gene delivery vectors with potential therapeutic applications. Previous studies of rearranged or fragmented centromeres of the human Y chromosome (4–6) identified an essential centromeric region containing ∼140 kb of alphoid DNA. In addition, in vitro studies in cultured mammalian cells support the idea that alphoid DNA plays a key role in centromere function. For example, constructs containing ∼100 kb of alphoid DNA form stable HACs with functional centromeres when transfected into cultured cells (7–15). Linear or circular alphoid DNA constructs formed HACs in these studies but the constructs were amplified 10–50-fold during propagation in transfected cells. Until now HAC formation has been investigated with only a few alphoid DNA arrays identified in bacterial artificial chromosome (BAC) and yeast artificial chromosome (YAC) libraries. Though it is obvious that HOR units in centromeric DNA are highly heterogeneous in nucleotide sequence and organization, and this heterogeneity may influence the efficiency of de novo centromere formation, analysis of structural requirements for de novo kinetochore formation is limited because, with few exceptions (12,16,17), the centromeric regions of human chromosomes remain to be isolated, adequately mapped and characterized. One of the problems is the difficulty of cloning and mapping repeated sequences. As a result, alphoid DNA sequences are under-represented in large-insert libraries (18).
In 1996 we described a novel recombinational cloning strategy called transformation-associated recombination (TAR) cloning (19). TAR cloning allows selective isolation of desirable large euchromatic chromosomal segments from complex genomes (20). The present report describes the use of TAR cloning to isolate large segments of highly repetitive human alphoid DNA without prior construction of genomic DNA libraries and time-consuming screening of thousands of random clones. The isolated segments were used for physical analysis of the centromere as well as for studying the structural requirements of de novo kinetochore formation.
MATERIALS AND METHODS
Construction of TAR cloning vectors
Two TAR vectors were designed for cloning human alphoid DNA arrays: pVC-Sat and pVC-Sat/Alu. pVC-Sat (SAT-CEN6-HIS3-SAT) was constructed using two oligonucleotides, 96 bp SAT1 (5′-ACACACACGGGCCCcatagagcagtttt gaaacactctttttgtagaatctgcaagtggatatttggaccgGCCGGCCCTG GGCCAACTTTTGGCG-3′) and 106 bp SAT2 (5′-ACAC ACACACGCGGCCGCcttctgtctagtttttatatgaagatattcccgtttcca acgaaggcctcaaagcggtccGGCCGGCCTTACGCCCCGCCCT GCCACT-3′). ApaI, FseI and NotI, which were used for cloning and linearization of a final vector, are underlined. These oligonucleotides are homologous to alphoid DNA in most human centromeres. Their positions correspond to positions 61–117 and 112–171 in the consensus sequence (21). The oligonucleotides were used to PCR-amplify the Cm gene from pBR325. The PCR product was treated with NotI and ApaI and cloned into the multiple cloning site of basic TAR vector pVC604 (20) to generate pVC-Sat. Similarly, SAT1 and 106 bp AluI (5′-acacacacacGCGGCCGCgcgcggtg gctcacgcctgtaatcccagcactttgggaggccgaggcgggcggatcacgaggt caggagaGGCCGGCCTTACGCCCCGCCCTGCCACT-3′) were used to construct the Alu-alphoid DNA TAR cloning vector pVC-Sat/Alu. AluI is homologous to the Alu consensus sequence at position 7–76. FseI linearized vector DNA was used for TAR cloning experiments.
We used the ARS-containing TAR vector pRS-Sat-Neo to isolate the alphoid DNA array of the ΔYp74 mini-chromosome because we failed to isolate the region with a pVC604-based vector. The vector was constructed as follows. A 2.7 kb fragment of the Neo gene was amplified by PCR using primers with NotI and XhoI restriction sites and BRV1 plasmid as template (20). The primers were NeonotR (5′-gcggatgaatggcagaaattcgat-3′) and NeoxhoF (5′-ccggctcgagctgtggaatgtgtgtcagttagg-3′). A 1.0 kb XmaI–BglII fragment was excised from the 2.7 kb Neo PCR product and cloned into SmaI–BamHI sites of pRS313 (22) to give pRS-Neo. The 1.0 kb fragment contains the Neo open reading frame but does not contain the SV40 promoter. A 117 bp alpha-satellite fragment was amplified by PCR using primers with SalI sites and human genomic DNA (Promega) as a template. The primers were Sat-Sal-Rev (5′-accgtcgactcacagagttgaa-3′) and Sat-Sal-For (5′-attcccgtttccaacgaagg-3′). The alpha-satellite DNA fragment was cloned into pCRII (Invitrogen), isolated as an EcoRI fragment, and cloned into the EcoRI site of pRS-Neo. The vector pRS-Sat-Neo was cut between the targeting sequences with SmaI to yield linear molecules with Sat and Neo hooks at the termini. The TAR vector was isolated with a Qiagen Plasmid Purification Kit.
Preparation of genomic chromosome-sized DNA in solid agarose plugs for TAR cloning experiments
For TAR cloning genomic DNA was prepared in agarose plugs (20). Agarose plugs (60 µl) containing ∼5 µg of high molecular weight human DNA were prepared from human fibroblasts MRC-5 (American Type Culture Collection), the GM06318C monochromosomal human–mouse hybrid cell line containing human chromosome 22 (Coriell Cell Repositories), the ΔYp74 hybrid (human–hamster) cell line containing a truncated human Y chromosome (23), and the DT40-11 monochromosomal human–chicken hybrid cell line containing human chromosome 11 (kindly provided by Dr Minoru Koi, Brunel University, UK).
Yeast strains and transformation
The highly transformable Saccharomyces cerevisiae strain VL6-48 (MAT alpha, his3–Δ1, trp1–Δ1, ura3-52, lys2, ade2-101, met14 ciro) (20) was used for transformations. Spheroplasts were prepared by as previously described (20). For transformation experiments, DNA-containing plugs were melted and treated with agarase. A standard lithium acetate procedure was used to introduce a BAC retrofitting vector into yeast cells. Yeast transformants were selected on synthetic complete medium plates lacking histidine or uracil.
Characterization of YAC clones
Chromosome size DNA was prepared from yeast transformants carrying circular YACs, separated by clamped homogeneous electrical field (CHEF) electrophoresis, blotted and hybridized with a 5.7 kb centromeric alphoid DNA probe or a Sat probe (see below). The size of circular YACs was estimated by exposing DNA to low dose gamma-rays (5 krad) before CHEF analysis. This dose of gamma rays linearizes ∼10% of YACs in the 100–400 kb size range (19).
DNA labeling
A 5.7 kb EcoRI alphoid DNA fragment from chromosome Y was labeled by nick-translation. A 56 bp oligonucleotide (Sat probe) was 5′-end-labeled. The Sat probe sequence was 5′-cattctcagaaacttctttgtgatgtgtgcattcaactcacagagttgaaccttcc-3′, corresponding to positions 1–56 in the alphoid DNA unit consensus.
Southern blot analysis
Southern blot hybridization was performed as described by Barnett et al. (24) with 32P-labeled probes. Blots were incubated for 2 h at 65°C in pre-hybridization solution (0.5 M Na-phosphate buffer containing 7% SDS and 100 µg/ml unlabeled salmon sperm carrier DNA). Labeled probe (20 µl) was heat denatured in boiling water for 5 min and snap cooled on ice. The probes were added to the hybridization buffer and allowed to hybridize overnight at 65°C. The Y centromere-specific probe was hybridized overnight at 78°C (25). Blots were washed twice in 2× SSC (300 mM NaCl, 30 mM sodium citrate, pH 7.0), 0.1% SDS for 30 min at room temperature, then three times in 0.1× SSC, 0.1% SDS for 30 min at 65°C. Blots were exposed to X-ray film for 24–72 h at –70°C.
