Abstract
Reverse transcription of HIV-1 RNA is primed by a tRNA3Lys molecule bound at the primer binding site (PBS). Complex intermolecular interactions were proposed between tRNA3Lys and the RNA of the HIV-1 Mal isolate. Recently, an alternative interaction was proposed between the TΨC stem of tRNA3Lys and a primer activation signal (PAS) of the Lai and Hxb2 RNAs, suggesting major structural variations in the reverse transcription complex of different HIV-1 strains. Here, we analyzed mutants of the Hxb2 RNA that prevent the interaction between the PAS and tRNA3Lys or/and a complementary sequence in the viral RNA. We compared the kinetics of reverse transcription of the wild type and mutant Hxb2 RNAs, using either tRNA3Lys or an 18mer oligoribonucleotide complementary to the PBS, which cannot interact with the PAS, as primers. We also used chemical probing to test the structure of the mutant and wild type RNAs, as well as the complex formed between the later RNA and tRNA3Lys. These experiments, together with the analysis of long term replication data of mutant viruses obtained by C. Morrow and coworkers (Birmingham, USA) that use alternate tRNAs as primers, strongly suggest that the interaction between the Hxb2 PAS and tRNA3Lys does not exist. Instead, the effects of the vRNA mutations on reverse transcription seem to be linked to incorrect folding of the mutant RNAs.
INTRODUCTION
Conversion of the single-stranded genomic RNA into a double-stranded DNA with duplicated long terminal repeats (LTR) by the viral reverse transcriptase (RT) is a key event in the retroviral replication cycle (1,2). This RNA- and DNA-dependent polymerase, which also harbors an RNase H domain, utilizes a cellular tRNA that is selectively encapsidated into the viral particles to prime reverse transcription (3–5). tRNA3Lys is the primer of most immunodeficiency viruses, including the type 1 human immunodeficiency virus (HIV-1), while tRNATrp and tRNAPro are used by most avian and murine retroviruses, respectively (3–5). In all cases, the 18 nt at the 3′ end of the primer anneal to a complementary region, the primer binding site (PBS), located in the 5′ region of the genomic RNA.
However, additional ‘virus-specific’ interactions have been described in several retroviruses and LTR containing retrotransposons including avian retroviruses (6–8), HIV-1 (9–17), HIV-2 (18,19), FIV (20), Ty1 (21,22) and Ty3 (23). These additional interactions are required for efficient replication (6,8,24–26), and in vivo initiation of reverse transcription (21,27,28).
In HIV-1, the existence of additional interactions between tRNA3Lys and the genomic RNA was initially demonstrated in vitro, using chemical and enzymatic probing (11,13,14) and site-directed mutagenesis (12,17,29). These data were used to construct secondary (11) and tertiary (14) structure models of the initiation complex of the HIV-1 Mal isolate (11). In these models, parts of the anticodon arm and of the variable loop of tRNA3Lys interact with viral sequences upstream of the PBS (Fig. 1A), in addition to the annealing of the 3′ part of tRNA3Lys to the PBS. The same interactions were observed when the primer–template complex was annealed by heat or by the nucleocapsid protein (30).
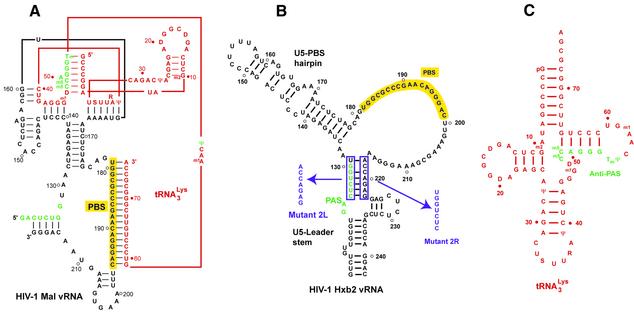
Figure 1.
Secondary structure models (A) of the binary complex formed between HIV-1 Mal vRNA and tRNA3Lys, (B) HIV-1 Hxb2 RNA and (C) tRNA3Lys. The vRNA molecules and the tRNA are drawn in black and red, respectively, except for the PAS and antiPAS sequences, which are green. The PBS is highlighted in yellow, and the sequences and the names of the substitutions introduced in the HIV-1 Hxb2 RNA are indicated in purple. Mutant 2LR is a double mutant containing both substitutions. The secondary structures of the complex formed between HIV-1 Mal vRNA and tRNA3Lys, and of HIV-1 Hxb2 RNA, were established in Isel et al. (11) and Beerens et al. (31), respectively.
Comparatively little structural data concerning the subgroup B isolates that prevail in Europe and North America are available. Functional analysis of HIV-1 mutants derived from the Hxb2 isolate that used tRNAHis or tRNAMet as primer suggested that the intermolecular interactions we proposed for the HIV-1 Mal RNA-tRNA3Lys complex also exist in the complex formed in vivo between HIV-1 Hxb2 RNA and the tRNA primer (24–26,28). However, recent probing data obtained on the free HIV-1 Lai RNA (31), which is identical to HIV-1 Hxb2 in the PBS domain, suggested significant structural differences with the free HIV-1 Mal RNA (11,13). In addition, Berkhout and co-workers proposed that, in the primer–template complex, the 5′ part of the TΨC arm of tRNA3Lys interacts with a complementary sequence located upstream of the PBS of the HIV-1 Lai RNA, which was named the primer activation signal (PAS) (Fig. 1B and C) (31–33). This proposal was initially based on the effects of mutations in the viral RNA (vRNA) on reverse transcription. Namely, mutations of two RNA stretches that have been proposed to form an intramolecular helix in the free vRNA (Fig. 1B) had different effects on reverse transcription, and introduction of compensatory designed-mutations did not restore reverse transcription (31,33). As the 5′ strand of this helix is complementary to the 5′ part of the TΨC arm of tRNA3Lys (Fig. 1C), the two RNA sequences were proposed to interact during initiation of reverse transcription (31,33).
