Abstract
Structural domains are considered as the basic units of protein folding, evolution, function and design. Automatic decomposition of protein structures into structural domains, though after many years of investigation, remains a challenging and unsolved problem. Manual inspection still plays a key role in domain decomposition of a protein structure. We have previously developed a computer program, DomainParser, using network flow algorithms. The algorithm partitions a protein structure into domains accurately when the number of domains to be partitioned is known. However the performance drops when this number is unclear (the overall performance is 74.5% over a set of 1317 protein chains). Through utilization of various types of structural information including hydrophobic moment profile, we have developed an effective method for assessing the most probable number of domains a structure may have. The core of this method is a neural network, which is trained to discriminate correctly partitioned domains from incorrectly partitioned domains. When compared with the manual decomposition results given in the SCOP database, our new algorithm achieves higher decomposition accuracy (81.9%) on the same data set.
INTRODUCTION
The concept of structural domains was first proposed by Wetlaufer in 1973 (1). Though there is still no established definition, domains are generally considered as compact, semi-independent units, each of which forms a structurally ‘separate’ region in a three-dimensional protein structure. Each domain should be able to exist and remain folded as a native-like structure if it is cleaved from the rest of the protein (2). Domains are considered as the basic units of protein folding, function, evolution and design.
Several databases of protein domains have been constructed, such as SCOP (3) and CATH (4), which provide a rich source of information for structural and functional prediction/annotation of proteins. One popular application of these domain databases is threading-based protein structure prediction (5–7). A clear advantage of domain-based threading, compared with protein chain-based threading, is its wider scope of applicability since a novel protein may share only a domain, rather than the whole structure, with a known protein structure.
Decomposition of multi-domain protein structures into individual domains has been traditionally done manually. As the rate of protein structure determination has increased significantly in the past few years and is expected to continue to go up, partly due to the world-wide Structural Genomics Projects (8), the manual process has become a bottleneck in keeping the domain databases up to date. There is apparently an urgent call for more efficient, reliable and fully automated methods for domain decomposition. A number of computer algorithms for domain assignment from protein structural coordinates have been proposed, which include PUU (9), DOMARK (10), DETECTIVE (11), STRUDL (12), DomainParser (13), etc. Whereas these algorithms/programs employ different computational approaches, they essentially follow one basic design principle: inter-residue interactions are denser within domains than between domains.
A systematic assessment by Jones et al. (14) was carried out on a test set of 55 protein structures on four popular domain decomposition programs: PUU (9), DETECTIVE (11), DOMARK (10) and the one developed by Islam et al. (15). The study showed that the most accurate program achieved an accuracy of 76% (PUU) when compared to human assignments by the original authors of the structures, while the other three fell between 67 and 76%. The four programs agreed on only 55.7% of the identified domains on a larger data set of 787 protein chains. Due to such limitations of the current computer programs, manual checking is usually needed for the construction of domain databases. For example, all protein domains in SCOP are defined manually by human experts (3); CATH uses both computer programs and human annotations for its domain definition. For each protein, CATH runs three domain decomposition programs, DETECTIVE, PUU and DOMARK. If a consensus is reached among the three programs, CATH adds the consensus result into its database; otherwise it relies on human assignments. For only about 53% of the proteins, CATH takes the computer-generated results (14).
Using a graph-theoretic approach, we have previously developed a program, DomainParser, for protein domain decomposition. On the same test set of 55 proteins, DomainParser assigned 78.2% of the proteins consistently with the manually identified domains (13). DomainParser formulates the domain decomposition problem as a network flow problem and employs the Ford–Fulkerson algorithm to solve it (16). Large-scale testing revealed a significant gap in its decomposition accuracy when DomainParser has or has no knowledge about the number of domains to be partitioned (13). This points to possible directions for further improvement of the program. We have observed that good domain partitions share certain common features, defined in terms of the number of segments and other parameters related to the partitioned domains, while bad partitions have different distributions of these parameters, suggesting that they could be distinguished through data clustering.
Although DomainParser performs equally well when compared with other automatic programs, it is in need of an effective way to assess the quality of a partition, which is a common problem among most automatic programs (10). Here we present a computational procedure that combines the structural information of a protein structure including hydrophobic moment profile and neural networks for quality checking of the identified domains. The hydrophobic moment, first introduced by Eisenberg (17), has been useful in measuring the amphiphilicity of α-helical protein structures. While the first-order moment can classify the helices into various groups, the second-order moment provides the spatial profiling of globular proteins hydrophobicity (18). Based on 30 relatively diverse protein structures, Silverman reported two important features of moment calculation. One is that when calculated from the interior to the exterior of the proteins, the overall profiles of the second-order ellipsoidal moments of all high resolution X-ray crystallographic structures follow a very similar pattern. This global feature is independent of the size and fold of the protein. Misfolded protein structures displayed irregular spatial profiles, which can be distinguished easily from the profiles of their native structure. The other feature is that the hydrophobic ratio Rt, the ratio of distances at which the second-order and zero-order moments vanish respectively, lies within a narrow range (18).
The hydrophobic moment profiles reflect the unique property of globular proteins; non-polar side chains tend to pack together to form a hydrophobic core, while hydrophilic side chains form an exterior shell. The working definition of a protein domain implies that each domain should contain an identifiable hydrophobic core (11), suggesting that each well-defined protein domain should also have the similar hydrophobic moment profile as that of the native globular proteins. It has been shown that individual domains have similar structures when solved either independently or as part of a large protein.