FISH analysis
YAC DNA for probe preparation was isolated from yeast cells according to a standard protocol (20). 5-Bromo-deoxyuridine was used to synchronize normal male peripheral lymphocytes, and metaphase spreads were prepared. Fluorescence in situ hybridization (FISH) was performed as described elsewhere (26,27). In brief, DNA probes were labeled with Spectrum Orange-dUTP (Vysis, Downers Grove, IL) by nick translation and ethanol-precipitated in the presence of 50× herring sperm DNA and 50× Cot-1 human DNA. Probes were dissolved in 50% Hybrisol and denatured at 78°C for 10 min, and 250 ng was applied to the slide. Slides were hybridized overnight in a humid chamber at 37°C and washed at 45°C 3× for 5 min in 50% formamide/2× SSC, 2× for 5 min in 0.1× SSC, and 1× for 2 min in 4× SSC/0.1% Tween-20 at room temperature. Slides were counterstained with 0.25 mg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI-antifade). Chromosomes were identified by converting DAPI-banding into G-simulated banding with the IPLab Image Software (Scanalytics Inc., Fairfax, VA).
Probes for BACs 14 and 5 were labeled by nick-translation with Spectrum Orange-dUTP and Spectrum Green-dUTP, respectively (Vysis, Downers Grove, IL). Probes were ethanol precipitated in the presence of 50× herring sperm DNA and 50× Cot-1 human DNA, dissolved in 50% Hybrisol, and denatured at 78°C for 10 min. FISH was carried out as described above. For fiber-FISH experiments, chromatin was stretched as previously described (28), and DNA hybridization and visualization were performed as above. Briefly, chromatin fibers were made from lymphoblast cells. Cells were washed three times in 1× PBS. The pellet was resuspended in 100 µl of PBS. Two drops (10 µl) of cell suspension were placed on the slide. Cells were incubated in lysis buffer (2 M MgCl2, 25 mM Tris–HCl pH 7.8 and 1% Triton X-100) for 30 min at room temperature. Slides were removed, air-dried in a vertical position for 20 min, and fixed in methanol:acetic acid (3:1) for 20 min.
HACs were identified by single or dual FISH to detect alphoid sequences and BRV vector sequences. One hundred and fifty metaphase spreads of untransfected HT1080 cells were screened as a negative control. Alphoid DNA was labeled with biotin-16-dUTP (Roche) and vector DNA with dig-11-dUTP by nick-translation (Roche). FISH hybridization and washing conditions were as described above. The biotinylated probe was detected with SA-FITC (Invitrogen) and the digoxigenin-labeled probe was detected with mouse anti-dig antibody (Roche) followed by Texas Red-conjugated goat anti-mouse antibody (Molecular Probes). The inter/intra Alu and pan-alphoid probes were generated essentially as described previously (11). Human rDNA was from a PAC clone isolated from a genomic library. DNA was labeled with dig-11-dUTP and detected as described above. The 22q Spectrum Orange paint probe was purchased from Vysis.
Retrofitting circular YACs into BACs and transfer into Escherichia coli
YACs were retrofitted into BACs with the yeast–bacteria–mammalian cell shuttle vector BRV1, which contained the F-factor origin of replication and the Neo gene (20). The standard lithium acetate transformation procedure was used. Yeast transformants were selected on synthetic complete medium plates lacking uracil. The retrofitted His+Ura+ YACs were moved to E.coli by electroporation. Low-melting-point agarose plugs were prepared from yeast His+Ura+ transformants by a standard method (20). One microliter of melted agarose plug was electroporated into 20 µl of E.coli DH10B-competent cells (Gibco BRL) with a Bio-Rad Gene Pulser (2.5 kV, 200 Ω and 25 µF). Colonies were selected on LB plates containing 12.5 µg/ml chloramphenicol.
Sequences analysis of alphoid DNA inserts
The sequence of the insert ends in YACs was determined as follows. DNA was purified from YAC-containing yeast cells and digested with EcoRI, HindIII, XbaI, BamHI or PstI, enzymes that do not cut the TAR vector. Linear DNA was circularized by ligation, transformed into E.coli by electroporation, and transformants were selected for ampicillin resistance. Cells carrying human DNA and TAR vector were recovered. DNA was prepared and sequenced with T3 and T7 primers.
SpeI fragments (2.8 and 2.9 kb) from Y chromosome alphoid DNA and a 1.8 kb SpeI fragment with inverted alphoid DNA were gel purified from SpeI-digested BAC DNA. The fragments were cloned into the SpeI site of pBluscript and sequenced. Divergent regions of alphoid DNA monomers were used to subclone smaller insert fragments and to design sequencing primers. The complete sequence of all three SpeI fragments was determined (accession nos AF522078 and AF533770).
Alphoid DNA fragments from BACs 11 and 5 were sequenced as follows. BACs were digested with EcoRI, and 2.1 and 2.8 kb fragments were subcloned into pBluescript. Forty-one randomly selected clones with 2.1 or 2.8 kb inserts were sequenced. Similar analysis was carried out for BAC 5 with gel purifed EcoRI fragments of 510 to ∼2.5 kb. EcoRI and XbaI fragments from BAC 25 were sequenced in a similar manner.
DNA sequencing was performed with a Rhodamine Dye Terminator Cycle Sequencing Kit (Perkin Elmer, Catalog No. 403 042) and a Perkin Elmer Model 377 automated DNA sequencer.
Sequence divergence between pairs of alpha satellite monomers was analyzed by the Kimura two-parameter method (29). Phylogenetic trees were constructed with the MEGA2 program (30) using the neighbor-joining algorithm and multiple alignments of nucleotide sequences (31). MEGA2 was used with default parameters. Bending and curvature were calculated with ‘BEND_TRI’ (32), a program that calculates the magnitude of local bending B(i) and macroscopic curvature C(i) at each position of a sequence. The average values of C(i) and B(i) were calculated for each sequence. NSITE (http://genomic.sanger.ac.uk/gf/gf.shtm) (33) was used to analyze similarity between consensus motifs and satellite DNA sequences. The BioEdit program (http://jwbrown.mbio.ncsu.edu/BioEdit/bioedit.html) was used to construct an entropy plot.
Mapping of yeast ARS-like sequence within an alphoid DNA unit
BAC11 was treated with EcoRI and the fragments were cloned into pVC604 (HIS3/CEN6). This vector lacks a yeast origin of replication site and transforms yeast cells with very low efficiency (5–10 colonies per µg DNA). Thirty-two recombinant plasmids were recovered carrying 2.1 or 2.8 kb alphoid DNA inserts, which transformed the yeast cells with high efficiency. Plasmid inserts were sequenced to confirm the presence of alpha satellite DNA. Plasmids were digested with HaeIII and fragments were subcloned into pVC604. Two autonomously replicating clones were isolated in yeast cells, transfered to E.coli, and sequenced with T3 and T7 primers. These clones carry 0.5 and 1.0 kb alphoid DNA inserts, respectively, which include a match to the ARS consensus sequence. An alphoid DNA monomer with a yeast ARS-like sequence was PCR-amplified from the plasmids with primers A/C-1 (5′-gatgtgtgcattcawcttacag-3′) and A/C-2 (5′-catcacaaagaagtttctcagaatg-3′). A yeast transformation assay suggested that the monomer included a functional ARS sequence.
