Abstract
In the ribosomal DNA (rDNA) of Saccharomyces cerevisiae replication forks progressing against transcription stall at a polar replication fork barrier (RFB) located close to and downstream of the 35S transcription unit. Forks blocked at this barrier are potentially recombinogenic. Plasmids bearing the RFB sequence in its active orientation integrated into the chromosomal rDNA in sir2 mutant cells but not in wild-type cells, indicating that the histone deacetylase silencing protein Sir2 (Sir2p), which also modulates the aging process in yeast, suppresses the recombination competence of forks blocked at the rDNA RFB. Orientation of the RFB sequence in its inactive course or its abolition by FOB1 deletion avoided plasmid integration in sir2 mutant cells, indicating that stalling of the forks in the plasmid context was required for recombination to take place. Altogether these results strongly suggest that one of the functions of Sir2p is to modulate access of the recombination machinery to the forks stalled at the rDNA RFB.
INTRODUCTION
Replication forks do not always proceed at a constant rate. On the contrary, the replication machinery often encounters obstacles that may cause it to pause or even stall. These replication fork pauses or barriers can occur by accident as in the case of a nick or any other lesion in the template. But in some cases, these barriers are genetically specified and play an important role in the regulation of DNA replication (1). In the Escherichia coli chromosome, a number of sites flanking the replication terminus act as polar replication fork barriers (RFBs) when a protein, called Tus, binds them (2,3). The DNA sequences that Tus binds are called Ter sites. Ter/Tus barriers ensure that replication forks progressing throughout each hemisphere of the chromosome do not invade the other hemisphere. These RFBs prevent collision of the replication forks with the RNA polymerases transcribing some of the most active genes (4). This type of collision is known to have deleterious effects (5). As a consequence, replication of the E.coli chromosome always terminates in a defined region of the genome, roughly 180° apart from the origin (6). Stalled forks are recombinogenic. This is attributed to the fact that they are prone to develop double-strand breaks (DSBs) (7) or, alternatively, reversed forks (8,9), which promote recombination.
In Saccharomyces cerevisiae, rDNA consists of 150–200 tandem repeats located on chromosome XII (10). Each repeat consists of the 35S gene and a region that contains the 5S gene flanked by two non-transcribed sequences, NTS1 and NTS2. The RFB is located in NTS1 and is actually formed by two closely spaced barriers that stall replication forks progressing against transcription (11–13). Consequently, replication of the rDNA locus occurs primarily in a unidirectional fashion by virtue of forks progressing in the same direction as transcription of the 35S gene.
Homologous recombination at the rDNA correlates with RFB activity. DNA sequences of the NTS1 that are necessary for recombination at ectopic places (HOT1) contain the RFB sequence (14). Moreover, a trans-acting factor necessary for RFB, Fob1p, is also necessary for HOT1 recombination (15). Fob1p stimulates recombination at the native rDNA locus too, as measured either by marker loss, marker duplication or formation of extrachromosomal rDNA circles (16–18). Finally, Fob1p is also necessary for expansion and contraction of the rDNA array (18). The DNA sequence containing the RFB is necessary but not sufficient for FOB1-dependent expansion of the rDNA array (19). Altogether these observations led to the idea that the RFB triggers or stimulates recombination within the rDNA array. A recent report, however, indicates that HOT1 recombination and the rDNA RFB are independent, even though they share common cis elements (20). A detailed analysis of the structure of the rDNA RFB revealed no single-stranded DNA regions prone to produce DSBs, suggesting that the rDNA RFB is less fragile than initially suspected (21). Additionally, no reversed forks were detected at the RFB by electron microsocopy. These observations suggest that the rDNA RFB either is not recombinogenic or its recombination competence is severely repressed in wild-type cells.
One protein involved in recombination repression, specifically at the rDNA, is Sir2p (22,23). Sir2p also silences genes integrated into the rDNA locus (24). Recombination repression and transcriptional silencing are thought to reflect the ability of Sir2p to promote a specialized closed chromatin structure at several regions of the rDNA locus (25). Availability of Sir2p in the nucleolus is limited and small changes in the amount of this protein affect rDNA silencing dramatically (26). Silencing and recombination levels at the rDNA also correlate with yeast longevity. Mutations in genes that reduce rDNA silencing or increase rDNA recombination shorten lifespan while mutations that increase silencing or repress recombination generally extend lifespan (16,27–31). Interestingly, calorie restriction, which is known to extend lifespan in yeast and mammals, also increases SIR2-dependent rDNA silencing in yeast (32). These observations suggest that maintenance of a particular chromatin structure at the yeast rDNA locus is important for extended longevity.