Since this interaction is not supported by the probing experiments carried out on the HIV-1 Mal isolate (11), the results obtained by Berkhout and co-workers using the HIV-1 Lai isolate might reflect differences between these two strains (31,33). Alternatively, the interaction proposed by Berkhout and coworkers (31,33) might only exist transiently, and hence cannot be detected by chemical probing. Finally, one cannot exclude that the effects of the vRNA mutations on reverse transcription have a different, presently unsuspected, structural origin. Here, we report detailed kinetic and structural analysis of vRNA mutants designed to address these possibilities.
MATERIALS AND METHODS
Primer, templates and RT
tRNA3Lys was purified from beef liver as described (34). Its sequence and post-transcriptional modifications are identical to those of human tRNA3Lys. After dephosphorylation with calf intestine phosphatase, it was labeled at its 5′ end with [γ-32P]ATP and phage T4 polynucleotide kinase, according to published procedures (14). An 18mer oligoribonucleotide (ORN) complementary to the PBS was obtained from Dharmacon Research (Lafayette, CO), deprotected according to the supplier’s protocol, and 5′ end-labeled using [γ-32P]ATP and phage T4 polynucleotide kinase.
Wild type vRNA, encompassing nucleotides 1–295 of Hxb2, was obtained by in vitro transcription of plasmid pHXB2(1–732) linearized with RsaI, using previously described conditions (35). Plasmid pHXB2(1–732) was obtained by cloning nucleotides 1–732 of the HIV-1 Hxb2 strain immediately downstream of the T7 RNA polymerase promoter. In order to obtain the templates for transcription of the 2L, 2R and 2LR mutant vRNAs, this plasmid was mutated with the QuickChange™ site-directed mutagenesis kit (Stratagene) using the protocols provided by the supplier and oligonucleotides 5′-CCG TCT GTT GTG TGA GAG ACC AAA CTA GAG ATC CCT C-3′ and 5′-GAG GGA TCT CTA GTT TGG TCT CTC ACA CAA CAG ACG G-3′ (mutants 2L and 2LR), and 5′-GAA AGC GAA AGG GAA TGG TCT CGA GCT CTC TCG ACG C-3′ and 5′-GCG TCG AGA GAG CTC GAG ACC ATT CCC TTT CGC TTT C-3′ (mutants 2R and 2LR).
The plasmid used for production of the heterodimeric HIV-1 RT was kindly provided to us by Dr Torsten Unge (Uppsala, Sweden), together with the protocols for protein overexpression and purification.
Annealing of the natural tRNA3Lys or the 18mer ORN to the vRNA
vRNA and purified tRNA3Lys or synthetic ORN were first denatured in water for 2 min at 90°C and chilled on ice. Annealing was performed at 70°C for 20 min in sodium cacodylate (pH 7.5) 50 mM, KCl 300 mM. An aliquot of each annealing reaction was analyzed on a native 8% polyacrylamide gel to determine the annealing efficiency. The reaction products were discarded if <95% of the tRNA3Lys or the ORN were annealed to the wild type or mutant templates.
(–) Strand strong stop DNA synthesis
In a standard experiment, vRNA (30 nM final concentration) was annealed with 32P-labeled tRNA3Lys or with 32P-labeled ORN (10 nM) as described above, and pre-incubated for 4 min with 25 nM RT in 50 mM Tris–HCl pH 8.0, 50 mM KCl, 6 mM MgCl2, 1 mM DTE. Reverse transcription was initiated by adding a mixture of the four deoxynucleotide triphosphates (50 µM each) in the same buffer. Formamide containing 50 mM EDTA was added to aliquots of the reaction mixture at times ranging from 15 s to 60 min, and the reaction products were analyzed on 15% (ORN priming) or 8% (tRNA3Lys priming) denaturing polyacrylamide gels, and quantified with a BioImager BAS 2000 (Fuji) using the MacBas software.
Chemical probing of vRNA
After hybridization, the vRNA/tRNA3Lys complexes were incubated at 20°C for 15 min in the annealing buffer supplemented with 5 mM MgCl2 before probing with dimethyl sulfate (DMS). Hybridization and probing buffers were the same for 1-cyclohexyl 3-(2 morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT), except that they contained 50 mM sodium borate (pH 8.0) instead of sodium cacodylate.
After addition of 1 µg of yeast total tRNA, RNA was modified by addition of 1 µl of 10-fold diluted DMS in ethanol for 5 or 10 min, or 2 µl of CMCT (40 mg/ml in water) for 10 or 20 min. RNA modification was stopped with 200 µl ethanol and 50 µl sodium acetate 0.3 M (pH 5.3) containing 1 µg glycogen. Modified bases were detected by primer extension with RT as previously described (36).
Chemical probing of tRNA
vRNA (12 pmol) and 50 000 c.p.m. of 5′ end-labeled tRNA3Lys (0.4 pmol) were hybridized as described above. Positions N-7 of adenines were modified with 2 µl of diethylpyrocarbonate (DEPC) for 20 and 40 min at 37°C in 50 mM sodium cacodylate pH 7.5, 300 mM KCl and 5 mM MgCl2. Reactions were stopped with 300 µl ethanol and 100 µl sodium acetate 0.3 M (pH 5.3) containing 1 µg glycogen. Aniline treatment of DEPC-modified RNA was performed as previously described (37). After ethanol precipitation, tRNA3Lys fragments were analyzed on 15% denaturing polyacrylamide gels.
RESULTS
Experimental strategy
The PAS–antiPAS interaction proposed by Berkhout and co-workers involves the complementary sequences (5′)48-m5Cm5CAGGGTmΨ-55(3′) in tRNA3Lys, named antiPAS, and (3′)130-GGUCUCAG-123(5′) in HIV Lai RNA, named PAS (31,33) (Fig. 1B and C). Notably, the PAS sequence is conserved in HIV Mal [(3′)128-GGUCUCAG-121(5′); Fig. 1A], and hence the same interaction can be proposed for this isolate. However, chemical and enzymatic probing of the HIV-1 Mal RNA-tRNA3Lys complex indicated that the PAS–antiPAS interaction is not present in the reverse transcription initiation complex formed by this isolate (11). Thus, taken together Berkhout’s and our data might indicate that different primer–template interactions take place in different HIV isolates.