Comparison of internal stability of alphoid DNA arrays in RAD+ and rad52 yeast host strains
To investigate whether yeast host recombination deficiency stabilized the inserts, we quantitatively evaluated YAC rearrangements during mitotic propagation in wild-type and rad52 mutant cells. For this purpose, several alphoid DNA arrays were internally marked by homologous recombination in yeast with the yeast counter-selectable marker TRP1. TRP1 was PCR-amplified from the pRS314 plasmid (22) was a 1.0 kb DNA fragment flanking by alphoid DNA sequences. The amplification was performed with the primer pair SBTRP-F (5′-GAATCTGCAAGTGGATATTTGGAGCCC TTTGAGGatccgatgctgacttgctgggtattatatgtg-3′) and SBTRP-R (5′-CATCACAAAGAAGTTTCTGAGAATGCTTCTGT CTAGgatccaccgcaggcaagtgcacaaacaatac-3′). Capital letters show the sequences homologous to alpha-satellite repeats. To simplify further analysis of targeted YACs, BamHI recognition sites (underlined) were placed between TRP1 and alpha-satellite sequences in each primer. Clones with the TRP1 gene inserted in approximately the middle of the alphoid DNA array were selected for further analysis. Counter-selection against the TRP1 gene was performed on media containing 0.05% 5-fluoro-2-aminobenzoic acid (5-FAA) as described by Toyn et al. (34). Loss of the TRP1 marker would presumably be due to homologous recombination between tandem repeat units that flank the marker in the insert. Use of a host strain with a conditional RAD52 allowed us to compare rates of TRP1 loop-out from the insert when RAD52 was switched on or off by transferring the cells from galactose-containing to glucose-containing media (35). In recombination-proficient cells the frequency of loss of TRP1 was ∼2.5 × 10–5. Physical analysis showed that all Trp– YAC clones had deletions of 30–70 kb (data not shown). The frequency of appearance of Trp– cells was four times decreased when RAD52 was not expressed. This result is in agreement with a previous report that inactivation of RAD52 partially stabilizes YACs with large blocks of alphoid DNA (36).
RESULTS
Isolation of human centromeric alphoid DNA by TAR cloning
This study examined whether TAR cloning can be used for isolating heterochromatic DNA. Let us first examine the possible limitations: (i) TAR cloning requires that the cloned DNA fragment carry at least one autonomously replicating sequence (ARS) that functions in yeast (19). ARS or ARS-like sequences are present on average every 20–40 kb in human euchromatic DNA (37–39), but their frequency in heterochromatic centromeres of human chromosomes may be much lower. (ii) Because heterochromatic DNA is highly repetitive and TAR cloning makes use of recombination events, it is possible that the cloned sequences derived from heterochromatic DNA will have smaller (or less stable) inserts than TAR clones containing euchromatin.
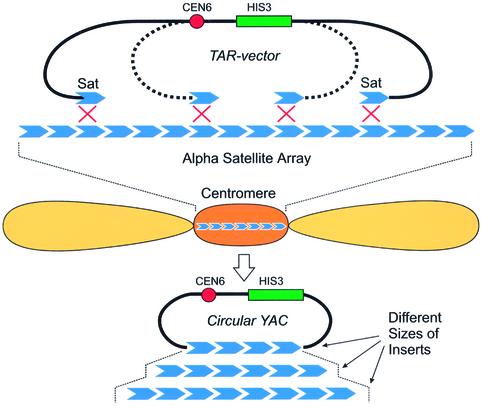
We constructed a circularizing TAR cloning vector with the structure SAT-CEN6-HIS3-SAT, where ‘SAT’ represents 56 and 60 bp fragments from the 5′ end of the alphoid DNA monomer (see Materials and Methods). The linear form of this vector has an alphoid DNA targeting sequence at each end. Figure 1 shows a mechanism by which the vector and an alphoid DNA genomic segment undergo homologous recombination to produce a YAC with an alphoid DNA-containing insert. The resulting YACs are expected to include either only alphoid DNA or alphoid plus non-alphoid DNA. A condition for survival of the clones is the presence of an ARS element that functions in yeast. TAR cloning of human centromeric DNA was carried out as follows (see Materials and Methods for details). High molecular weight genomic DNA was prepared from MRC-5 human fibroblasts. Five micrograms of genomic DNA, 1 µg of FseI-linearized TAR vector, and 2 × 109 spheroplasts were combined, incubated, and plated under selection as described. The average yield was 20–30 transformants, which is similar to the yield observed in the isolation of single copy genes from mammalian genomic DNA by TAR cloning (20). One hundred and thirty His+ transformants were selected randomly and tested for inserts containing alphoid DNA by dot-hybridization (see Materials and Methods). Ninety-eight (75%) of 130 transformants reacted with the alphoid DNA probe, indicating a high selectivity for TAR cloning.
Figure 1.
Isolation of centromeric regions by TAR cloning. A linearized TAR vector carrying yeast selectable marker (HIS3), a yeast centromere (CEN6), and two targeting satellite sequences were co-incubated with human genomic DNA and yeast spheroplasts. Homologous recombination occurred between targeting sequences in the vector and human centromeric satellite DNA and produced circular YACs.
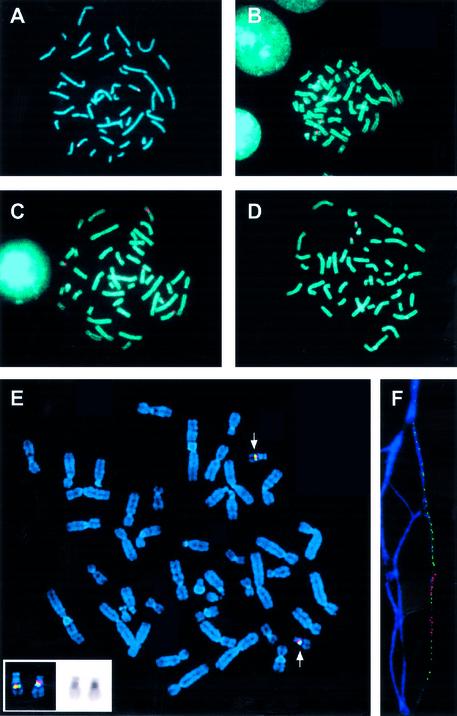
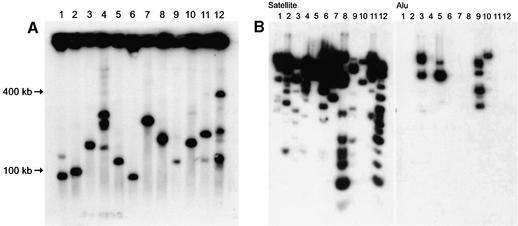
Sixty His+ clones with strong hybridization signals were partially sequenced and further characterized. Sequence analysis showed that the YAC ends consisted exclusively of alphoid DNA monomers. Results of analysis of representative TAR isolates are shown in Table 1. FISH analysis was carried out with total DNA isolated from each yeast clone. All YAC isolates contained DNA sequences that mapped at or near human centromeres (Fig. 2A–D and Table 1). In most cases, more than one signal was observed, which is consistent with the observation that some satellite sequences cross-hybridize with centromeric DNA sequences on several chromosomes. The size of YAC inserts was determined by CHEF gel electrophoresis and visualized with an alphoid DNA probe. The size of the YACs varied from 50 to 400 kb (Fig. 3A and Table 1). Most of the alphoid DNA TAR isolates were structurally stable during propagation in yeast. The isolates were also stable in E.coli cells (the clones were transferred into bacterial cells after retrofitting of YACs into BACs). Some isolates had more than one band, suggesting that satellite DNA is unstable in yeast cells. Mitotically unstable clones were partially stabilized by transferring YACs into strains with a conditional RAD52 gene by kar1 mating (see Materials and Methods). The frequency of internal deletions in YACs decreased approximately four times when RAD52 was not expressed. DNA from yeast isolates was also digested by HindIII, EcoRI or XbaI, separated by gel electrophoresis, and hybridized with Alu-, LINE-, MIR- or alphoid DNA probes. Eighty-five percent (51/60) of the clones hybridized only with alphoid DNA. Nine isolates hybridized with non-alphoid probes (Alu and/or LINE or MIR; Fig. 3B). These nine isolates probably have inserts from pericentromeric regions where the alphoid DNA array is frequently interrupted with non-alphoid DNA insertions. This suggestion is supported by data from the partial sequencing of the inserts (data not shown). For example, clone 25 reacted with MIR and LINE probes, and clone 25 exactly matches a contig from the 2p11.1 pericentromeric region of chromosome 2 (contig NT_022171.6; positions 1665802–1665119).