One possibility is that replication forks stalled at the rDNA RFB are recombinogenic but Sir2p represses this potential. This has already been suggested by genetic studies but no molecular evidence supporting this notion is yet available (27,33).
Here we show that plasmids bearing the RFB sequence in the orientation that stalls replication forks integrate into the genome at the chromosomal rDNA locus in sir2 mutant cells but not in wild-type cells. This observation indicates that in sir2 mutants forks stalled at the RFB are recombinogenic. The observation that elimination of the barrier in the plasmid, by either placing the RFB sequence in its inactive orientation or deleting FOB1, avoids plasmid integration in sir2 mutant cells, points out that fork stalling in the plasmid context is necessary for integration to take place. Altogether, these observations strongly suggest that Sir2p suppresses the recombination competence of replication forks stalled at the rDNA RFB.
MATERIALS AND METHODS
Yeast strains and plasmids
The CT711 yeast strain (MATa, leu2-3, -113, his3Δ1, trp1, ura3-52, ade2-101, can1) was used throughout the study. PCR-mediated deletion was used to obtain SIR2Δ::LEU2 in which the entire SIR2 ORF was removed. FOB1 was deleted by replacing the entire ORF with pRS303 (FOB1Δ::HIS3). Deletions and disruptions were confirmed by Southern blot analysis. pBB6-RFB+ and pBB6-RFB– are described in Brewer et al. (12). To obtain pBB6-RFB+(del) the PCR product of an rDNA region spanning from the EcoRI site at NTS1 to 100 bp downstream of the HpaI site was inserted at the EcoRI site of pBB6 in the orientation that stalls replication forks (see Fig. 4B). Plasmids were introduced into yeast cells by the lithium acetate method (34).
Figure 4.
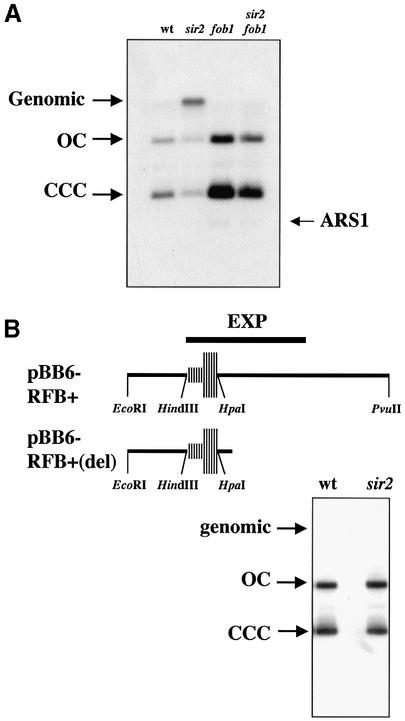
(A) Deletion of FOB1 avoided integration of pBB6-RFB+ in sir2 cells. DNA isolated from wild-type (CT711), sir2 (sir2Δ::LEU2), fob1 (fob1Δ::HIS3) and sir2 fob1 double mutants transformed with pBB6-RFB+ were digested with XhoI and XbaI and subjected to electrophoresis and Southern blot hybridization. To avoid cross-hybridization with bacterial sequences, a chromosomal 237 bp HindIII–BglII ARS1 fragment was used as probe. The restriction enzymes used do not cut pBB6-RFB+. A signal co-migrating with genomic DNA was detected only in sir2 cells. The weak band running faster than CCCs corresponded to the endogenous ARS1 fragment (ARS1). (B) Elimination of part of the region downstream of the HpaI site avoided integration in sir2 cells. The maps on top depict the most relevant features of the rDNA RFB and adjacent sequences in pBB6-RFB+ and pBB6-RFB+(del). pBB6-RFB+(del) was obtained as described in Material and Methods and used to transform both wild-type (CT711) and sir2 (sir2Δ::LEU2) strains. Undigested DNA was subjected to electrophoresis and Southern blot hybridization using pBR322 as probe. No hybridization signal was observed co-migrating with genomic DNA.