Several parameters related to a specific partition of a protein and the resulting domains can be used to evaluate the specific partition. A neural network is a natural tool to incorporate all the information available for the domain identification. In this report, we describe how we incorporated the hydrophobic moment profiling and other valuable data into DomainParser. DomainParser with new capabilities showed improved performance over the original DomainParser. The new program is available at http://compbio.ornl.gov/structure/domainparser2.
MATERIALS AND METHODS
Our overall approach can be summarized as follows. For a given protein structure, we first employ a ‘top-down’ approach to partition the structure into domains using DomainParser with modified parameters to avoid possible undercutting. This step is followed by a domain quality checking step to assess whether an individual domain is overcut through a ‘bottom-up’ approach.
In the first step, DomainParser first bi-partitions the structure into two substructures with the smallest (minimum) bottleneck, then tries to recursively solve the minimum cut problem on each substructure until the stopping criteria are met. The stopping criteria used in this study are slightly more relaxed than the one in the previous version of DomainParser in order to minimize the number of undercut. So our focus in quality assessment will be mainly on the possibility of overcutting of domains. The result of the recursive application of the Fork–Fulkerson algorithm is a set of predicted domain structures. For each of them, we employ a neural network approach with various parameters to assess its quality being like a native structure of a protein domain. The parameters include: (i) the fitness of the hydrophobic moment profile of the predicted domain with the general hydrophobic moment profile of experimental structures; (ii) the number of non-consecutive sequence segments in a predicted domain; (iii) the compactness of the domain; (iv) the size of the interface between two domains; (v) relative motion between two domains; (vi) the size of the predicted domain.
A neural network is trained to distinguish correctly partitioned domains from incorrectly partitioned, specifically overcut, domains. If a predicted domain receives a bad quality score from the trained neural network, the program then undoes the partition (merge) of the domain and goes back one level up in the recursion, and then checks the quality of the predicted domain at that level. This evaluation process stops when all the predicted domains receive good quality scores.
Network flow representation of the domain partition problem
The domain decomposition problem has been formulated as a network flow problem (13). In this formulation, each residue is represented as a node of a connected network and each residue–residue contact is represented as an edge if the distance between the atoms is within a cut-off value. In the current DomainParser program, the cut-off is set at 4 Å (9,13). The capacity of an edge is defined by the type of the interaction between the two involved residues (13). We use the following function to assign the capacity of an edge c(u,v) between residues u and v:
c(u,v) = ku,v + kbu,vωb + kβu,vωβ + keu,vωe1
where ku,v is the number of atom–atom contacts between residues u and v. kbu,v is the number of backbone–backbone atom contacts between residues u and v, which adds extra weights to backbone–backbone atom contacts. kβu,v is used to prevent a β-sheet from being decomposed into different domains. kβu,v = 1 if u and v form a backbone–backbone hydrogen bond across a β-sheet, otherwise it is 0. keu,v = 1 if u and v belong to the same β-strand, or it is 0. ωb, ωβ, and ωe are scaling factors. The values of ωb and ωβ are obtained through training (ωb = 5; ωβ = 12) and ωe (= 1000) is determined arbitrarily as described (13).
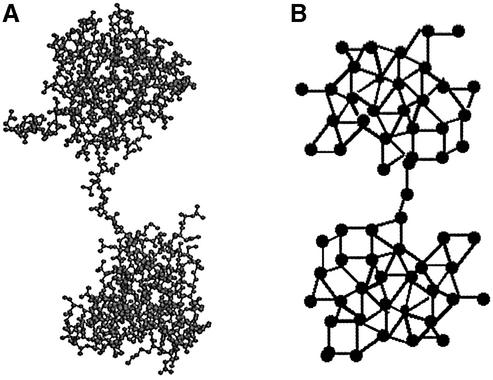
Each interface between two domains is modeled as a minimum flow cut (or bottleneck) of the network. For proteins with multiple domains, our program runs on each of the partitioned substructures recursively until the appropriate stopping criteria are met. Figure 1 shows an example of a two-domain protein and a schematic of its network representation.
Figure 1.
Protein structure of 2bb2 (β B2-crystallin) and its schematic representation of a flow network. (A) The ball–stick representation of the protein structure. (B) Schematic representation of the flow network of 2bb2.
To find the minimum cut or the bottleneck of a flow network, we need to find a connection between two subunits where the total edge capacity across it is minimized. This minimum cut problem can be solved efficiently by finding the maximum flow of the network with the Ford–Fulkerson algorithm (16). Using a representation by Picard and Queyranne (19), we can efficiently enumerate all minimum cuts of the network.
Stopping criteria and initial evaluation
In our current implementation, the stopping criteria are defined as follows. (i) A domain should have at least 35 residues. (ii) A β-sheet with more than two residues in each strand should belong to only one domain, which generally keeps a β-sheet intact; at most one β-strand can be cut at the interface between two domains. (iii) A domain must be compact enough to satisfy the following condition (9,13):
(∑i,jPi,j)/na ≥ gm2
where i and j represent any two atoms separated by at least three residues on the sequence; Pi,j = 1 if the distance between i and j is 4 Å or less, otherwise Pi,j = 0; na is the number of atoms in the domain. (iv) The interface between two domains should be small enough to satisfy
(∑inter-domainPi,j)/(∑intra-domainPi,j) ≤ fm3
(v) The number of segments and the number of residues in a domain, D, should satisfy the following:
[r(D)]/[s(D)] ≥ ls4
where r(D) and s(D) are the numbers of residues and segments in a domain D, respectively. The original values of gm, fm and ls (0.54, 0.52 and 35) were obtained through training on a small set of proteins (13). In our current implementation, the following values are adopted: gm = 0.52; fm = 0.56. These values are chosen to reduce the number of undercut.