Table 1. Characterization of TAR clones containing alphoid DNA.
| TAR isolate | Size of YAC in yeast (kb) | FISH mapping | Alphoid DNA at both ends of insert | Size of BACin E.coli (kb) | Non-alphoid DNA repeats in insert |
|---|---|---|---|---|---|
| Chr 22a | |||||
| 3 | 50 | 22/cen | + | 50 | – |
| 5 | 140 | 22/cen | + | 140 | – |
| 6 | 120 | 22/cen | + | 120 | – |
| 10 | 80 | 22/cen | + | 80 | – |
| 11 | 60, 140 | 22/cen | + | 60, 140 | – |
| 14 | 50, 100, 200 | 22/cen | + | 50, 100 | – |
| 15 | 100 | 22/cen | + | 100 | – |
| 19 | 70, 110 | 22/cen | + | 70, 110 | – |
| 20 | 60 | 22/cen | + | 60 | – |
| 29 | 70, 200 | 22/cen | + | 140 | – |
| 35 | 60 | 22/cen | + | 60 | – |
| Chr 11b | |||||
| 2 | 75, 150, 400 | 11/cen | + | 75, 125, 200 | – |
| MRC5c | |||||
| 8 | 75 | X, 22, 15/cen (5/tel) | + | 75 | – |
| 11 | 140 | 8/cen | + | 110 | + |
| 13 | 140, 220, 270 | 13, 21/cen | + | 100 | – |
| 16 | 100 | 19, 20, Y, 14 | + | 100 | + |
| 25 | 220 | 2, 10/cen | + | 220 | + |
| 26 | 140 | 15/cen | + | 100 | – |
| 41 | 150 | 8/cen | + | 150 | + |
| 59 | 175 | 5, 19/cen | + | 175 | – |
cen, FISH signal is in the centromeric region; tel, FISH signal signal is in the telomeric region.
aClones were isolated from monochromosomal hybrid cell line containing chromosome 22.
bClone was isolated from monochromosomal hybrid cell line containing chromosome 11.
cClones were isolated from MRC5 human normal fibroblasts.
Figure 2.
FISH mapping of YAC clones carrying human alphoid DNA. The satellite-positive isolates mapped at or near the human centromere. Some satellite sequences cross-hybridized with more than one centromere. (A) YAC clone hybridized to centromere of chromosome 15 (no. 26 in Table 1). (B) YAC clone hybridized to centromeres of chromosomes 13 and 21 (no. 13 in Table 1). (C) YAC clone hybridized to centromere of chromosome 11 (no. 2 in Table 1) (multiple signals are due to cross-hybridization with other centromeres). (D) YAC clone hybridized (nos 5 and 11 in Table 1) to the centromere of chromosome 22. (E) Two-color FISH was carried out with probes from BAC 11 (Spectrum Orange) and BAC 5 (Spectrum Green). Both probes hybridized to the centromere of chromosome 22. (F) Fiber-FISH using the same probes (bottom) as in (E).
Figure 3.
(A) Physical characterization of YACs in yeast. Chromosome-size DNA was isolated from yeast transformants, exposed to a low dose of gamma rays, separated by CHEF gel electrophoresis, and blot-hybridized with a Sat probe. Lanes 1–12 correspond to 12 independent yeast isolates. The size of inserts varied from 50 to 400 kb. Lanes 2, 3, 4, 5, 7, 8, 10, 11 and 12 on the gel correspond to YACs 8, 11, 13, 16, 25, 26, 41, 59 and 2 in Table 1. Lanes 1, 6 and 9 correspond to additional independent YAC isolates. (B) YAC DNAs were digested with HindIII, EcoRI or XbaI, gel-separated, and blot-hybridized with either a Sat or Alu probe.
The high selectivity of alphoid DNA isolation was also observed with genomic DNA prepared from monochromosomal hybrid cell lines. Thirty-nine of 100 transformants with inserts from human–mouse hybrid GM06318C contained large blocks of alphoid DNA from the centromere of human chromosome 22. Results of analysis of representative TAR isolates are shown in Table 1. The inserts varied from 50 to 200 kb and hybridized to human centromere 22 when used as a FISH probe (Fig. 2D). Seven of 38 YAC isolates were Alu-positive, suggesting that they were derived from the pericentromeric region of centromere 22. Similar cloning results were obtained with genomic DNA from human–chicken hybrid DT40-11 cells containing human chromosome 11 (data not shown).
The alphoid DNA array from a human mini-chromosome ΔYq74 (5,23) was also isolated by TAR cloning from a hybrid cell line. ΔYq74 is a 12 Mb mini-chromosome generated by telomere-directed breakage of the human Y chromosome; ΔYq74 includes ∼140 kb of alphoid DNA. Because the Y chromosome was truncated by a telomere-containing vector, the alphoid DNA array in ΔYq74 must be physically linked to the Neo gene. This region was isolated with the TAR vector pRS-Sat-Neo, which has an alphoid DNA fragment and a Neo gene fragment as targeting sequences (see Materials and Methods). Yeast spheroplasts were co-transformed with a SmaI-linearized TAR vector and genomic DNA from ΔYq74 human–mouse hybrid cell line, and transformants were selected for His+. A set of circular His+ YACs was isolated with alphoid DNA inserts of from 50 to 140 kb. The largest insert was approximately the same size as the entire alphoid DNA array on the mini-chromosome. FISH hybridization with human metaphase chromosomes indicated that all the clones hybridized strongly with centromeric sequences from the human Y chromosome (data not shown).
In summary, our results demonstrated that the repetitive nature of alphoid DNA did not prevent its isolation by in vivo recombination in yeast. The high selectivity of alphoid DNA isolation by TAR cloning provides the opportunity to construct centromere-specific libraries that will greatly simplify the physical characterization of centromeres.