In order to confirm the existence of the PAS–antiPAS interaction and to gain insight into its role in the reverse transcription process, we introduced some of the mutants studied by Berkhout and co-workers in the HIV-1 Hxb2 isolate, which is identical to the Lai isolate in the PBS domain (Fig. 1B). Mutant 2L disrupts the proposed PAS–antiPAS interaction, as well as an intramolecular helix that has been proposed to be formed in the free vRNA (Fig. 1B) (31,33). Mutant 2R disrupts the intramolecular interaction of the PAS in the free form of the vRNA but does not prevent its base-pairing with the antiPAS in the binary primer/template complex. On the contrary, mutant 2LR, which combines the two previous mutations, was designed to restore the intramolecular base-pairing, while preventing the PAS–antiPAS interaction. These mutants were used to perform kinetics of (–) strand strong stop DNA synthesis and structural probing on the vRNAs and vRNA/tRNA complexes (31,33).
Kinetics of (–) strand strong stop DNA synthesis from tRNA3Lys
The product resulting from the extension of a primer bound to the PBS up to the 5′ end of the genomic RNA is named the (–) strand strong stop DNA. Beerens et al. used the amount of (–) strand strong stop DNA synthesized after 30 min, or the efficiency of single nucleotide incorporation, to evaluate the effect of the 2L, 2R and 2LR mutations on reverse transcription (31,33). In order to obtain more detailed information, we monitored the time course of (–) strand strong stop DNA synthesis over a 60 min time course (Fig. 2). In addition, we used 5′ end-labeled tRNA, instead of incorporation of radioactive nucleotides, in order to follow incorporation of the first nucleotides with increased sensitivity.
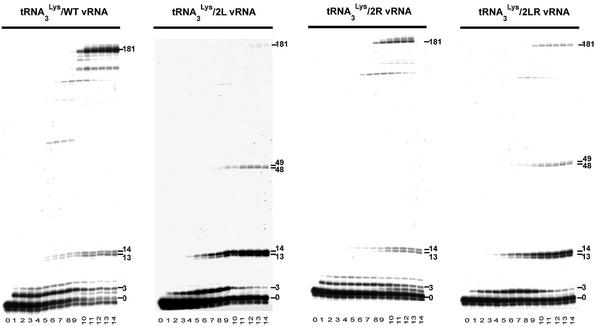
Figure 2.
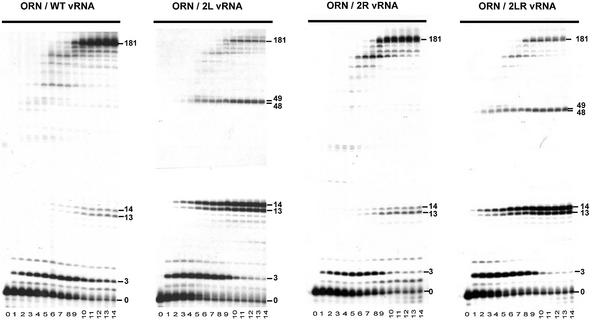
Kinetics of (–) strand strong stop DNA synthesis from the tRNA3Lys primer. Radiolabeled tRNA3Lys was hybridized either to wild type, 2L, 2R or 2LR vRNA and (–) strand strong stop DNA synthesis was initiated by addition of a mixture of the four deoxyribonucleotide triphosphates and RT. The reaction was stopped after 0, 15, 30 and 45 s, 1, 2, 3, 4, 5, 10, 20, 30, 40, 50 and 60 min (lanes 0 to 14, respectively), and the reaction products were analyzed on an 8% denaturing polyacrylamide gel. The products corresponding to extension of the primer by 3, 13, 14, 48, 49 and 181 nt are indicated on the right hand side of the gels. The larger product is the (–) strand strong stop DNA.
The kinetics of DNA synthesis indicated that the amount of (–) strand strong stop DNA was almost constant between 20 and 60 min for all vRNA templates (Fig. 2). Quantification revealed a 2-fold (mutant 2R), 4-fold (mutant 2LR) and 5-fold (mutant 2L) decrease of (–) strand strong stop DNA synthesis, as compared with the wild type template (Table 1). These results present both strong similitude and a divergence with those previously published (31). In agreement with Beerens et al. (31), we found that synthesis of (–) strand strong stop DNA was strongly reduced in mutants 2L and 2LR. However, while we noticed a 2-fold reduction of the (–) strand strong stop DNA synthesis with the 2R vRNA, Beerens et al. observed a 2.5-fold increase, as compared with the wild type vRNA (31). This difference most likely originates from variations of the experimental protocols, which include the presence of competitor tRNA and lower dNTP concentrations in the study by Berkhout and co-workers (31). Nevertheless, the main results, i.e. that mutant 2L was more severely affected than mutant 2R and that (–) strand strong stop DNA synthesis was not restored in mutant 2LR, were found in both studies.
Table 1. Synthesis of (–) strand strong stop DNA on the wild type and mutant templates using either the natural tRNA3Lys or a synthetic 18mer ORN as primers.