Yeast DNA isolation
Yeast DNA used for plasmid integration analysis was obtained from 5 ml of saturated yeast cultures grown in synthetic medium without uracil and containing 2% glucose (SC-URA). DNA was obtained by breaking the cells with acid-washed glass beads in 500 µl lysis buffer (100 mM Tris, pH 8.0, 50 mM EDTA, 1% SDS). Ammonium acetate (2.5 M, pH 7.0) was added to the liquid phase and incubated for 5 min at 65°C plus 5 min on ice. Chloroform (1 vol) was added and the aqueous phase was precipitated with 1 ml isopropanol. The pellet was washed with 70% ethanol, dried and dissolved in 40 µl H2O. DNA was electrophoresed in 0.8% agarose in 1× TAE buffer and subjected to Southern hybridization. Yeast DNA used for 2-dimensional (2D) agarose gel electrophoresis was prepared as described by Huberman et al. (35).
Purification of rDNA
DNA isolated from yeast cells was analyzed by density centrifugation in CsCl gradients as described by Huberman et al. (35). The rDNA-enriched band was removed, extracted with isopropanol, purified through a Microcon-30 column (Millipore) and precipitated with ethanol.
2-Dimensional agarose gel electrophoresis
Neutral/neutral 2D agarose gel electrophoresis (36) was performed to analyze pBB6-RFB+ integration. The first dimension was run at 1 V/cm in a 0.4% agarose (SeaKem; FMC Bioproducts) gel in 1× TBE buffer for 22 h at room temperature. The second dimension was run at 5 V/cm in a 1% agarose gel in 1× TBE/0.3 µg/ml ethidium bromide for 12 h at 4°C. After electrophoresis gels were subjected to Southern hybridization.
Pulsed field electrophoresis
Preparation of samples for pulsed field electrophoresis was carried out as follows. Yeast cells (wild-type or sir2 mutants) transformed with pBB6-RFB+ were grown to saturation in 30 ml SC-URA, washed with 50 mM EDTA (pH 7.5) and resuspended in 1 ml CPES buffer [40 mM citric acid, 120 mM Na2HPO4, 1.2 M sorbitol, 20 mM EDTA, 5 mM DTT (pH 6.0)]. Next, 1.6 ml melted 1% low melting point agarose containing 1% zymolase 20T (ICN Biomedicals) was added to the cell suspension and poured into 200 µl block formers that were placed at room temperature for 30 min followed by another 30 min at 4°C. Blocks were removed and incubated at 30°C for 1 h in 2 ml 40 mM citric acid, 120 mM Na2HPO4, 20 mM EDTA (pH 6.0) (CPE). Next, blocks were incubated for 24 h at 50°C in 1 mg/ml proteinase K in 10 mM Tris–HCl (pH 8.0), 0.45 M EDTA, 1% lauryl sarcosine. Blocks were stored in 0.5 M EDTA (pH 9) at 4°C. Prior to electrophoresis, blocks were washed twice in 10 mM Tris–HCl (pH 8.0), 1 mM EDTA and cut in 2 mm slices. Pulsed field electrophoresis was performed on a CHEF-DR® II (Bio-Rad) apparatus at 14°C in a 1% agarose gel (Pulsed-field Certified; Bio-Rad) in 0.5× TBE buffer for 27 h at 6 V/cm using a 120° included angle with a 6.75–158 s switch time ramp. Gels were stained with 1 µg/ml ethidium bromide and subjected to Southern hybridization.
Southern hybridization
Agarose gels were transferred to nylon membranes (Zeta-Probe; Bio-Rad) and hybridized with probes radiolabeled by random priming. For 2D gel hybridizations, the Random Primer Fluorescein Labeling Kit (NEN™; Life Science Products) was used. Probes (600 ng) were labeled by random priming with fluorescein-labeled nucleotides and detection was performed using an anti-fluorescein–alkaline phosphatase conjugate.