Physical characterization of TAR clones containing alphoid DNA arrays
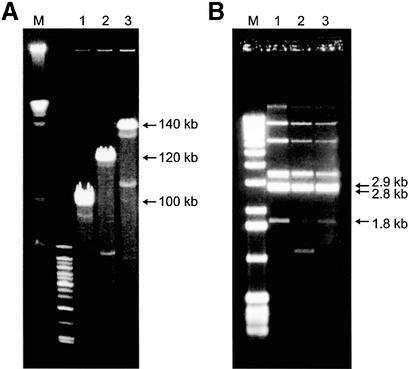
Three circular YACs containing alphoid DNA arrays from mini-chromosome ΔYq74 (insert sizes 100, 120 and 140 kb) and 11 YACs with alphoid DNA from chromosome 22 were retrofitted into YAC/BACs by recombination in yeast and transferred to E.coli (see Materials and Methods). BAC DNA was isolated from 10 independent E.coli transformants for each YAC/BAC, digested with NotI, and analyzed by CHEF gel electrophoresis. Most of the BACs were identical in size to the corresponding YAC (Table 1), indicating that the alphoid DNA containing BACs were reasonably stable in E.coli. Approximately 5% of the BACs had visible deletions (Fig. 4A). Tyler-Smith and Brown (40) showed that alphoid DNA from the centromere of the human Y chromosome is organized into repeating units ∼5.7 kb long. Each unit includes 34 tandemly repeated 171 bp alphoid DNA monomers and has a single EcoRI site and two XbaI sites. TAR clones carrying alphoid DNA from ΔYq74 have a similar organization. The structure of the ΔYq74-specific BACs was determined by digestion with EcoRI or XbaI, gel electrophoresis, and hybridization with a 5.7 kb alphoid DNA fragment from a human Y chromosome. BACs have a major 5.7 kb EcoRI fragment as well as the expected 4.8 and 0.9 kb XbaI fragments (41) (data not shown). BACs also have 2.9 and 2.8 kb repeats, as shown by SpeI digestion (Fig. 4B), composed of 16 or 18 alphoid DNA monomers, respectively. (Other minor fragments visible on the gel corresponded to vector sequences and sequences corresponding to the junction between the vector and the insert.) SpeI digestion of the BACs also identified an additional 1.8 kb fragment containing alphoid DNA (Fig. 4B). Because we failed to detect this fragment in a SpeI digest of male genomic DNAs (data not shown), it was generated during chromosome truncation. This fragment contains two blocks of alphoid DNA (seven and four monomers) organized as palindromes. The blocks are separated by two incomplete monomers. Previous sequence analysis showed that the seven-monomer block corresponds to a targeting alphoid DNA in the truncation vector (23). The sequence of the four-monomer block is 99% homologous to the sequence of the 5.7 kb alphoid DNA repeat unit in ΔYq74 determined in this study (see below). Because this palindrome is localized at the end of alphoid DNA array in TAR YAC isolates, i.e. physically linked to the Neo gene sequence (data not shown), the truncated centromere of the mini-chromosome ΔYq74 is structurally similar to the centromere of the human Y chromosome, where the major centromeric alphoid array is not palindromic (41).
Figure 4.
(A) CHEF electrophoresis of NotI-linearized BACs with inserts of 100, 120 and 140 kb alphoid DNA fragments from ΔYq74. Minor bands represent deleted BACs generated during propagation in E.coli. Lane 1 is a size marker. (B) SpeI digestion of the same BACs. SpeI fragments (2.8 and 2.9 kb) co-migrated during gel electrophoresis. These two fragments formed a 5.7 kb alphoid DNA unit with 34 copies of a ∼171 bp alphoid DNA monomer. An additional 1.8 kb fragment containing alphoid DNA was generated during chromosome truncation. This fragment contained two blocks of alphoid DNA (seven and four monomers) organized as a palindrome. Other upper fragments visible on the gel correspond to vector sequences and sequences corresponding to the junction between the vector and the insert. Lane 1 is a size marker.
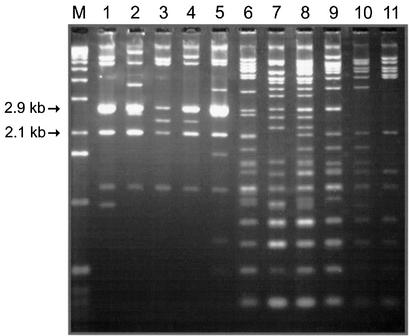
TAR clones carrying alphoid DNA from chromosome 22 were also characterized. YACs with an alphoid DNA array of up to 175 kb were efficiently transferred to E.coli after being retrofitted by a BRV1 BAC vector (Table 1). YACs with larger inserts underwent deletions. McDermid et al. (42) showed that alphoid DNA in the centromere of chromosome 22 is organized into 2.1 or 2.8 kb tandemly repeating units, which include 12 or 16 alphoid DNA monomers, respectively. These units are flanked by EcoRI sites. The structures of some but not all chromosome 22-specific BACs were consistent with the observations of McDermid et al. (42). BAC clones 11, 14, 19, 29 and 35 had two major EcoRI fragments of 2.1 and 2.8 kb (Fig. 5), indicating that these DNA inserts have higher order alphoid repeats. BAC clones 3, 5, 6, 10, 15 and 20 had more divergent alphoid DNA repeats as indicated by multiple EcoRI fragments with a periodicity of 171 bp (Fig. 5). Probes corresponding to BAC clone 11, which has HORs, and BAC clone 5, which does not, were differentially labeled for use in FISH analysis. These probes produced overlapping hybridization signals on the centromeric region of chromosome 22 (Fig. 2E). Fiber-FISH high-resolution mapping was performed to confirm that the location of the BAC sequences overlapped (Fig. 2F). One, or perhaps two, regions of hybridization were observed for the BAC 11 probe (Spectrum Orange); these regions fall within the region of BAC 5 hybridization (Spectrum Green), which shows homology with the extended region of the centromere. Thus, analysis of several clones from a human centromere 22 library has shown that they derived from at least two different blocks of alphoid array.
Figure 5.
BACs with different levels of divergence of alphoid DNA arrays from chromosome 22 were digested with EcoRI. Lanes 1–11 correspond to YAC nos 14, 19, 35, 29, 11, 10, 5, 6, 15, 20 and 3 in Table 1. Lane M is a size marker.
Sequence analysis of alphoid DNA
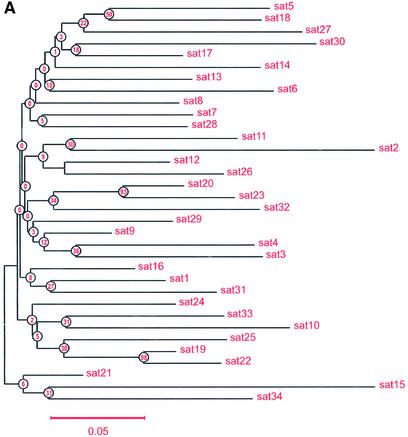
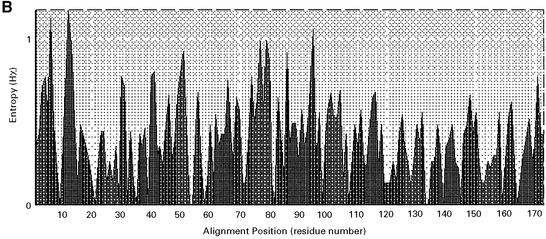
Only a partial DNA sequence was previously reported for the 5.7 kb alphoid DNA repeat unit on the human Y chromosome (40). In our study, the complete sequence of the 5.7 kb repeat was determined (accession no. AF522078). The sequences of the 171 bp monomers in this repeat unit were aligned and compared. Values of divergence between pairs of monomers were calculated for all 34 monomers. All were type A, possessing pJ-alpha binding sites and lacking CENP-B binding sites typical for type B monomers (Table 2), as reported previously by Cooper et al. (41) and Tyler-Smith et al. (5). These monomers are highly diverged with an average divergence from consensus of 0.116. This result suggests that homogenization events are infrequent for these sequences and that they are not subject to concerted evolution. The same mode of evolution (birth-and-death evolution) has been observed in tandemly organized polyubiquitin genes and some other multigene families (43). A neighbor-joining phylogenetic tree (Fig. 6A) shows that a few monomers may have been duplicated relatively recently (e.g. pairs sat19– sat22 and sat20–sat23). The high level of divergence of these repeats (12–30%) may explain why these YACs and BACs propagate stably in yeast and E.coli. An entropy plot was constructed for monomers from the 5.7 kb alphoid DNA unit with BioEdit (Fig. 6B). Lower Hx values correspond to lower variability. Interestingly, the region corresponding to the pJ-alpha box, located at the end of the sequence, does not have the lowest Hx value.