| Template | Primer |
||||
|---|---|---|---|---|---|
| tRNA3Lys |
18mer ORN |
||||
| PAS–antiPAS interaction | Relative (–) strand strong stop DNA synthesis | Rate of primer extension (s–1) | PAS–antiPAS interaction | Relative (–) strand strong stop DNA synthesis | |
| Wild type | Possible | 100% | 0.016 ± 0.003 | Not possible | 100% |
| Mutant 2L | Not possible | 20% | 0.016 ± 0.001 | Not possible | 20% |
| Mutant 2R | Possible | 50% | 0.0038 ± 0.0005 | Not possible | 53% |
| Mutant 2LR | Not possible | 26% | 0.010 ± 0.001 | Not possible | 25% |
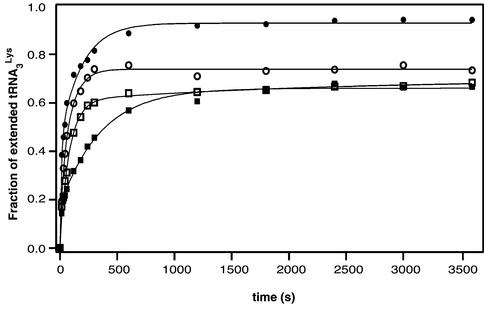
Since we used 5′ end-labeled tRNA3Lys, we were able to measure the rate of primer extension by quantifying the band corresponding to the unextended tRNA (Fig. 3). After 60 min, ∼90% of the tRNA3Lys annealed to the wild type vRNA was extended, as compared with ∼70% for mutant 2L, and ∼60% for mutants 2R and 2LR. We checked that incomplete primer extension was not due to incomplete primer annealing (see Materials and Methods). It most likely reflected structural heterogeneity of the initiation complexes. More important, the initial primer extension rate was similar with wild type and 2L RNAs, while it was slightly reduced with 2LR RNA, and severely affected with 2R RNA (Table 1). Notably, there was no correlation between the initial primer extension rate and the efficiency of (–) strand strong stop DNA synthesis: mutant 2R was the most affected as far as tRNA3Lys extension was concerned, yet it was the least affected mutant when considering (–) strand strong stop DNA synthesis (Table 1).
Figure 3.
Kinetics of tRNA3Lys extension. The gels shown in Figure 2 were quantified, and the radioactivity of all extension products divided by the total radioactivity was plotted versus time for the wild type vRNA (closed circles) and mutants 2L (open circles), 2R (closed squares) and 2LR (open squares). The extension rates determined by fitting the experimental data with a single exponential equation are listed in Table 1.
Furthermore, we noticed that decreased (–) strand strong stop DNA synthesis in mutants 2L and 2LR correlated with pausing of RT after addition of 48 and 49 nt, which was not observed with wild type and 2R templates, and enhanced pausing after addition of 13 and 14 nt (Fig. 2A and Table 1). The latter pause occurred in the A-stretch located in the internal loop of the U5-top hairpin upstream of the PBS (Fig. 1B). The former pause took place at the junction between the U5-top hairpin and the upper part of the U5-Leader stem, i.e. just before RT reached the PAS (Fig. 1B). Importantly, this pause was only observed in mutants 2L and 2LR, in which the PAS–antiPAS cannot take place. Furthermore, only one of these mutants (2LR) can form the intramolecular helix between nucleotides 125–131 and 217–223 (Fig. 1B). Thus, RT pausing after addition of 48 and 49 nt can be linked neither to the PAS–antiPAS interaction nor to the existence of the intramolecular helix constituting the upper part of the U5-Leader stem.
This observation raised the possibility that mutation of the PAS resulted in an aberrant folding of the vRNA, irrespective of the simultaneous mutation of nucleotides 217–223. In order to test this possibility we performed chemical probing of the wild type and mutant vRNAs.
Chemical probing of the wild type and mutant vRNAs
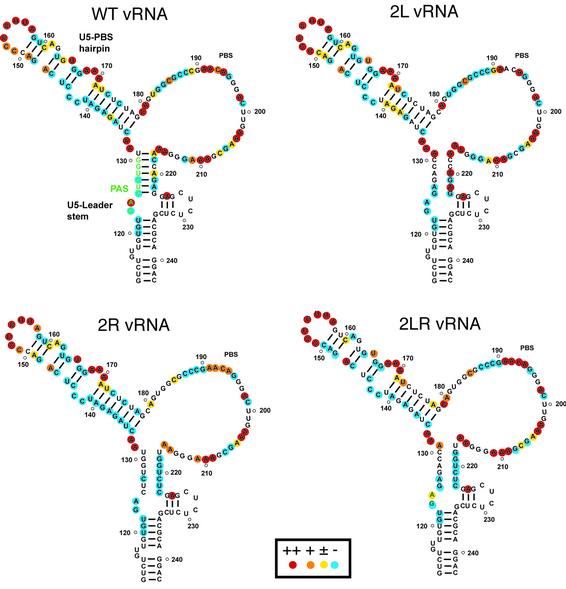
We tested the conformation of nucleotides 120–230 of the free wild type, 2L, 2R and 2LR vRNAs with DMS, which reacts with the N1 position of adenines and the N3 position of cytosines, and with CMCT, which modifies the N1 position of guanines and the N3 position of uridines, when those positions are not engaged in hydrogen bonds (37). The modified nucleotides were detected by a primer extension assay (37). Examples of probing gels are available as Supplementary Material, and the results are summarized in Figure 4.
Figure 4.
Structural probing of the wild type and mutant vRNAs. Summary of the DMS and CMCT probing data. The color code used to indicate highly, moderately, marginally and unreactive nucleotides is indicated in the insert. The absence of symbol indicates that the reactivity of the corresponding nucleotide could not be determined, because of the presence of a band in the control lane.
Overall, our data support the conformation of the U5-PBS hairpin proposed by Berkhout and co-workers on the basis of DMS, DEPC and enzymatic probing data (31) (Fig. 4). However, C142 and C143 were unreactive (Fig. 4), suggesting that these nucleotides might either be stacked on the adjacent helices or be involved in a tertiary interaction. In addition, probing data suggest that nucleotides 187–197 fold into a short stem–loop with nucleotides 190-GAACA-194 in the loop, as proposed for the HIV-1 Mal isolate (11). Our data also suggest that nucleotides 125–131 do base-pair with nucleotides 217–223 as proposed in the secondary structure model (Fig. 4). However, the structure of the lower part of the U5-Leader stem remains uncertain, since the strong reactivity of A225 that we systematically observed is not compatible with the proposed structure (Fig. 4).
In mutant 2L, A217, A220 and A222 became strongly reactive towards DMS (Fig. 4), supporting the fact that, in the wild type vRNA, these adenines are base-paired with the mutated sequence. However, neither G221 and G223 nor G125 and A126 were reactive in mutant 2L. In addition, A124 was strongly reactive in wild type vRNA, but unreactive in 2L vRNA. Our data suggest nucleotides 125–131 undergo a structural rearrangement in 2L vRNA. This conclusion holds true for mutant 2R. In this mutant, none of the substituted nucleotides became reactive towards DMS or CMCT, and C127 was also unreactive. As for mutant 2L, A124 was not modified by DMS in 2R vRNA (Fig. 4).