Hydrophobic moment profile analysis
The hydrophobic moment profile of each partitioned domain is analyzed using the method of Silverman (18) with some modifications for our specific application. The Eisenberg hydrophobicity consensus scale is used for the calculation (arginine –1.76, lysine –1.1, aspartic acid –0.72, glutamine –0.69, asparagine –0.64, glutamic acid –0.62, histidine –0.4, serine –0.26, threonine –0.18, proline –0.07, tyrosine 0.02, cysteine 0.04, glycine 0.16, alanine 0.25, methionine 0.26, tryptophan 0.37, leucine 0.53, valine 0.54, phenylalanine 0.61, isoleucine 0.73) (20). The average hydrophobicity per residue [Hd0(d)] and the value of the second-order moment per residue [Hd2(d)] collected within a spherical surface with increasing distance d from the centroid of a protein domain is calculated as described by Silverman except that instead of an ellipsoidal representation of the protein, a spherical representation is employed.
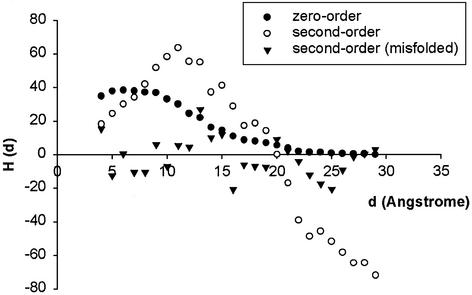
Figure 2 shows an example of profiling one protein structure, 2i1b. For a native protein structure, the second-order moment, [Hd2(d)], increases to a peak value then decreases before becoming negative. The zero-order moment, [Hd0(d)], has a similar profile except that it vanishes around zero.
Figure 2.
The zero- and second-order spherical moment profiles of the protein 2i1b and the second-order spherical moment profile of the decoy structure of 2i1b. The zero-order moment shown has been multiplied by 30.
To calculate the hydrophobic ratio Rt = d–/d0, we adopt a different protocol from the one used by Silverman. In Silverman’s approach, d– is chosen when all the values of the second-order moment per residue [Hd2(d)] are negative at larger values of d compared to d– and hydrophobic ratios cannot be assigned to any decoy structures or other non-native structures since they have irregular second-order moment profiles, as shown in Figure 2. For our purpose to use neural networks, we assign an Rt value for each partitioned domain along with three other parameters describing the second-order moment profile. The d– value is chosen after the second-order moment per residue turns negative. For native protein structures, our protocol does not affect the Rt value since in most cases [Hd2(d)] only intercepts with the x-axis once. For a decoy protein or an overcut protein domain, the hydrophobic moment profile generally fluctuates a lot and may intercept the x-axis multiple times. In our study, three additional parameters, Md, Ud and Od, are calculated. Md represents the number of points of [Hd2(d)] that are positive after it becomes negative for the first time. Ud is a measure of the number of intercepts with the x-axis of the moment profile, which reflects the irregularity of the profile. For a native protein, zero is expected for Ud, while it could be any positive number for a decoy or an overcut protein domain. Od reflects the relationship between the total number of residues and the maximum value of d of a protein structure. These three parameters are used for the neural network trained to distinguish native from non-negative (including decoy and incorrectly cut domains) structures.
Other parameters related to domain partition and the resulting domains
In addition to the above parameters, we have used five other parameters to help assess the quality of a predicted domain. They include Sd, Fd, Cd, Id and Td for each partitioned domain. Sd is the size of a single domain in terms of the number of residues; Fd is the number of segments in a partitioned domain; Cd represents the compactness of a partitioned domain; Id is a measure of the ‘size’ of a domain interface relative to the ‘volume’ of the domain as discussed earlier. The last parameter, Td, is a measure of the relative motion between compact domains, which is decided by the strength of non-bonded interactions across the interface (21).
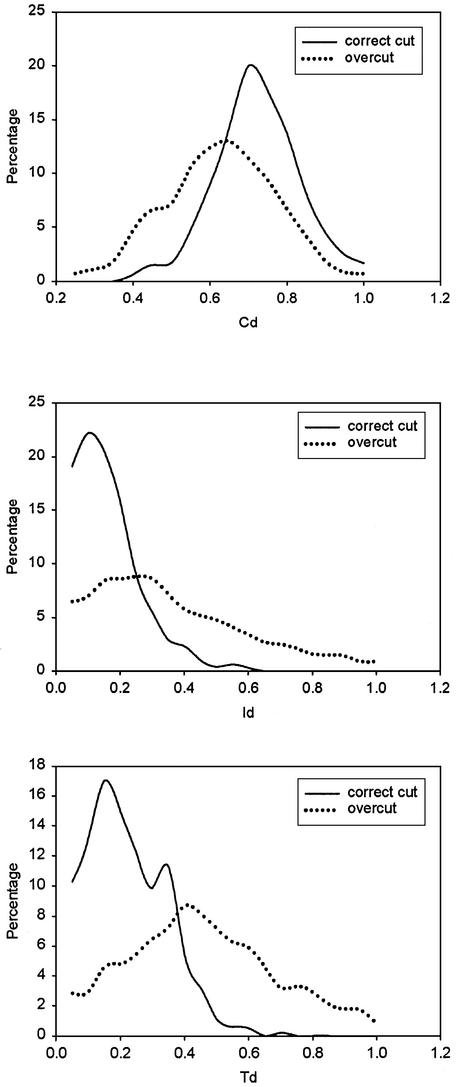
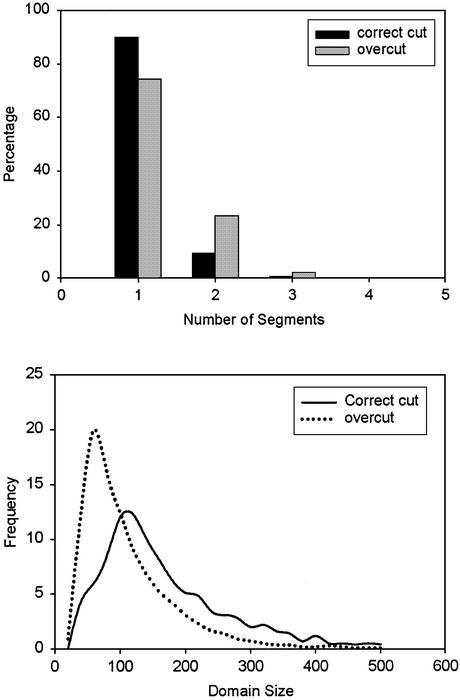
The parameters from good domain partitions and bad partitions have different distributions, as shown in Figures 3 and 4. The generation of the results for incorrect domain partition is described in the next section. It is obvious that domains resulting from overcut partitions have more fragments, higher Id and Td values and smaller Cd values. Domain size Sd is also an important parameter of a protein domain. Studies have shown that the sizes of protein domains have a narrow distribution (Fig. 4) (22).