Table 2. Distribution of pJ-alpha and CENP-B boxes in different alphoid DNA arrays.
| Alphoid DNA construct | Chromosome origin | Size of alphoid DNA insert (kb) | HOR | Frequency of pJ-alpha boxes | Frequency of CENP-B boxes |
|---|---|---|---|---|---|
| YAC/BAC5 | 22 | 140 | Unregular | 17/54 | 11/54 |
| YAC/BAC11 | 22 | 140 | 2.1 kb, 2.8 kb | 59/105 | 0/105 |
| YAC/BAC3 | ΔYq74 | 140 | 5.7 kb | 372/400 | 0/400 |
A satellite unit is assumed to be box-positive if the number of mismatches between pJ-alpha/CENP-B box in a unit and pJ-alpha and CENP-B consensus sequences (YTTCGTTGGAARCGGGA and CTAYGGTGRAAAAGGAA) is ≤3.
Figure 6.
(A) Unrooted neighbor-joining tree analysis of alphoid DNA monomers from the 5.7 kb alphoid DNA unit; each taxon is an alpha satellite monomer. The scale is based on the number of nucleotide substitutions per site. Sequences were compared pairwise and divergence calculated by the Kimura two-parameter method (30). The numbers are bootstap values (1000 pseudoreplicates) used to estimate the confidence of internal branches (46). The alpha number represents the order in which a 171 bp unit comes out in tandem repeat spanning a 5.7 kb alphoid DNA unit. Five monomers were identical and appeared as one. (B) Entropy plot for monomers from the 5.7 kb alphoid DNA unit. A smaller Hx value corresponds to lower variability.
Chromosome 22-specific BAC clone11 also has primarily type A alphoid DNA monomers (Table 2). In contrast, alphoid DNA from BAC clone 5 has type A and B monomers (Table 2). Similar patterns of sequence organization have been observed in autosomal alpha satellite DNA (reviewed in 44).
A condition for TAR cloning is the presence of an ARS-like sequence in the fragment to be cloned (19). Indeed, alphoid DNA arrays obtained in TAR clones contain ARS-like sequences. Analysis of sequences of BAC clone 11 identified several potential ARS-like elements in positions 126–141 (Fig. 7). These putative ARS sequences had two mismatches with the consensus WWWTTTAYRTTTWDTT (45), but other potential ARS-like sequences with three or more differences with the consensus sequence were also identified. These sequences were sufficient to confer high transformation efficiency to a yeast CEN vector lacking an ARS element, indicating that alphoid DNA monomers can initiate DNA replication in yeast. However, putative ARS sequences were not present in each alphoid DNA monomer.
Figure 7.
ARS-like sequences in 171 bp satellite monomer units are underlined. The ARS consensus sequence is shown above the centromeric sequences.
Functional analysis of alphoid DNA arrays isolated by TAR
In previous studies, four fragments of alphoid DNA arrays from chromosomes 17, 21, X and Y were examined for their ability to form a functional centromere (7–15). These studies suggested that autosomal core A/B arrays from chromosomes 17 or 21 are more proficient at de novo centromere assembly than A-type arrays from the Y chromosome; in contrast, diverged A/B-type arrays are probably not competent for HAC formation and de novo centromere formation (9,14,15).
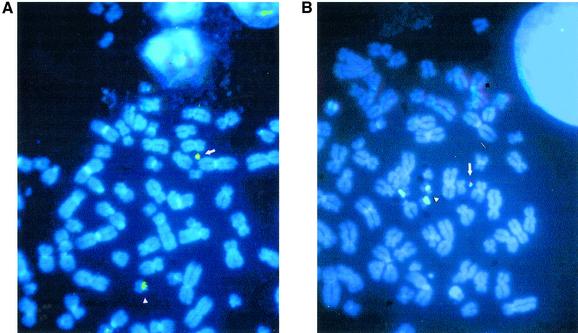
In the work reported here, experiments were carried out to characterize the ability of the chromosome 22 type A and type A/B alphoid DNA arrays to form a de novo centromere. HT1080 cells were transfected with BAC clone 11 (type A) and BAC clone 5 (type A/B). The transformants were screened for G418 resistance. Sixteen G418-resistant transfectants were analyzed; one contained a HAC in 20% of the nuclei and one (22_3) contained a HAC in 80% of the nuclei (Table 3). One or two HACs of the expected size were observed per nucleus. FISH analysis showed that these HACs contained alphoid DNA and vector DNA (Fig. 8). HAC size was estimated to be between one-tenth to one-twentieth of chromosome 22, indicating amplification of the input DNA. A more extensive analysis was carried out on clone 22_3. Cells carrying this HAC were grown in culture in the absence of selection for 30 days, during which time the HAC remained mitotically stable, suggesting the presence of a functional centromere. FISH analysis was carried out with four probes: a 22q-arm paint, rDNA, a pan-alphoid probe and an inter/intra Alu probe. No hybridization was observed, indicating that the HAC had not captured any large host chromosomal fragments (data not shown). This result indicates that some regions of centromeric alphoid A-type arrays from human autosomes are sufficient for de novo centromere formation. In agreement with a previous observation that diverged alphoid arrays are not competent for de novo centromere formation (9), no HACs were observed with BAC clone 5 containing type A/B monomers (data not shown).
Table 3. Efficiency of HAC formation by different alphoid DNA arrays.
| Alphoid DNA construct | Chromosome origin | Size of array (kb) | Type of array | Frequency of HAC-positive colonies | % of cells with HAC in the colony |
|---|---|---|---|---|---|
| YAC/BAC11 | 22 | 140 | Aa | 2/16 | 20; 80b |
| YAC/BAC3 | ΔYq74 | 140 | Aa | 0/28 | |
| Pac_7c5c | 21 | 70 | A+Ba | 8/10 | 80d |
aArrays with HOR structures.
bEach individual colony contains either 20 or 80% of cells with HAC.
cPac_7c5 construct was used as a positive control (11).
dThe eight positive colonies with ≥80% of cells with HAC. Pac_7c5 HACs were not validated.
Figure 8.
FISH of HACs from BAC 11 (centromeric DNA from chromosome 22 is characterized by higher order alphoid repeats). The green signal is the alphoid probe, which hybridizes to chromosome 22 (arrowhead) and a HAC (arrow). The red or yellow/red signal is the BRV1 vector probe, which hybridizes to a HAC. (A) and (B) correspond to different metaphase plates.
HAC formation was also investigated using the alphoid DNA array from mini-chromosome ΔYq74 (140 kb BAC clone 3). We did not observe HAC formation with this alphoid DNA array (Table 3). ΔYq74 contains the smallest alphoid DNA array that supports proper mini-chromosome segregation (5,23). These results suggest that the structural requirements for de novo centromere formation may be different for naked and chromosomal DNA.