RESULTS
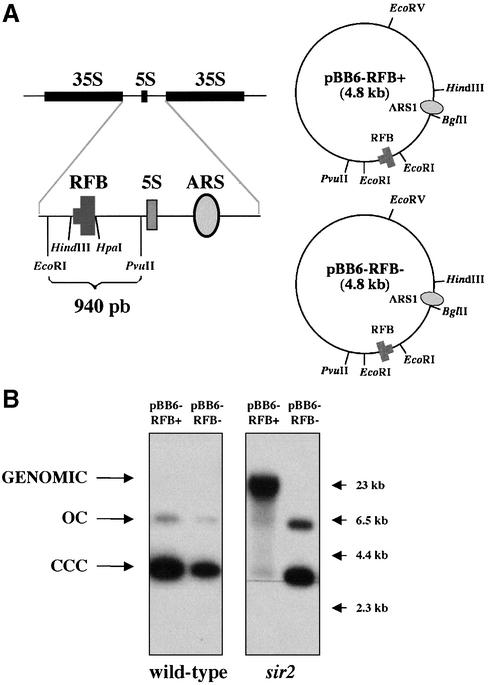
To explore the recombination competence of yeast rDNA RFB, we studied the stability of plasmids bearing this barrier. pBB6-RFB+ and pBB6-RFB– are described in Brewer et al. (12). These plasmids contain a 940 bp EcoRI–PvuII rDNA fragment, with the RFB sequence plus some adjacent rDNA sequences, inserted at the EcoRI site of pBB6 (Fig. 1A). pBB6-RFB+ contains the barrier in its active orientation so that it blocks forks initiated at ARS1 progressing clockwise. In pBB6-RFB–, the RFB sequence is in the opposite inactive orientation so that in this plasmid replication forks initiated at ARS1 progressing clockwise have no impediment to go through. Stalling of replication forks at the RFB in pBB6-RFB+ but not in pBB6-RFB– was confirmed by 2D agarose gel electrophoresis (12). CT711 yeast cells (37) were transformed with these plasmids and undigested DNA was electrophoresed and analyzed by Southern hybridization. pBR322 DNA was used as a probe to avoid any cross-hybridization with endogenous yeast sequences. Both plasmids occurred as extra-chromosomal monomers in wild-type cells (Fig. 1B, left panels). Two bands, corresponding to covalently closed circles (CCC) and open circles (OC), were detected in each case. pBB6 behaved similarly (data not shown). These results indicate that plasmids bearing an active or inactive rDNA RFB are stable in wild-type cells and suggest that forks stalled at the RFB do not recombine in these cells.
Figure 1.
The different behavior of pBB6-RFB+ in sir2 cells. (A) Schematic representation of two rDNA repeats. The 940 bp EcoRI–PvuII fragment, which contains the RFB, was inserted at the EcoRI site of pBB6 in both orientations to yield pBB6-RFB+ and pBB6-RFB–. (B) Stability of pBB6-RFB+ and pBB6-RFB– in wild-type (CT711) and sir2 (sir2Δ::LEU2) strains. Undigested yeast DNA was electrophoresed and subjected to Southern blot hybridization using radiolabeled pBR322 as a probe. Arrows on the right-hand side point to λ-HindIII size markers. A high molecular weight band that co-migrated with genomic DNA was detected only in the case of pBB6-RFB+ isolated from sir2 cells.
Sir2p is involved in maintenance of a repressive chromatin structure at the S.cerevisiae rDNA locus avoiding recombination between repeats (25). In fact, Sir2p specifically protects a region located within the NTS (24,25). To test if Sir2p also avoids recombination of replication forks stalled at the RFB in plasmids, we deleted SIR2 in CT711 and transformed these sir2 mutants with pBB6-RFB+ and pBB6-RFB–. DNA from these transformants was analyzed as above. The results obtained were strikingly different (Fig. 1B, right panel). In sir2 mutant cells pBB6-RFB– behaved as in the case of wild-type CT711. But pBB6-RFB+ migrated as a high molecular weight band together with the bulk of sheared genomic DNA. The same result was obtained in five different experiments.
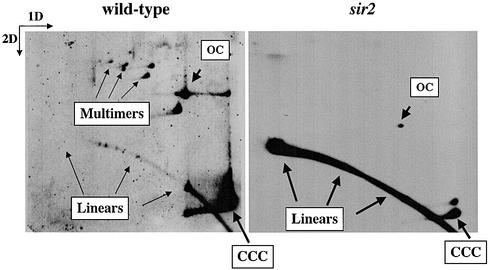
To discriminate between multimerization and integration we analyzed intact forms of pBB6-RFB+ obtained from wild-type and sir2 mutant cells by 2D agarose gel electrophoresis (36,38,39). As clearly seen in Figure 2, pBB6-RFB+ occurred as monomeric and multimeric CCC and OC forms in wild-type cells. In sir2 mutants, however, the only prominent signal corresponded to sheared linear forms. Circular extrachromosomal forms were negligible. These data strongly suggest that in most sir2 mutants pBB6-RFB+ became integrated into the genome, probably by homologous recombination at the rDNA locus. This observation implied that replication forks stalled at the rDNA RFB recombined efficiently in sir2 mutant cells. Moreover, the observation that pBB6-RFB– remained extra-chromosomal in these mutants strongly suggests that recombination was triggered by fork stalling in the plasmid context, although sir2 mutation affected the chromosomal RFB copies as well. It should be noted that although the 940 bp rDNA fragment containing the RFB is functional on a plasmid, its efficiency drops significantly if compared to the chromosomal context (12). Nevertheless, it only took approximately 40–60 generations (from transformation to DNA isolation) to get almost 90% of the episomes integrated in sir2 mutant cells (Fig. 1B). Another aspect that should be considered is the stability of native as well as integrated copies in the chromosomal context. ARS1 is known to be more active than the native rDNA ARS, something that could affect replication and stability of the locus. The analysis of rDNA stability, however, needs to be addressed specifically and is outside the scope of the present report.