Figure 3.
The distributions of the values of parameters Cd, Id and Td in well-partitioned domains and overcut domains. (Top) Cd, compactness of a domain. (Middle) Id, interface size relative to the domain volume. (Bottom) Td, relative motion between domains of each partition.
Figure 4.
Domain size and the number of segments of each domain in well-partitioned and overcut domains. (Top) Distributions of the number of segments. (Bottom) Distributions of domain sizes.
Neural network training and performance evaluation
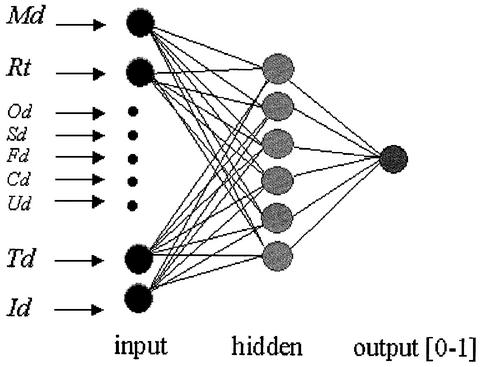
Figures 2–4 show that the parameters have different distributions between correctly partitioned domains and incorrectly partitioned domains. We have trained a neural network to separate such correct from incorrect domain partitions. Our training set consists of 633 correctly partitioned domains and 928 incorrectly partitioned domains from a total of 1619 protein structures. The correct domain partitions are based on the annotations of the SCOP database. The incorrect domain partitions (overcut domains) are generated using the old version of DomainParser, which can specify the number of domains to be decomposed. The number of domains used for DomainParser to generate incorrect domain partitions is larger than the number of domains assigned by SCOP. For training, each partitioned domain (correct or incorrect) is represented as a vector of nine values of the parameters described above, plus a categorical value to indicate which classes the domain belongs to. We have divided all partitioned domains into two categories: correctly cut (represented as 1) and overcut (represented as 0). A domain is considered as ‘correctly cut’ if it is at least 85% in consistency with the annotated result in SCOP. Our neural network has nine input nodes, for the nine parameter values respectively, and one output node representing the category a predicted domain belongs to (Fig. 5).
Figure 5.
Neural network architecture for evaluation of decomposed individual domains. This network has nine input nodes, six hidden nodes and one output node. The nine input parameters are shown on the left.
Several neural network architectures were trained and tested using SNNS (Stuttgart Neural Network Simulator) version 4.3. Various numbers of the hidden nodes and learning functions, including the standard back-propagation learning algorithm, were tested. A total of 20 different networks with different architectures have been trained and evaluated. The best performing network architecture is the one with one hidden layer of six nodes along with nine input nodes and one output node, as shown in Figure 5. RPROP, which stands for ‘resilient propagation’ (23), was selected as the learning function. The RPROP algorithm was developed to overcome the inherent disadvantages of pure gradient-descent. It performs a local adaptation of the weight updates according to the behavior of the error function. The RPROP algorithm generally provides faster convergence than most other algorithms. SNNS generates a C code of the trained neural network, which was included as a subroutine in the new DomainParser program.
Confidence estimation
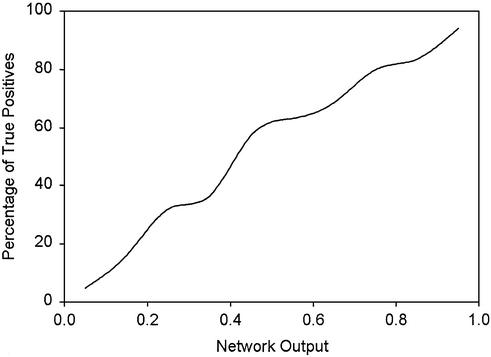
One of the most important evaluation criteria on the partitioned domains is their reliability. The output from the neural network is a value between 0 and 1, which does not imply its direct use as a probability value. To better estimate the reliability of the results based on the output of the neural network, the same testing and training set for the neural network was used to assign the confidence to various output levels from the network. The results from both the training and testing sets were combined and sorted by the network output value. The frequency of true positives was obtained for different ranges. The confidence of each partitioned domain was determined as the frequency of true positives as described in Materials and Methods. Figure 6 shows the confidence level of different ranges of the network output.