DISCUSSION
The mammalian centromere plays an essential role in chromosome segregation during mitosis and meiosis, but it remains relatively poorly characterized on a molecular level. At least in part, characterization of centromeric regions is impeded by the difficulty in cloning large blocks of repetitive DNA. When centromeric DNA is digested by a restriction endonuclease, the resulting fragments are often too small or too large for efficient cloning in BAC or YAC libraries. The TAR method overcomes this problem because it does not require enzymatic treatment of genomic DNA before cloning. In this study, clones of human alphoid DNA up to ∼400 kb in length were isolated by TAR cloning (20) from total genomic DNA. The main advantage of the TAR method over standard BAC and YAC library construction is selectivity of cloning that excludes the time-consuming step of screening thousands of random clones for clones containing the centromeric regions. The work reported here demonstrates that centromere-specific sequences can be selectively cloned from rodent–human monochromosomal hybrid cell lines. This excludes uncertainty in the identification of chromosome-specific clones and provides an opportunity to build a single centromere contig.
In this work, centromeric segments of eight human chromosomes were recovered. FISH mapping and physical analysis showed that the majority of the TAR clones carried centromeric and pericentromeric regions. The DNA contained in most of the centromeric clones was exclusively alphoid, organized in some cases in HOR arrays. Approximately 15% of the isolates contained alphoid DNA arrays interrupted with non-alphoid DNA sequences, including Alu and LINE repeats. TAR clones carrying alphoid DNA are reasonably stable during propagation in yeast and E.coli, indicating that these clones can be used as a starting material to construct physical maps of centromeric regions. Because fragments isolated by TAR cloning must carry an ARS-like sequence (19), some regions of the centromere may not be isolated by the original TAR method. This problem can be resolved by using a recently modified TAR cloning system (V.Noskov, manuscript submitted for publication), in which an ARS sequence is incorporated into the TAR vector. Negative–positive selection is used in this system to reduce the background due to vector re-circularization. With this system the yield of clones containing alphoid DNA is >80%. Combination of two versions of TAR method could facilitate isolation of centromeric fragments to assemble a contig covering an entire human centromere.
It is clear that the process of kinetochore formation is central for efficient HAC formation. In previous studies, HACs were recovered after transfecting YACs or BACs into human cells when they carried large arrays of alphoid DNA (7–15). HACs formed efficiently with constructs containing alphoid DNA from chromosomes 17, 21 and X. All these alphoid DNA arrays have homogeneous monomers that form characteristic HORs. Alphoid DNA from chromosomes 17 and X and the 21-I domain are also rich in CENP-B-box motifs. In contrast, alphoid DNA from chromosome 21, which is not homogeneous and lacks CENP-B boxes, was inefficient in forming HACs. Homogeneous arrays from the Y chromosome that lack CENP-B boxes were also poor HAC-forming sequences (8,9,15). These studies suggested that homogeneous monomers and a high density of CENP boxes are required for de novo kinetochore formation. Our results suggest that CENP-B binding sites may not be strongly required for de novo kinetochore formation. For example, chromosome 22 alphoid DNA consisting of HOR monomers has no detectable CENP-B boxes but formed HACs when transfected into HT1080 cells. At the same time HACs were not observed with alphoid DNA containing type A/B monomers, possibly because of the high divergence of the alphoid array. A region of a truncated centromere of mini-chromosome ΔYq74 (23) was also investigated. ΔYq74 was generated by two rounds of telomere-directed breakage of the human Y chromosome (24). One break occurred in the centromeric alphoid DNA and deleted the long arm of the chromosome, producing an acrocentric chromosome with 140 kb of alphoid DNA and the short arm of the Y chromosome. The 140 kb alphoid DNA array of ΔYq74 could not be truncated further without loss of mini-chromosome stability, indicating that ΔYq74 carries the minimal essential components of a mini-chromosome centromere. Physical analysis of TAR alphoid DNA isolates showed that the truncated centromere of the mini-chromosome ΔYq74 is composed of type A HOR monomers. Alphoid DNA isolated from ΔYq74 was not effective in HAC formation. That may suggest that the requirements for recognizing a functional centromere on an endogenous chromosome and for forming a de novo centromere are distinct and are not yet understood.
In conclusion, a systematic comparative analysis of different types of alphoid DNA arrays is required to elucidate the role of alphoid DNA divergence in HAC formation. We believe that the further analysis of a representative collection of TAR clones from the centromere of chromosome 22 contained in this work may help to address this question.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Albert Ly for his contribution to some of these experiments. This research was partially supported by the Biological and Environmental Research Program (BER), US Department of Energy, Interagency Agreement No. DE-AI02-01ER63079.
DDBJ/EMBL/GenBank accession nos AF522078, AF533770
REFERENCES
- 1.Durfy S.J. and Willard,H.F. (1989) Patterns of intra- and interarray sequence variation in alpha satellite from the human X chromosome: evidence for short-range homogenization of tandemly repeated DNA sequences. Genomics, 4, 810–821. [DOI] [PubMed] [Google Scholar]
- 2.Romanova L.Y., Deriagin,G.V., Mashkova,T.D., Tumeneva,I.G., Mushegian,A.R., Kisselev,L.L. and Alexandrov,I.A. (1996) Evidence for selection in evolution of alpha satellite DNA: the central role of CENP-B/pJ alpha binding region. J. Mol. Biol., 26, 334–340. [DOI] [PubMed] [Google Scholar]
- 3.Choo K.H.A. (1997) The Centromere. Oxford University Press, Oxford.
- 4.Brown K.E., Barnett,M.A., Burgtorf,C., Shaw,P., Buckle,V.J. and Brown,W.R.A. (1994) Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum. Mol. Genet., 3, 1227–1237. [DOI] [PubMed] [Google Scholar]
- 5.Tyler-Smith C., Oakey,R.J., Larin,Z., Fisher,R.B., Crocker,M., Affara,N.A., Ferguson-Smith,M.A., Muenke,M., Zuffardi,O. and Jobling,M.A. (1993) Localization of DNA sequences required for human centromere function through an analysis of rearranged Y chromosomes. Nature Genet., 5, 368–375. [DOI] [PubMed] [Google Scholar]
- 6.Yang J.W., Pendon,C., Yang,J., Haywood,N., Chand,A. and Brown,W.R. (2000) Human mini-chromosomes with minimal centromeres. Hum. Mol. Genet., 9, 1891–1902. [DOI] [PubMed] [Google Scholar]
- 7.Harrington J.J., Van Bokkelen,G., Mays,R.W., Gustashaw,K. and Willard,H.F. (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nature Genet., 15, 345–355. [DOI] [PubMed] [Google Scholar]
- 8.Ikeno M., Grimes,B., Okazaki,T., Nakano,M., Saitoh,K., Hoshino,H., McGill,N.I., Cooke,H. and Masumoto,H. (1998) Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol., 16, 431–439. [DOI] [PubMed] [Google Scholar]
- 9.Masumoto H., Ikeno,M., Nakano,M., Okazaki,T., Grimes,B., Cooke,H. and Suzuki,N. (1998) Assay of centromere function using a human artificial chromosome. Chromosoma, 107, 406–416. [DOI] [PubMed] [Google Scholar]