Importantly, our probing data suggest that the wild type secondary structure was not restored in mutant 2LR. The lack of chemical modification of nucleotides 217–223 in 2LR vRNA cannot be taken as evidence for an interaction with nucleotides 125–131, since those nucleotides were also unreactive in 2R vRNA (Fig. 4). On the contrary, A131 was modified by DMS, arguing against this interaction. Finally, A124, which was strongly reactive in wild type vRNA and unreactive in 2L and 2R vRNAs, was only marginally modified in 2LR vRNA. Therefore, a difference in the structure rather than in the sequence might explain the reduced (–) strand strong stop DNA synthesis on the 2LR template, as compared with the wild type RNA.
Interestingly, we observed a decreased reactivity of U165 and an increased reactivity of U172 in mutants 2L and 2LR, as compared with wild type and 2R vRNAs, suggesting a subtle structural distortion of this region in the mutants showing decreased (–) strand strong stop DNA synthesis (Fig. 4). This observation is likely to be significant since U165 and U172 flank the pausing site in the A stretch that is exacerbated in mutants 2L and 2LR.
Chemical probing of the wild type Hxb2 vRNA–tRNA3Lys complex
In order to directly test the existence of the PAS–antiPAS interaction, we compared the DMS modification profiles of the wild type vRNA in its free form and in complex with tRNA3Lys. If the PAS interacts with the TΨC loop of tRNA3Lys, A217, A220 and A222 should become reactive in the primer–template complex, as observed for the free form of 2L vRNA.
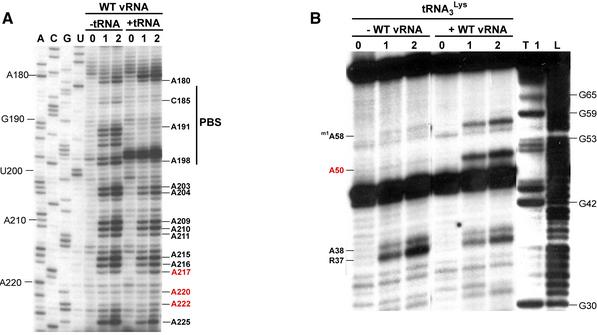
As expected, we observed a complete protection of the PBS nucleotides C185, A191, A192, C193, A194 and A198 in the Hxb2 vRNA–tRNA3Lys complex (Fig. 5A). However, the reactivity of adenines 217, 220, 222 did not increase upon annealing of tRNA3Lys, strongly arguing against the existence of the proposed PAS–antiPAS interaction (Fig. 5A).
Figure 5.
Structural probing of the HIV-1 Hxb2 RNA-tRNA3Lys complex. (A) Comparison of the DMS modification profiles of nucleotides 180–225 of wild type Hxb2 vRNA, in its free form and engaged in the primer–template complex. Lanes marked 0, 1 and 2 correspond to a mock reaction without DMS and modification for 5 and 10 min, respectively. Lanes ACGU correspond to sequencing reactions. (B) Comparison of the DEPC modification profile of tRNA3Lys, either free or involved in the primer–template complex. Lanes marked 0, 1 and 2 correspond to a mock reaction without DEPC and modification for 20 and 40 min, respectively. Lanes T1 and L1 correspond to a sequencing reaction with RNase T1, which is specific for G residues, and a ladder generated by mild alkaline hydrolyis, respectively. Nucleotides whose reactivity in the primer-template complex are inconsistent with the existence of the PAS–antiPAS interaction are indicated by red labels.
Next, we analyzed the reactivity of N7 position of the tRNA3Lys adenines using DEPC. A large collection of examples showed that DEPC only modifies unstacked adenines, whereas those located inside or at the termini of helices are not modified by DEPC (37). Hence, DEPC can be used to identify nucleotides that do not form Watson–Crick base pairs.
In the free tRNA3Lys, A38, located in the anticodon loop, was modified by DEPC, and its reactivity decreased when tRNA3Lys was annealed to the vRNA (Fig. 5B). Conversely, A50 and m1A58 were modified in the vRNA–tRNA3Lys complex, as we previously observed in the Mal vRNA-tRNA3Lys complex (11), but not in the free form of the primer (Fig. 5B). In the free tRNA3Lys, A50 is base-paired in the TΨC arm whereas, due to the tertiary interactions between the D and TΨC loops, m1A58 is stacked between C61 and G18 and forms a Hoogsteen base-pair with Tm54 that directly protects its N7 position (34). The reactivity of A50 in the vRNA–tRNA3Lys complex strongly suggests that the PAS–antiPAS interaction does not exist since, if it did, A50 would be base-paired with U128 in the vRNA (Fig. 1), and hence it should not be modified by DEPC.
Taken together, our probing data on the vRNA and tRNA3Lys engaged in the primer–template complex strongly argue against the existence of the PAS–antiPAS interaction before initiation of DNA synthesis takes place. However, these data do not exclude the possibility that such an interaction could take place during DNA synthesis.
Kinetics of (–) strand strong stop DNA synthesis from an 18mer ORN annealed to the PBS
To test the possibility of a transient PAS–antiPAS interaction taking place during the reverse transcription process, we studied the kinetics of (–) strand strong stop DNA synthesis, using an 18mer ORN annealed to the PBS as primer. This primer does not allow any PAS–antiPAS interaction to take place, the latter sequence being absent from the ORN.
If the differences we observed between the wild type and mutant vRNAs when using the natural tRNA3Lys as primer were due to a transient PAS–antiPAS interaction, the wild type and mutant vRNAs should all be reverse transcribed with the same efficiency when using the ORN as primer. Conversely, if the mutations we introduced in the vRNA primarily exert their effects by modifying the structure of the reverse transcription complex independently of any PAS–antiPAS interaction, the relative amount of (–) strand strong stop DNA obtained with the wild type and mutant templates should be the same with both primers.