Figure 2.
pBB6-RFB+ integrated into the genome of sir2 cells. Undigested DNA from wild-type (CT711) and sir2 (sir2Δ::LEU2) strains were subjected to 2D agarose gel electrophoresis and Southern blot hybridization using radiolabeled pBR322 as a probe. Monomeric and multimeric extra-chromosomal forms of pBB6-RFB+ were clearly detected in wild-type cells. In sir2 mutants, however, the only prominent signal corresponded to sheared linear forms.
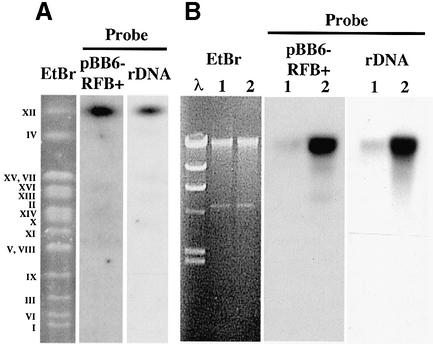
To test if plasmid integration indeed took place at the rDNA locus, we used pulsed field electrophoresis to analyze yeast chromosomes of the sir2 mutant strain transformed with pBB6-RFB+. Plasmid DNA integrated into chromosome XII of S.cerevisiae (Fig. 3A), where the rDNA array is located (10). No hybridization signal was detected in wild-type cells transformed with the same plasmid (data not shown). Moreover, pBB6-RFB+ from sir2 mutants co-purified with the rDNA fraction isolated using CsCl density gradients (Fig. 3B). These results indicated that in sir2 mutant cells pBB6-RFB+ integrated specifically at or close to the rDNA locus.
Figure 3.
In sir2 cells pBB6-RFB+ integrated into the rDNA locus at chromosome XII. (A) Undigested DNA from sir2 cells transformed with pBB6-RFB+ was subjected to pulsed field electrophoresis. The gel was stained with ethidium bromide (EtBr lane), to separate the different yeast chromosomes (Roman numerals on the left-hand side) and subjected to Southern blot hybridization using pBR322 as probe (pBB6-RFB+ lane). The stripped blot was re-hybridized with a yeast rDNA probe (rDNA lane). Note that both probes hybridized to a single band that co-migrated with chromosome XII. (B) Yeast DNA obtained from sir2 cells transformed with pBB6-RFB+ was subjected to ultracentrifugation in a CsCl gradient in the presence of Hoescht 33258. Two fractions enriched for bulk DNA (lanes designated 1) or rDNA (lanes designated 2) were isolated from the CsCl gradient, subjected to electrophoresis and hybridized using pBR322 (pBB6-RFB+) or a yeast rDNA fragment (rDNA) as probes. The EtBr panel corresponds to the gel stained with ethidium bromide (λ = lambda/HindIII size marker). In both cases probes hybridized mainly to the rDNA-enriched sample.
Fob1p is necessary for rDNA RFB (15). To confirm that stalling of the replication forks was needed for integration of plasmid DNA in sir2 mutant cells we deleted FOB1 in both wild-type and sir2 mutants and analyzed the recombination competence of pBB6-RFB+ in fob1 mutants as well as in sir2 fob1 double mutants. If forks stalling at the RFB were needed for recombination, elimination of the RFB would avoid integration. No integration was observed either in fob1 cells or in sir2 fob1 double mutants (Fig. 4A). This observation strengthens the notion that forks stalled at the RFB in the plasmid promote recombination in sir2 mutant cells. It should be noted that digestion of genomic rDNA with restriction enzymes that do not cut within the plasmid would still produce a high molecular weight band if the inserted plasmid molecules occur as tandem repeats (Fig. 4A).