- 10.Henning K.A., Novotny,E.A., Compton,S.T., Guan,X.Y., Liu,P.P. and Ashlock,M.A. (1999) Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc. Natl Acad. Sci. USA, 96, 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebersole T.E., Ross,A., Clark,E., McGill,N., Schindelhauer,D., Cooke,H. and Grimes,B. (2000) Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet., 9, 1623–1631. [DOI] [PubMed] [Google Scholar]
- 12.Schueler M.G., Higgins,A.W., Rudd,M.K., Gustashaw,K. and Willard,H.F. (2001) Genomic and genetic definition of a functional human centromere. Science, 294, 109–115. [DOI] [PubMed] [Google Scholar]
- 13.Mejia J.E., Willmott,A., Levy,E., Earnshaw,W.C. and Larin,Z. (2001) Functional complementation of a genetic deficiency with human artificial chromosomes. Am. J. Hum. Genet., 69, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mejia J.E., Alazami,A., Willmott,A., Marschall,P., Levy,E., Earnshaw,W.C. and Larin,Z. (2002) Efficiency of de novo centromere formation in human artificial chromosomes. Genomics, 79, 297–304. [DOI] [PubMed] [Google Scholar]
- 15.Grimes B.R., Rhoades,A.A. and Willard,H.F. (2002) Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol. Ther., 5, 798–805. [DOI] [PubMed] [Google Scholar]
- 16.Jackson M.S., Rocchi,M., Thompson,G., Hearn,T. Crosier,M., Guy,J., Kirk,D., Mulligan,L., Ricco,A., Piccininni,S., Marzella,R., Viggiano,L. and Archidiacono,N. (1999) Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet., 2, 205–215. [DOI] [PubMed] [Google Scholar]
- 17.Horvath J.E., Viggiano,L., Loftus,B.J., Adams,M.D., Archidiacono,N., Rocchi,M. and Eichler,E.E. (2000) Molecular structure and evolution of an alpha satellite/non-alpha satellite junction at 16p11. Hum. Mol. Genet., 9, 113–123. [DOI] [PubMed] [Google Scholar]
- 18.Osoegawa K., Mammoser,A.G., Wu,C., Frengen,E., Zeng,C., Catanese,J.J. and de Jong,P.J. (2001) A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res., 11, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larionov V., Kouprina,N., Graves,J. and Resnick,M.A. (1996) Highly selective isolation of human DNAs from rodent-human hybrid cells as circular YACs by TAR cloning. Proc. Natl Acad. Sci. USA, 93, 13925–13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouprina N. and Larionov,V. (1999) Selective isolation of mammalian genes by TAR cloning. In Dracopoli,N.C., Haines,J.L., Korf,B.R., Moir,D.T., Morton,C.C., Seidman,C.E., Seidman,J.G., Smith,D.R. and Boyle,A.L. (eds), Current Protocols in Human Genetics. John Wiley and Sons, New York, Vol. I, pp. 5.17.1–5.17.21. [DOI] [PubMed]
- 21.Vissel B. and Choo,K.H. (1987) Human alpha-satellite DNA-consensus sequence and conserved regions. Nucleic Acids Res., 15, 6751–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heller R., Brown,K.E., Burgtorf,C. and Brown,W.R. (1996) Mini-chromosomes derived from the human Y chromosome by telomere directed chromosome breakage. Proc. Natl Acad. Sci. USA, 93, 7125–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnett M.A., Buckle,V.J., Evans,E.P., Porter,A.C., Rout,D., Smith,A.G. and Brown,W.R. (1993) Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res., 21, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oakey R. and Tyler-Smith,C. (1990) Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics, 7, 325–330. [DOI] [PubMed] [Google Scholar]
- 26.Pinkel D., Straume,T. and Gray,J.W. (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl Acad. Sci. USA, 83, 2934–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pack S.D., Zbar,B., Pak,E., Ault,D.O., Humphrey,J.S., Pham,T., Hurley,K., Weil,R.J., Park,W.S., Kuzmin,I., Stolle,C., Glenn,G., Liotta,L.A., Lerman,M.I., Klausner,R.D., Linehan,W.M. and Zhuang,Z. (1999) Constitutional von Hippel-Lindau (VHL) gene deletions detected in VHL families by fluorescence in situ hybridization Cancer Res., 59, 5560–5564. [PubMed] [Google Scholar]
- 28.Haaf T. and Ward,D.C. (1994) High resolution ordering of YAC contigs using extended chromatin and chromosomes. Hum. Mol. Genet., 3, 629–633. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol., 16, 111–120. [DOI] [PubMed] [Google Scholar]
- 30.Kumar S.K., Tamura,K. and Nei,M. (1994) MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci., 10, 189–191. [DOI] [PubMed] [Google Scholar]
- 31.Saitou N. and Nei,M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 32.Goodsell D.S. and Dickerson,R.E. (1994) Bending and curvature calculations in B-DNA. Nucleic Acids Res., 22, 5497–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solovyev V.V., Rogozin,I.B., Salikhova,A.K., Seledtsov,I.A., Salamov,A.A. and Kel,A.E. (1992) ‘Context’ package of computer programs for analysis of DNA, RNA and protein sequences. In Ratner,V.A. and Kolchanov,N.A. (eds), Modelling and Computer Methods in Molecular Biology and Genetics. Nova Science Publishers, New York, pp. 125–130.
- 34.Toyn J.H., Gunyuzlu,P.L., White,W.H., Thompson,L.A. and Hollis,G.F. (2000) A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast, 16, 553–560. [DOI] [PubMed] [Google Scholar]
- 35.Kouprina N., Nikolasishvili,N., Graves,J., Koriabine,M., Resnick,M.A. and Larionov,V. (1999) Integrity of human YACs during propagation in recombination-deficient yeast strains. Genomics, 56, 262–273. [DOI] [PubMed] [Google Scholar]
- 36.Neil D.L., Villasante,A., Fisher,R.B., Vetrie,D., Cox,B. and Tyler-Smith,C. (1990) Structural instability of human tandemly repeated DNA sequences cloned in yeast artificial chromosome vectors. Nucleic Acids Res., 18, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stinchomb D.T., Thomas,M., Kelly,I., Selker,E. and Davis,R.W. (1980) Eukaryotic DNA segments capable of autonomous replication in yeast. Proc. Natl Acad. Sci. USA, 77, 4559–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Bravant A.J., Fangman,W.L. and Brewer,B.J. (1999) Active role of human genomic insert in replication of a yeast artificial chromosome. Mol. Cell. Biol., 19, 4231–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noskov V., Kouprina,N., Leem,S.H., Koriabine,M., Barrett,J.C. and Larionov,V. (2002) A genetic system for direct selection of gene-positive clones during recombinational cloning in yeast. Nucleic Acids Res., 30, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyler-Smith C. and Brown,W.R. (1987) Structure of the major block of alphoid satellite DNA on the human Y chromosome. J. Mol. Biol., 195, 457–470. [DOI] [PubMed] [Google Scholar]
- 41.Cooper K.F., Fisher,R.B. and Tyler-Smith,C. (1993) The major centromeric array of alphoid satellite DNA on the human Y chromosome is non-palindromic. Hum. Mol. Genet., 2, 1267–1270. [DOI] [PubMed] [Google Scholar]
- 42.McDermid H.E., Duncan,A.M., Higgins,M.J., Hamerton,J.L., Rector,E., Brasch,K.R. and White,B.N. (1986) Isolation and characterization of an alpha-satellite repeated sequence from human chromosome 22. Chromosoma, 94, 228–234. [DOI] [PubMed] [Google Scholar]
- 43.Nei M., Rogozin,I.B. and Piontkivska,H. (2000) Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc. Natl Acad. Sci. USA, 97, 10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexandrov I., Kazakov,A., Tumeneva,I., Shepelev,V. and Yurov,Y. (2001) Alpha-satellite DNA of primates: old and new families. Chromosoma, 110, 253–266. [DOI] [PubMed] [Google Scholar]
- 45.Theis J.F. and Newlon,C.S. (1997) The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl Acad. Sci. USA, 94, 10786–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39, 783–791. [DOI] [PubMed] [Google Scholar]