The kinetics of (–) strand strong stop DNA synthesis from the ORN are shown in Figure 6 and the relative amount of final products are listed in Table 1. As previously observed with tRNA3Lys, mutations 2L and 2LR strongly decreased the amount of (–) strand strong stop DNA synthesized from the ORN primer, while mutation 2R had an intermediate phenotype (Fig. 6). Hence, the relative efficiency of (–) strand strong stop DNA synthesis on the wild type and mutant vRNAs was independent of the primer used to initiate reverse transcription (Table 1). Furthermore, the pausing pattern generated by extension of ORN was essentially the same as the one obtained with tRNA3Lys (compare Figs 2 and 6). Noticeably, enhanced pausing at positions +13 and +14, and pausing at positions +48 and +49 that was not apparent with wild type and 2R vRNAs, were observed during reverse transcription of 2L and 2LR RNAs (Fig. 6). Thus, comparison of the reverse transcription kinetics obtained with tRNA3Lys and ORN indicate that if a PAS–antiPAS interaction took place during this process, it was not functionally significant.
Figure 6.
Kinetics of (–) strand strong stop DNA synthesis from the ORN primer. Radiolabeled ORN was hybridized either to wild type, 2L, 2R or 2LR vRNA and (–) strand strong stop DNA synthesis was initiated by addition of a mixture of the four deoxyribonucleotide triphosphates and RT. The reaction was stopped after 0, 15, 30 and 45 s, 1, 2, 3, 4, 5, 10, 20, 30, 40, 50 and 60 min (lanes 0 to 14, respectively), and the reaction products were analyzed on 15% denaturing polyacrylamide gel. The products corresponding to extension of the primer by 3, 13, 14, 48, 49 and 181 nt are indicated.
Analysis of the PAS–antiPAS interaction in mutant viruses using tRNAHis or tRNAMet as primer
Morrow and co-workers obtained mutant viruses stably using either tRNAHis (24,26,28,38) or tRNAMet (25) during long term cell culture, by simultaneously mutating the PBS and the A-rich loop that is complementary to the anticodon loop of tRNA3Lys in the wild type Hxb2 strain. Both tRNAHis and tRNAMet contain sequence variations, as compared with tRNA3Lys, that destabilize the PAS–antiPAS interaction by 4.5 kcal/mol (Fig. 7).
Figure 7.
Potential interactions between the Hxb2 PAS and the antiPAS sequence of tRNA3Lys, tRNAMet and tRNAHis. The stability of the interactions at 37°C was calculated using the 2 state hybridization server of the mfold algothrithm on Michael Zuker’s web site (http://www.bioinfo.rpi.edu/applications/mfold/).
We looked at whether the mutant viruses selected mutations in the PAS that would restore the stability of the PAS–antiPAS interaction upon long term culture. Sequencing data of the PAS upon long term cell culture of the tRNAMet adapted clones can be found in the initial publication by Morrow and co-workers (25). Analysis of the published data did not reveal any mutation in the PAS sequence of the adapted virus. The PAS sequences of mutants using tRNAHis as primer were not included in the corresponding publications (24,26,28,38). However, sequencing of plasmids, kindly provided by C. Morrow (Birmingham, USA), containing the PAS sequence of those viruses before and after long term cell culture revealed no change in the PAS. Thus, even though the mutant viruses selected several mutations in the PBS domain that enhanced viral replication upon long term cell culture (24–26,28,38), none was located in the PAS.
DISCUSSION
This study was designed to obtain further evidence for the existence, in the Hxb2 isolate, of the PAS–antiPAS interaction initially proposed by Berkhout and co-workers (31–33), and hence to demonstrate potential structural differences in the reverse transcription initiation complex of different HIV isolates. When we used tRNA3Lys as primer and followed the kinetics of (–) strand strong stop DNA synthesis, we confirmed two main findings made by Beerens et al. (31). First, mutant 2L was more severely affected than mutant 2R, and second, (–) strand strong stop DNA synthesis was not restored in mutant 2LR. However, other experimental data do not support the conclusion that these effects are explained by an interaction between a PAS and tRNA3Lys.
We noticed that the kinetics of (–) strand strong stop DNA synthesis did not correlate with the kinetics of primer extension, but were strongly affected by RT pausing during DNA elongation. Since the pausing sites could be linked neither to the existence of the PAS–antiPAS interaction nor to the presence of the upper helix of the U5-Leader stem, these results suggested that the mutations induced aberrant folding of the vRNA.
Probing of the wild type Hxb2 RNA with DMS and CMCT supports several important features of the secondary structure model proposed by Berkhout and co-workers (31), including the U5-PBS structure and the upper part of the U5-Leader stem. However, our probing data on the 2LR vRNA showed that its structure was different from that of the wild type RNA, even though it was designed to restore the native structure. Therefore, a structural alteration could explain the low efficiency of the (–) strand strong stop DNA synthesis observed with this mutant, without the need to invoke any PAS–antiPAS interaction. Probing of the 2L and 2R vRNAs indicated that the substituted sequences did not remain fully single-stranded and implied that structural rearrangements also took place in these mutants. Interestingly, we observed a subtle distortion of the U5-PBS hairpin that correlated with increased pausing after addition of 13 and 14 nt and reduced (–) strand strong stop DNA synthesis.
Comparison of the chemical probing profiles of the wild type vRNA and tRNA3Lys in their free form and in the Hxb2 vRNA-tRNA3Lys complex brought direct evidence against the PAS–antiPAS interaction. The viral sequence complementary to the PAS did not become reactive upon formation of this complex, while in the tRNA3Lys, A50 that was expected to base pair in the PAS–antiPAS interaction became single stranded.
Finally, we observed the same effects of mutations 2L, 2R and 2LR on reverse transcription when using either tRNA3Lys or an ORN as primers, even though the latter lacks the antiPAS sequence. These results strongly argue against any functional role for the proposed PAS–antiPAS interaction. At the opposite, Berkhout and co-workers observed no effect of the 2L, 2R and 2LR mutations when using an 18mer DNA primer complementary to the PBS (31). However, extension of DNA primers by RT dramatically differs from extension of RNA primers (9,10,29,39,40). Whereas DNA extension is fast and highly processive, extension of RNA primers is slow and distributive (39,40). Therefore, defects due to mutations in the vRNA that are apparent when looking at the inefficient extension of RNA primers could be masked during the efficient extension of DNA primers, and comparison between RNA and DNA primers might not allow to draw conclusions about putative extended primer–template interactions.