Figure 6.
The frequency of true positive assignments plotted against the output of the neural network.
RESULTS
Test set selection
A list of 3242 protein chains was selected from the latest release (June 16, 2002) of FSSP (24). These protein chains share <25% sequence identity when aligned with each other. The SCOP domain database was used as reference domain definitions because it is defined by human experts and therefore more reliable (3). The SCOP protein domain database (release 1.59, May 2002) was searched for the 3242 protein chains, which resulted in a list of 2936 protein chains that have domain definitions in SCOP. Some of the protein chains are defined as multi-domain folds by SCOP without boundary assignments. Among the 2936 protein chains, 2002 are single-domain chains, 720 are two-domain proteins, 146 are three-domain proteins and 68 have more than three domains. Then 1619 of the 2936 protein chains were selected for neural network training. The rest were used for testing. The testing set included 860 single-domain proteins, 330 two-domain proteins, 99 three-domain proteins and 28 four-domain proteins (see Table 3).
Table 3. Domain assignment using different programs.
| SCOP | DomainParser (old version) | DomainParser (new version) | |
|---|---|---|---|
| Single-domain | 860 | 752 | 800 |
| Two-domain | 330 | 182 | 218 |
| Three-domain | 99 | 38 | 52 |
| Four-domain | 28 | 9 | 8 |
Neural network performance
The neural net architecture shown in Figure 5 was used with the learning function of RPROP. On both the training and the testing data, the selected network achieved an accuracy of ∼86% (Table 1). The advantage of using SNNS v.4.3 is that the network chosen can be easily converted to a C function and incorporated into the main program.
Table 1. Neural network performance.
| Sets | Category | Performance |
|---|---|---|
| Training (1561 domains) | Correct cut | 87.6% |
| Overcut | 86.0% | |
| Testing (1677 domains) | Correct cut | 85.1% |
| Overcut | 86.0% |
New DomainParser performance
Using default parameters, we tested the performance of the previous version of DomainParser on the test set of 1317 protein chains. According to the SCOP protein domain definitions, 860 protein chains are single-domain proteins, 330 are two-domain proteins, 99 are three-domain proteins and 28 have four domains (see Table 3). For the purpose of comparison, we used the definition of ‘correct’ by Jones et al. (14). The domain decomposition is considered as ‘correct’ if the number of decomposed domains is the same and the residue assignment is at least 85% in agreement with the domain structure reported. DomainParser correctly assigned 752 single-domain protein chains and 229 multi-domain protein chains (Table 3). Among the incorrect decompositions, 224 domains are overcut, 94 are undercut. The overall accuracy is ∼74.5% (Table 2).
Table 2. Comparison of the domain decomposition between old and new DomainParser.
| DomainParser (old version) | DomainParser (new version) | |
|---|---|---|
| Overcut | 224 | 110 |
| Undercut | 94 | 109 |
| <85% assignment | 18 | 20 |
| Correct assignment | 74.5% | 81.9% |
Upon applying the quality checking ability with the neural network to DomainParser, 800 single-domain protein chains and 278 multi-domain protein chains were assigned correctly (Table 3). The overall accuracy improved to 81.9 from 74.5% (Table 2). The results are available at http://compbio.ornl.gov/structure/domainparser2/testing.html. The old version of DomainParser tends to overcut protein chains into domains using default parameters. The number of overcut is reduced substantially from 17 to 8.4% with the quality check capability. The number of undercut is only slightly increased from 7.1 to 8.3%.
We also tested the performance of the new version of DomainParser on the set of 55 proteins used by several programs (13,14). Among the 55 proteins, 30 are single-domain proteins, 20 are two-domain proteins, two are three-domain proteins and three are four-domain proteins. The performance of the new version of DomainParser on this set is 80%, while the old version has an accuracy of 78.2% (13) according to the domain assignments from the literature. The new version correctly assigned 28 one-domain proteins out of 30, while the old one missed three of them (13). On the other hand, both the old and the new versions of DomainParser assigned 16 multi-domain proteins correctly (Table 4) (13). However, if we use the protein domain definitions by SCOP as references, the performance of our new program is 83.6% (Table 4).
Table 4. Protein PDB codes, residue ranges of domains assigned by the literature (‘/’ is used to separate domains), residue ranges of domains assigned by the old version of DomainParser, residue ranges of domains assigned by the new version of DomainParser and the definition by the SCOP domain database.