The rDNA RFB is located within the HindIII–HpaI fragment of NTS1 (20). However, to achieve full RFB activity in pBB6-RFB+, the adjacent EcoRI–HindIII fragment to the left is also needed (12). Interestingly, Kobayashi et al. (19) found that a fragment called EXP (Fig. 4B, top), containing the HindIII–HpaI fragment plus ∼400 bp situated to the right, is necessary for expansion of the rDNA locus, indicating that in addition to the RFB other sequences are required for rDNA recombination. These ∼400 bp DNA sequences adjacent to the HindIII–HpaI fragment, however, are dispensable for stalling replication forks (20). pBB6-RFB+ contains a 940 bp rDNA fragment that includes the whole EXP region. We showed earlier that forks stalled at the barrier were necessary for recombination to take place in sir2 mutant cells. To find out if the ∼400 bp region was also needed, we prepared a new plasmid, pBB6-RFB+(del), where a fragment lacking part of the EXP region was inserted in pBB6 in the RFB active orientation. The new plasmid still contained the EcoRI– HindIII–HpaI fragment needed for full RFB activity, plus only ∼100 bp to the right (Fig. 4B, top). Integration of this plasmid occurred neither in wild-type nor in sir2 mutants (Fig. 4B, bottom). This observation confirmed that in addition to stalled forks, the ∼400 bp region to the right of the RFB sequence is also needed for recombination to occur in sir2 mutant cells. Identical results were obtained for a plasmid containing just the EcoRI–HindIII–HpaI fragment (data not shown).
DISCUSSION
The results reported here showed that in S.cerevisiae, the recombination competence of replication forks stalled at the rDNA RFB was repressed by Sir2p, thus implicating a silencing protein in the stabilization of stalled replication forks. A region adjacent to the RFB sequence was also necessary for recombination in sir2 mutant cells. This region contains DNA sequences that are needed for the expansion of rDNA repeats but are dispensable for stalling forks at the RFB (19). Using sensitivity assays to micrococcal nuclease and dam methyltransferase, Fritze and co-workers (25) determined that Sir2p creates a closed chromatin structure specifically in this region, called SIR2 Responsive Region (SRR) 1. These data suggest that forks stalled at the rDNA RFB are not recombinogenic per se, but may serve as the initial substrate for recombination proteins that would be recruited to the SRR1 region when Sir2p is absent or its activity is repressed. In mutants for the RNA polymerase I transcription factor UAF, some variants are able to grow normally using RNA polymerase II for rDNA transcription (40). This polymerase switch also requires the FOB1-dependent expansion of the rDNA locus. In sir2 mutant cells the frequency of switching increases, probably due to stimulation in the rate of rDNA expansion (41). We propose that one of the functions of Sir2p at the rDNA locus is to modulate recombination of the forks stalled at the rDNA RFB and consequently the expansion and contraction of the rDNA array in response to environmental or metabolic changes. Under conditions requiring a change in the number of rDNA repeats, Sir2p activity would be inhibited, facilitating access of recombination proteins to the SRR1 region to process the forks stalled at the RFB.
Using DNA combing and single molecule imaging, Pasero et al. (42) found that in the yeast chromosomal context functional rDNA origins are clustered and interspersed with large domains where initiation is silenced. This repression appears to be mediated by Sir2p. Based on this observation they claim recombination could increase in sir2 mutants because more forks would become stalled at the RFB in these cells. But in our plasmid integration assay we found that recombination in sir2 mutant cells was triggered by fork stalling in the plasmid context. This observation implies that the number of forks stalled at the RFB in the chromosomal context cannot explain by itself the elevated recombination rates observed in sir2 mutants.
Interestingly, deletion of FOB1, which is necessary for RFB formation, does not completely suppress the rDNA hyper-recombination phenotype of sir2 mutant cells (16,27). This observation suggests that Sir2p also suppresses recombination at other regions in the rDNA locus besides the RFB. Actually, Fritze et al. (25) showed that Sir2p creates a closed chromatin structure at the 35S transcription unit (SRR2). It remains to be shown how Sir2p modulates recombination in this region and if it also depends on replication fork pausing or blocking. Although no other RFB is detected in the rDNA of wild-type cells by 2D gel electrophoresis, progression of replication forks through the locus requires specialized helicases to bypass specific pausing sites (43). It is plausible that Sir2p also suppresses or modulates recombination at these pausing sites, where replication forks could eventually stall. It is worth noting that in a screen for mutants that affect rDNA silencing several genes involved in DNA replication were recovered (44), reinforcing the possible connection between chromatin silencing, recombination and rDNA replication.