Analysis of the sequence of mutant viruses using either tRNAMet or tRNAHis as primer did not reveal any variation in the PAS upon long term cell culture that would restore the stability of the wild type PAS–antiPAS interaction. This lack of adaptation of the PAS to primer sequence indicates that if the PAS–antiPAS interaction does exist in the wild type Hxb2 strain, it has minor influence on its replication capacity. The effects of mutations that render the PBS and PAS sequences complementary to tRNA1,2Lys have been studied on reverse transcription in vitro (32). It was found that simultaneous adaptation of the PAS and PBS slightly increased reverse transcription. However, reverse transcription of this tRNA1,2Lys adapted RNA was far less efficient than that of the wild type RNA, indicating that the putative PAS–antiPAS interaction is not a major determinant of the efficiency of the HIV-1 initiation of reverse transcription. Our observations are also in line with those of Beerens and Berkhout (32), who found that mutations in the tRNA3Lys antiPAS sequence cannot compensate the negative effects of mutations in the PAS.
One argument put forward in support of an interaction between HIV-1 RNA and the TΨC loop of tRNA3Lys is that similar interactions have been proposed for HIV-2 (19), avian retroviruses (6–8), and Ty1 (21,22), and can be drawn for some members of all retrovirus genera (32). However, RTs of HIV-2, SIV, FIV, EIAV, AMV and MLV do not efficiently initiate tRNA3Lys primed reverse transcription of HIV-1 RNA (10,29). These results suggest that the efficiency of the initiation of HIV-1 reverse transcription is mainly governed by virus-specific rather than general structural features. In addition, even though an interaction could potentially exist between the TΨC loop of tRNAPro and the MLV genomic RNA (32), extensive in vitro structural probing does not support it (41).
As far as the PAS–antiPAS interaction is concerned, this study revealed no difference between the reverse transcription initiation complexes formed the by Mal and Hxb2 vRNAs. However, we cannot exclude that other differences might exist in the initiation complexes formed by these two strains. Work is in progress to clarify this issue.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to G. Bec and G. Keith for the purification of tRNA3Lys and to P. Walter for the gift of HIV-1 RT. We thank Catherine Isel, Jean-Christophe Paillart and Pascale Romby for critical reading of the manuscript. This work was supported by the ‘Agence Nationale de Recherches sur le SIDA’ (ANRS).
REFERENCES
- 1.Baltimore D. (1970) Viral RNA-dependent DNA polymerase. Nature, 226, 1209–1211. [DOI] [PubMed] [Google Scholar]
- 2.Temin H.M. and Mizutani,S. (1970) RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature, 226, 1211–1213. [DOI] [PubMed] [Google Scholar]
- 3.Arts E.J. and Le Grice,S.F.J. (1998) Interaction of retroviral reverse transcriptase with template–primer duplexes during replication. Prog. Nucleic Acid Res. Mol. Biol., 58, 339–393. [DOI] [PubMed] [Google Scholar]
- 4.Mak J. and Kleiman,L. (1997) Primer tRNAs for reverse transcription. J. Virol., 71, 8087–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marquet R., Isel,C., Ehresmann,C. and Ehresmann,B. (1995) tRNAs as primer of reverse transcriptase. Biochimie, 77, 113–124. [DOI] [PubMed] [Google Scholar]
- 6.Aiyar A., Cobrinik,D., Ge,Z., Kung,H.J. and Leis,J. (1992) Interaction between retroviral U5 RNA and the TΨC loop of the tRNATrp primer is required for efficient initiation of reverse transcription. J. Virol., 66, 2464–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobrinik D., Soskey,L. and Leis,J. (1988) A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J. Virol., 62, 3622–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris S., Johnson,M., Stavnezer,E. and Leis,J. (2002) Replication of avian sarcoma virus in vivo requires an interaction between the viral RNA and the TΨC loop of the tRNATrp primer. J. Virol., 76, 7571–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arts E.J., Ghosh,M., Jacques,P.S., Ehresmann,B. and Le Grice,S.F.J. (1996) Restoration of tRNA3Lys-primed (–) strand DNA synthesis to an HIV-1 reverse transcriptase mutant with extended tRNAs. Implications for retroviral replication. J. Biol. Chem., 271, 9054–9061. [DOI] [PubMed] [Google Scholar]
- 10.Arts E.J., Stetor,S.R., Li,X.G., Rausch,J.W., Howard,K.J., Ehresmann,B., North,T.W., Wohrl,B.M., Goody,R.S., Wainberg,M.A. and Le Grice,S.F.J. (1996) Initiation of (–) strand DNA synthesis from tRNA3Lys on lentiviral RNAs: implications of specific HIV-1 RNA-tRNA3Lys interactions inhibiting primer utilization by retroviral reverse transcriptases. Proc. Natl Acad. Sci. USA, 93, 10063–10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isel C., Ehresmann,C., Keith,G., Ehresmann,B. and Marquet,R. (1995) Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA3Lys (template/primer) complex. J. Mol. Biol., 247, 236–250. [DOI] [PubMed] [Google Scholar]
- 12.Isel C., Keith,G., Ehresmann,B., Ehresmann,C. and Marquet,R. (1998) Mutational analysis of the tRNA3Lys/HIV-1 RNA (primer/template) complex. Nucleic Acids Res., 26, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isel C., Marquet,R., Keith,G., Ehresmann,C. and Ehresmann,B. (1993) Modified nucleotides of transfer-RNA3Lys modulate primer/template loop–loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem., 268, 25269–25272. [PubMed] [Google Scholar]
- 14.Isel C., Westhof,E., Massire,C., Le Grice,S.F.J., Ehresmann,C., Ehresmann,B. and Marquet,R. (1999) Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J., 18, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanchy J.M., Isel,C., Keith,G., Le Grice,S.F., Ehresmann,C., Ehresmann,B. and Marquet,R. (2000) Dynamics of the HIV-1 reverse transcription complex during initiation of DNA synthesis. J. Biol. Chem., 275, 12306–12312. [DOI] [PubMed] [Google Scholar]