| Protein | Literature | DomainParser (old version) | DomainParser (new version) | SCOP |
|---|---|---|---|---|
| 2 domains | ||||
| 1ezm | 1–134/135–298 | 1–133/134–298 | 1–133/134–298 | 2 domain |
| 1fnb | 19–161/162–314 | 19–152/153–314 | 19–159/160–314 | 19–154/155–314 |
| 1gpb | 19–489/490–841 | 19–63/64–484;828–841/558–648; 712–792/485–557;649–711;793–827 | 19–484;828–841/485–827 | 2 domain |
| 1lap | 1–150/171–484 | 1–173/174–484 | 1–147/148–484 | 1–159/160–484 |
| 1pfka | 0–138;251–301/139–250;302–319 | 0–137;254–319/138–253 | 0–137;254–303/138–253;304–319 | 2 domain |
| 1ppn | 1–10;112–208/21–111;209–212 | 1 domain | 1 domain | 1 domain |
| 1rhd | 1–158/159–293 | 1–63;74–157/64–73;158–293 | 1–157/158–293 | 1–149/150–293 |
| 1sgt | 22–123;234–245/129–233 | 1 domain | 1 domain | 2 domain |
| 1vsga | 1–29;92–251/42–75;266–362 | 1–32;86–255/33–85;256–362 | 1–32;86–255/33–85;256–362 | 1 domain |
| 1bksb | 9–52;86–204/53–85;205–393 | 90–189/9–89;190–393 | 90–189/3–89;190–394 | 2 domain |
| 2cyp | 3–145;266–294/164–265 | 2–144;273–294/145–272 | 2–144;273–294/145–272 | 2 domain |
| 2had | 1–155;230–310/156–229 | 1 domain | 1 domain | 1 domain |
| 3cd4 | 1–98/99–178 | 1–98/99–178 | 1–98/99–178 | 1–97/98–178 |
| 1g6na | 1–129/139–208 | 1 domain | 1 domain | 7–137/138–206 |
| 3pgk | 1–185;403–415/200–392 | 0–188;402–415/189–401 | 0–188;402–415/189–401 | 2 domain |
| 4gcr | 1–83/84–174 | 1–83/84–174 | 1–83/84–174 | 1–85/86–174 |
| 5fbpa | 6–201/202–335 | 1 domain | 1 domain | 2 domain |
| 8adh | 1–175;319–374/176–318 | 1–173;321–374/174–320 | 1–173;321–374/174–320 | 1–174;325–374/175–324 |
| 8atca | 1–137;288–310/144–283 | 1–130;292–310/131–291 | 1–130;292–310/131–291 | 1–150/151–310 |
| 8atcb | 8–97/101–152 | 8–97/101–153 | 8–97/101–153 | 8–100/101–153 |
| 3 domains | ||||
| 1phh | 1–175/176–290/291–394 | 32–124/180–268/1–31;125–179;269–394 | 1 domain | 1–173;276–394/174–275 |
| 3grs | 8–157;294–364/158–293/365–478 | 8–161;290–368/162–289/369–478 | 18–161;290–368/162–289/369–478 | 18–165;291–363/166–290/364–478 |
| 4 domains | ||||
| 1atna | 1–32;70–144; 338–372/33–69/145–180;270–337/181–269 | 0–33;97–147;337–372/34–96/148–180;273–336/181–272 | 0–147;337–372/148–336 | 1–146/147–372 |
| 3pmga | 1–188/192–315/325–403/408–561 | 1–188/189–303/304–406/407–561 | 1–190/191–303/304–406/407–561 | 1–190/191–303/304–420/421–561 |
| 8acn | 2–200/201–317/320–513/538–754 | 2–530/531–754 | 2–530/531–754 | 2–528/529–754 |
Eight protein chains are defined as two-domain folds without boundary assignments.
We further tested our program on a non-redundant set of 90 protein chains with author assignments maintained at http://www.bmm.icnet.uk/∼domains/test/dom-rr-all-c.html by Islam et al. (15), and achieved an accuracy of 90%. Among the 90 proteins, each of which represents a distinct fold in FSSP (24), 23 are multi-domain proteins. The result is available at http://compbio.ornl.gov/structure/domainparser2/islamset.html.
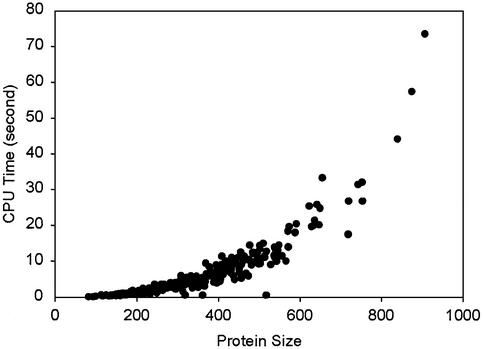
Even though new capabilities were added to DomainParser, it runs as efficiently as DomainParser alone. Figure 7 shows the computational time on 278 protein chains that resulted in two domains. It took less than 25 s of CPU time to complete the decomposition for all the protein chains that have 600 residues or less (Fig. 7). It may take a little longer for the new DomainParser since the initial partition may result in more than two domains.
Figure 7.
CPU time of running new DomainParser as a function of the number of residues for 278 two-domain chains in FSSP.
General functionality of DomainParser
DomainParser takes a protein structure in PDB (25) format as input and generates one set of domains. In most cases, running DomainParser using defaults should give satisfactory partitions. For each predicted domain, DomainParser provides a value indicating the confidence. In addition, several options offered in DomainParser can provide a partition that a user desires or correct some overcut/undercut partitions. For example, if a user has some knowledge about the possible number of domains a protein chain may have, he/she can input this number, k, as a constraint. DomainParser will generate the best k-domain partition. The user can also define the minimum number of residues (r) in a domain (default r = 35) and the maximum number of β-strands (b1) that allow bridging two different domains (default b1 = 1).
DISCUSSION
On the test set of 1317 protein chains, the new DomainParser program performed significantly better than the previous version. About 81.9% of the results from new DomainParser are in agreement with the SCOP assignments, while the old one achieved 74.5% consistency.