Sir2p also affects the aging process in yeast (27,28). Deletion of SIR2 shortens yeast replication lifespan while its overexpression extends it. The observation that mutations in other genes that affect rDNA silencing or recombination also affect yeast lifespan (16,27–31) further supports this notion. Altogether, these observations suggest that maintenance of rDNA stability is important for cell longevity. It is worth noting that a similar role in stabilizing stalled replication forks or, alternatively, in their correct processing by recombination has also been proposed for the Werner syndrome protein (WRNp) and other RecQ helicases (45–47). Absence of a functional WRNp causes a premature aging phenotype in humans (48) and, like Sir2p, this helicase is also located in the nucleolus (49,50). These observations suggest that Sir2p and WRNp might have a similar role and a common mechanism in modulating the aging process in yeast and human cells, respectively.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Bonita Brewer and S. M. Jazwinski for plasmids and Ernesto García for his advice on pulsed field electrophoresis. We acknowledge also María Luisa Martínez-Robles and Pilar Robles for technical assistance. A.B. held successive post-doctoral fellowships from the Comunidad Autónoma de Madrid and the Spanish Ministerio de Ciencia y Tecnología. This work was partially supported by grants PGC PB98-048 from the Spanish Comisión Interministerial de Ciencia y Tecnología, SAF2001-1740 from the Spanish Ministerio de Ciencia y Tecnología and 08.5/0057/2001.1 and 08.1/0067/2001.1 from the Comunidad Autónoma de Madrid.
REFERENCES
- 1.Hernández P., Martín-Parras,L., Martínez-Robles,M.L. and Schvartzman,J.B. (1993) Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J., 12, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastia D. and Mohanty,B.K. (1996) Mechanisms for completing DNA replication. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, New York, NY, pp. 177–215.
- 3.Hill T.M., Pelletier,A.J., Tecklenburg,M.L. and Kuempel,P.L. (1988) Identification of the DNA sequence from E. coli terminus region that halts replication forks. Cell, 55, 459–466. [DOI] [PubMed] [Google Scholar]
- 4.Brewer B.J. (1988) When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell, 53, 679–686. [DOI] [PubMed] [Google Scholar]
- 5.Olavarrieta L., Hernández,P., Krimer,D.B. and Schvartzman,J.B. (2002) DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol., 322, 1–6. [DOI] [PubMed] [Google Scholar]
- 6.Rothstein R., Michel,B. and Gangloff,S. (2000) Replication fork pausing and recombination or “gimme a break”. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- 7.Michel B., Ehrlich,S.D. and Uzest,M. (1997) DNA double-strand breaks caused by replication arrest. EMBO J., 16, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olavarrieta L., Martínez-Robles,M.L., Sogo,J.M., Stasiak,A., Hernández,P., Krimer,D.B. and Schvartzman,J.B. (2002) Supercoiling, knotting and replication fork reversal in partially replicated plasmids. Nucleic Acids Res., 30, 656–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postow L., Ullsperger,C., Keller,R.W., Bustamante,C., Vologodskii,A.V. and Cozzarelli,N.R. (2001) Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem., 276, 2790–2796. [DOI] [PubMed] [Google Scholar]
- 10.Petes T.D. (1979) Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl Acad. Sci. USA, 76, 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brewer B.J. and Fangman,W.L. (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell, 55, 637–643. [DOI] [PubMed] [Google Scholar]
- 12.Brewer B.J., Lockshon,D. and Fangman,W.L. (1992) The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell, 71, 267–276. [DOI] [PubMed] [Google Scholar]
- 13.Linskens M.H.K. and Huberman,J.A. (1988) Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 8, 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang G.S. and Keil,R.L. (1995) Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics, 141, 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T. and Horiuchi,T. (1996) A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells, 1, 465–474. [DOI] [PubMed] [Google Scholar]
- 16.Defossez P.A., Prusty,R., Kaeberlein,M., Lin,S.J., Ferrigno,P., Silver,P.A., Keil,R.L. and Guarente,L. (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell, 3, 447–455. [DOI] [PubMed] [Google Scholar]
- 17.Johzuka K. and Horiuchi,T. (2002) Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S.cerevisiae. Genes Cells, 7, 99–113. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi T., Heck,D.J., Nomura,M. and Horiuchi,T. (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev., 12, 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi T., Nomura,M. and Horiuchi,T. (2001) Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward T.R., Hoang,M.L., Prusty,R., Lau,C.K., Keil,R.L., Fangman,W.L. and Brewer,B.J. (2000) Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: shared sequences but independent activities. Mol. Cell. Biol., 20, 4948–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruber M., Wellinger,R.E. and Sogo,J.M. (2000) Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol. Cell. Biol., 20, 5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasser S.M. and Cockell,M.M. (2001) The molecular biology of the SIR proteins. Gene, 279, 1–16. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb S. and Esposito,R.E. (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell, 56, 771–776. [DOI] [PubMed] [Google Scholar]