- 16.Skripkin E., Isel,C., Marquet,R., Ehresmann,B. and Ehresmann,C. (1996) Psoralen crosslinking between human immunodeficiency virus type 1 RNA and primer tRNA3Lys. Nucleic Acids Res., 24, 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldschmidt V., Rigourd,M., Ehresmann,C., Le Grice,S.F.J., Ehresmann,B. and Marquet,R. (2002) Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription. J. Biol. Chem. 277, 43233–43242. [DOI] [PubMed] [Google Scholar]
- 18.Boulme F., Freund,F., Gryaznov,S., Nielsen,P.E., Tarrago-Litvak,L. and Litvak,S. (2000) Study of HIV-2 primer-template initiation complex using antisense oligonucleotides. Eur. J. Biochem., 267, 2803–2811. [DOI] [PubMed] [Google Scholar]
- 19.Freund F., Boulme,F., Litvak,S. and Tarrago-Litvak,L. (2001) Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNALys3 duplex. Nucleic Acids Res., 29, 2757–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J.T., Ehresmann,B., Hubscher,U. and Le Grice,S.F. (2001) A novel interaction of tRNALys,3 with the feline immunodeficiency virus RNA genome governs initiation of minus strand DNA synthesis. J. Biol. Chem., 276, 27721–27730. [DOI] [PubMed] [Google Scholar]
- 21.Friant S., Heyman,T., Bystrom,A.S., Wilhelm,M. and Wilhelm,F.X. (1998) Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNAiMet primer are required for initiation of reverse transcription in vivo. Mol. Cell. Biol., 18, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friant S., Heyman,T., Wilhelm,M.L. and Wilhelm,F.X. (1996) Extended interactions between the primer tRNAiMet and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res., 24, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabus C., Ficheux,D., Rau,M., Keith,G., Sandmeyer,S. and Darlix,J.L. (1998) The yeast Ty3 retrotransposon contains a 5′-3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J., 17, 4873–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakefield J.K., Kang,S.-M. and Morrow,C.D. (1996) Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J. Virol., 70, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang S.M., Zhang,Z.J. and Morrow,C.D. (1997) Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNAMet. J. Virol., 71, 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z.J., Kang,S.M., Li,Y. and Morrow,C.D. (1998) Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA, 4, 394–406. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Yu,Q., Kang,S.M., Buescher,J. and Morrow,C.D. (1998) Preferential completion of human immunodeficiency virus type 1 proviruses initiated with tRNA3Lys rather than tRNA1,2Lys. J. Virol., 72, 5464–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Kang,S.M. and Morrow,C.D. (1998) Genetic evidence of the interaction between tRNALys,3 and U5 facilitating efficient initiation of reverse transcription by human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses, 14, 979–988. [DOI] [PubMed] [Google Scholar]
- 29.Isel C., Lanchy,J.M., Le Grice,S.F.J., Ehresmann,C., Ehresmann,B. and Marquet,R. (1996) Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J., 15, 917–924. [PMC free article] [PubMed] [Google Scholar]
- 30.Brulé F., Marquet,R., Rong,L., Wainberg,M.A., Roques,B.P., Le Grice,S.F.J., Ehresmann,B. and Ehresmann,C. (2002) Structural and functional properties of the HIV-1 RNA-tRNA3Lys primer annealed by the nucleocapsid protein: Comparison with the heat annealed complex. RNA, 8, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beerens N., Groot,F. and Berkhout,B. (2001) Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem., 276, 31247–31256. [DOI] [PubMed] [Google Scholar]
- 32.Beerens N. and Berkhout,B. (2002) Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS. RNA, 8, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beerens N. and Berkhout,B. (2002) The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J. Virol., 76, 2329–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bénas P., Bec,G., Keith,G., Marquet,R., Ehresmann,C., Ehresmann,B. and Dumas,P. (2000) The crystal structure of HIV reverse transcription primer tRNALys,3 shows a canonical anticodon loop. RNA, 6, 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marquet R., Baudin,F., Gabus,C., Darlix,J.L., Mougel,M., Ehresmann,C. and Ehresmann,B. (1991) Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res., 19, 2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baudin F., Marquet,R., Isel,C., Darlix,J.L., Ehresmann,B. and Ehresmann,C. (1993) Functional sites in the 5′ region of human immunodeficiency virus type-1 RNA form defined structural domains. J. Mol. Biol., 229, 382–397. [DOI] [PubMed] [Google Scholar]
- 37.Ehresmann C., Baudin,F., Mougel,M., Romby,P., Ebel,J.P. and Ehresmann,B. (1987) Probing the structure of RNA in solution. Nucleic Acids Res., 15, 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Kang,S.M., LeBlanc,A., Hajduk,S.L. and Morrow,C.D. (1996) Nucleotide sequences within the U5 region of the viral RNA genome are the major determinants for an human immunodeficiency virus type 1 to maintain a primer binding site complementary to tRNA(His). Virology, 226, 306–317. [DOI] [PubMed] [Google Scholar]
- 39.Lanchy J.M., Ehresmann,C., Le Grice,S.F.J., Ehresmann,B. and Marquet,R. (1996) Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J., 15, 7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 40.Lanchy J.M., Keith,G., Le Grice,S.F.J., Ehresmann,B., Ehresmann,C. and Marquet,R. (1998) Contacts between reverse transcriptase and the primer strand govern the transition from initiation to elongation of HIV-1 reverse transcription. J. Biol. Chem., 273, 24425–24432. [DOI] [PubMed] [Google Scholar]
- 41.Fossé P., Mougel,M., Keith,G., Westhof,E., Ehresmann,B. and Ehresmann,C. (1998) Modified nucleotides of tRNAPro restrict interactions in the binary primer/template complex of M-MLV. J. Mol. Biol., 275, 731–746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.