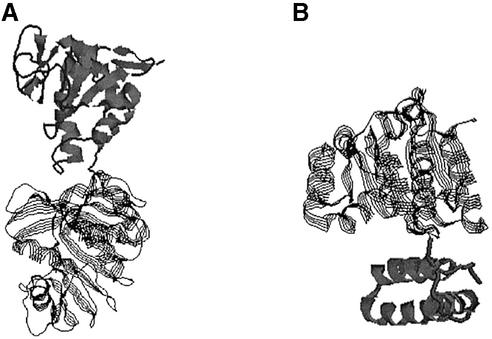
Among the assignments inconsistent with SCOP, some may not necessarily indicate the inaccuracy of the assignments by DomainParser. The discrepancies may simply be the result of the lack of a standard definition of a domain, as discussed by several studies (11,12,26). Manual assignments could sometimes be subjective, as evidenced by the inconsistent domain assignments for the same protein by different experts (13). We further examined the protein chains in our test set that do not agree with the assignments by SCOP. These proteins fall into several groups. Among the 110 protein chains that are overcut by our program with the SCOP definitions as standard, we believe that the majority are decomposed correctly by DomainParser from the view point of structural domains. Figure 8 shows several examples. DomainParser assigns two domains for both 1hzda (74–279 and 280–339) and 2adma (21–243 and 244–413), while SCOP assigns them as single-domain proteins. Clearly, they are multi-domain proteins from a structural point of view. It is not clear why SCOP lists them as single-domain proteins. One possible explanation is that experts take the functions of proteins and evolution information into consideration when they examine the protein structures for domain identifications (27). Usually, active sites are located in the clefts between domains (9). In these cases, one structural domain may not be a full functional domain. Several proteins defined as two-domain proteins by SCOP are assigned as three- or four-domain proteins by DomainParser. Though the domain numbers are different, they do agree on some partitioned domains. Holm and Sander (21) argued that there could be alternative, equally reasonable ways to decompose a large protein into smaller domains.
Figure 8.
Domain decompositions of 2adma and 1hzda by DomainParser. SCOP assigns both 2adma and 1hzda as single-domain proteins. The thick ribbons and thin strands show different domains. (A) 2adma (21–243/244–413). (B) 1hzda (74–279/280–339).
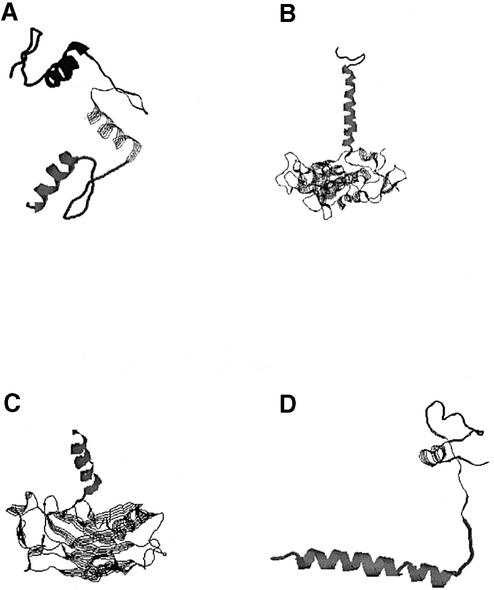
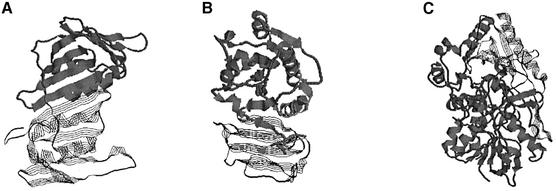
Some discrepancies in the undercut category are the results of the rules used by DomainParser and other domain decomposition programs. A compact, near globular structure with a hydrophobic core is expected for a protein domain. Figure 9 shows four examples, which are assigned as multiple domains by SCOP but single-domain structures by DomainParser. Domains 1–36 of 6prch, 31–66 and 67–100 of 1zmec and 334–349 of 1d0ab, which are separated domains in SCOP, are too simple to be treated as single domains (some of them are just an α-helix) and are rejected by DomainParser, on the basis of the rules discussed above and the minimum size requirement of 35 amino acids (see Materials and Methods). Using recurrence information to divide the polypeptide chain into domains, Holm and Sander found that a single long helix is one of the most dominant domain fold types (21). However, this is not consistent with the general rule of domain decomposition, i.e. each domain should be in general compact and globular. In almost all the undercut cases, if we require that the number of decomposed domains is two or more (one of the options provided by DomainParser), the results are in high agreement with the SCOP assignments. One of the rules DomainParser uses is that a β-sheet can belong to only one domain, which resulted in the assignments of 1plq and 1ig3a as single-domain protein chains. Apparently, this rule is not strictly applied in the SCOP assignments. SCOP assigns two domains (1–126 and 127–258) to 1plq. However, residues 46–53 in one domain and residues 244–250 in the other domain are parts of a β-sheet (Fig. 10). By relaxing this rule, DomainParser assigns the domains of this protein in consistency with SCOP. In protein chain 1ig3a that has two domains defined by SCOP (10–178 and 179–263), two β-sheets are split since strands 172–177 and 179–184 and strands 22–25 and 189–193 belong to different domains. Some of the domain assignments by SCOP, which are undercut by DomainParser into single-domain proteins, are structurally close. The two domains assigned by SCOP for 1bs8a (319–450 and 9–318;451–506) are very close (Fig. 10C). The above discrepancies are apparently the result of how a ‘domain’ is defined. A reasonable comparison of the performance of automatic domain partition programs is possible only when we have a rigorous definition of a protein domain.
Figure 9.
Single α-helix and simple structures are assigned as separate domains by SCOP. (A) 1aaya (103–131/132–159/160–187). (B) 6prch (1–36/37–258). (C) 1d0ab (150–501/334–349). (D) 1zmec (31–66/ 67–100). DomainParser defines them as single-domain proteins.