- 24.Smith J.S. and Boeke,J.D. (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev., 11, 241–254. [DOI] [PubMed] [Google Scholar]
- 25.Fritze C.E., Verschueren,K., Strich,R. and Easton-Esposito,R. (1997) Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J., 16, 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith J.S., Brachmann,C.B., Pillus,L. and Boeke,J.D. (1998) Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics, 149, 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaeberlein M., McVey,M. and Guarente,L. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev., 13, 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S., Benguria,A., Lai,C.Y. and Jazwinski,S.M. (1999) Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell, 10, 3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merker R.J. and Klein,H.L. (2002) hpr1{Delta} affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae. Mol. Cell. Biol., 22, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy N. and Runge,K.W. (2000) Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol., 10, 111–114. [DOI] [PubMed] [Google Scholar]
- 31.Sinclair D.A., Mills,K. and Guarente,L. (1997) Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science, 277, 1313–1316. [DOI] [PubMed] [Google Scholar]
- 32.Lin S.J., Kaeberlein,M., Andalis,A.A., Sturtz,L.A., Defossez,P.A., Culotta,V.C., Fink,G.R. and Guarente,L. (2002) Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature, 418, 344–348. [DOI] [PubMed] [Google Scholar]
- 33.Guarente L. (1999) Diverse and dynamic functions of the Sir silencing complex. Nature Genet., 23, 281–285. [DOI] [PubMed] [Google Scholar]
- 34.Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huberman J.A., Spotila,L.D., Nawotka,K.A., el-Assouli,S.M. and Davis,L.R. (1987) The in vivo replication origin of the yeast 2 microns plasmid. Cell, 51, 473–481. [DOI] [PubMed] [Google Scholar]
- 36.Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- 37.Saavedra R.A. and Huberman,J.A. (1986) Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell, 45, 65–70. [DOI] [PubMed] [Google Scholar]
- 38.Martín-Parras L., Lucas,I., Martínez-Robles,M.L., Hernández,P., Krimer,D.B., Hyrien,O. and Schvartzman,J.B. (1998) Topological complexity of different populations of pBR322 as visualized by two-dimensional agarose gel electrophoresis. Nucleic Acids Res., 26, 3424–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schvartzman J.B., Adolph,S., Martín-Parras,L. and Schildkraut,C.L. (1990) Evidence that replication initiates at only some of the potential origins in each oligomeric form of Bovine Papillomavirus Type 1 DNA. Mol. Cell. Biol., 10, 3078–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vu L., Siddiqi,I., Lee,B.S., Josaitis,C.A. and Nomura,M. (1999) RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA, 96, 4390–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oakes M., Siddiqi,I., Vu,L., Aris,J. and Nomura,M. (1999) Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol., 19, 8559–8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasero P., Bensimon,A. and Schwob,E. (2002) Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev., 16, 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivessa A.S., Zhou,J.Q. and Zakian,V.A. (2000) The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell, 100, 479–489. [DOI] [PubMed] [Google Scholar]
- 44.Smith J.S., Caputo,E. and Boeke,J.D. (1999) A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol., 19, 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Constantinou A., Tarsounas,M., Karow,J.K., Brosh,R.M., Bohr,V.A., Hickson,I.D. and West,S.C. (2000) Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep., 1, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karow J.K., Constantinou,A., Li,J.L., West,S.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen J. and Loeb,L.A. (2001) Unwinding the molecular basis of the Werner syndrome. Mech. Ageing Dev., 122, 921–944. [DOI] [PubMed] [Google Scholar]
- 48.Epstein C.J., Martin,G.M., Schultz,A.L. and Motulsky,A.G. (1966) Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine, 45, 177–221. [DOI] [PubMed] [Google Scholar]
- 49.Marciniak R.A., Lombard,D.B., Johnson,F.B. and Guarente,L. (1998) Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl Acad. Sci. USA, 95, 6887–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki T., Shiratori,M., Furuichi,Y. and Matsumoto,T. (2001) Diverged nuclear localization of Werner helicase in human and mouse cells. Oncogene, 20, 2551–2558. [DOI] [PubMed] [Google Scholar]