Figure 10.
Domain assignments of 1plq, 1ig3a and 1b8sa by SCOP. All three are defined as two-domain proteins. Thick ribbons and thin strands show different domains assigned by SCOP. (A) 1plq (1–126/127–258). (B) 1ig3a (179–263/10–178). (C) 1b8sa (319–450/9–318;451–506).
Since we used the hydrophobic moment profile to assess the quality of partitioned domains, and we are dealing with individual protein chains, some domains that are decomposed correctly by DomainParser are missed by the quality checking step. This could be the result of the unique properties of those proteins. Some protein structures are solved with co-factors or hetero molecules, which may affect the hydrophobic moment profiling of the partitioned domains.
All these discrepancies between our assignments and SCOP could provide us with ideas for possible improvements of DomainParser. Incorporation of functional and evolutionary information may be the next step to take in generating the next version of DomainParser. With its efficient and accurate computation, we believe that the program should become a valuable tool for automated domain decomposition at large scale, which will be particularly useful for keeping the domain databases up to date in a timely fashion. We plan to construct a domain database using DomainParser to complement other protein domain databases.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Victor Olman, Manesh Shah and Kyle Ellrott for their help in many ways. This work was supported by the Office of Biological and Environmental Research, US Department of Energy, under contract DE-AC05-00OR22725 managed by UT-Battelle, LLC.
REFERENCES
- 1.Wetlaufer D.B. (1973) Nucleation, rapid folding and globular intrachain regions in proteins. Proc. Natl Acad. Sci. USA, 70, 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson J.S. (1981) The anatomy and taxonomy of protein structure. Adv. Protein Chem., 34, 167–339. [DOI] [PubMed] [Google Scholar]
- 3.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 4.Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) CATH—a hierarchic classification of protein domain structures. Structure, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- 5.Jones D.T. (1999) GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol., 287, 797–815. [DOI] [PubMed] [Google Scholar]
- 6.Kelley L.A., MacCallum,R.M. and Sternberg,M.J. (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol., 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y. and Xu,D. (2000) Protein threading using PROSPECT: design and evaluation. Proteins, 40, 343–354. [PubMed] [Google Scholar]
- 8.National Institute of General Medical Sciences (1999) Pilot Projects for the Protein Structure Initiative (Structural Genomics). National Institute of General Medical Sciences, Bethesda, MD.
- 9.Holm L. and Sander,C. (1994) Parser for protein folding units. Proteins, 19, 256–268. [DOI] [PubMed] [Google Scholar]
- 10.Siddiqui A.S. and Barton,G.J. (1995) Continuous and discontinuous domains: an algorithm for the automatic generation of reliable protein domain definitions. Protein Sci., 4, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swindells M.B. (1995) A procedure for detecting structural domains in proteins. Protein Sci., 4, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wernisch L., Hunting,M. and Wodak,S.J. (1999) Identification of structural domains in proteins by a graph heuristic. Proteins, 35, 338–352. [PubMed] [Google Scholar]
- 13.Xu Y., Xu,D. and Gabow,H.N. (2000) Protein domain decomposition using a graph-theoretic approach. Bioinformatics, 16, 1091–1104. [DOI] [PubMed] [Google Scholar]
- 14.Jones S., Stewart,M., Michie,A., Swindells,M.B., Orengo,C. and Thornton,J.M. (1998) Domain assignment for protein structures using a consensus approach: characterization and analysis. Protein Sci., 7, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam S.A., Luo,J. and Sternberg,M.J. (1995) Identification and analysis of domains in proteins. Protein Eng., 8, 513–525. [DOI] [PubMed] [Google Scholar]
- 16.Ford L.R. and Fulkerson,D.R. (1962) Flows in Networks. Princeton University Press, Princeton, NJ.
- 17.Eisenberg D., Weiss,R.M. and Terwilliger,T.C. (1982) The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature, 299, 371–374. [DOI] [PubMed] [Google Scholar]
- 18.Silverman B.D. (2001) Hydrophobic moments of protein structures: spatially profiling the distribution. Proc. Natl Acad. Sci. USA, 98, 4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picard J.C. and Queyranne,M. (1980) On the structure of all minimum cuts in a network and applications. Math. Programming Study, 13, 8–16. [Google Scholar]
- 20.Eisenberg D., Weiss,R., Terwilliger,T. and Wilcox,W. (1982) Hydrophobic moments and protein structure. Faraday Symp. Chem. Soc., 17, 109–120. [Google Scholar]
- 21.Holm L. and Sander,C. (1998) Dictionary of recurrent domains in protein structures. Proteins, 33, 88–96. [DOI] [PubMed] [Google Scholar]
- 22.Wheelan S.J., Marchler-Bauer,A. and Bryant,S.H. (2000) Domain size distributions can predict domain boundaries. Bioinformatics, 16, 613–618. [DOI] [PubMed] [Google Scholar]
- 23.Riedmiller M. and Braun,H. (1993) A direct adaptive method for faster backpropagation learning: the RPROP algorithm. In Proceedings of the International Conference on Neural Networks. IEEE, San Francisco, CA, pp. 586–591.
- 24.Hobohm U., Scharf,M., Schneider,R. and Sander,C. (1992) Selection of representative protein data sets. Protein Sci., 1, 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.E.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- 26.Taylor W.R. (1999) Protein structural domain identification. Protein Eng., 12, 203–216. [DOI] [PubMed] [Google Scholar]
- 27.Lo Conte L., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]