Abstract
Polyubiquitination marks proteins for degradation by the 26S proteasome and is carried out by a cascade of enzymes that includes ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s). The anaphase-promoting complex or cyclosome (APC/C) comprises a multisubunit ubiquitin ligase that mediates mitotic progression. Here, we provide evidence that the Saccharomyces cerevisiae RING-H2 finger protein Apc11 defines the minimal ubiquitin ligase activity of the APC. We found that the integrity of the Apc11p RING-H2 finger was essential for budding yeast cell viability, Using purified, recombinant proteins we showed that Apc11p interacted directly with the Ubc4 ubiquitin conjugating enzyme (E2). Furthermore, purified Apc11p was capable of mediating E1- and E2-dependent ubiquitination of protein substrates, including Clb2p, in vitro. The ability of Apc11p to act as an E3 was dependent on the integrity of the RING-H2 finger, but did not require the presence of the cullin-like APC subunit Apc2p. We suggest that Apc11p is responsible for recruiting E2s to the APC and for mediating the subsequent transfer of ubiquitin to APC substrates in vivo.
INTRODUCTION
Protein ubiquitination is accomplished through a complex process involving ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and, in some cases, specificity-conferring ubiquitin ligases (E3s) (Hershko and Ciechanover, 1998). Ubiquitin ligases are loosely defined as proteins or protein complexes that mediate the transfer of ubiquitin from E2s to substrates. Some E3s, such as the HECT (homologous to E6AP C-terminus) domain family, have been shown to interact with E2s and to form thioester linkages with ubiquitin before modifying substrates (Huibregtse et al., 1995; Scheffner et al., 1995). Others do not act as ubiquitin carriers, but are proposed to function by bringing E2s into proximity with substrates and by providing a favorable environment for the transfer of ubiquitin. Such E3s thus serve as bridging molecules, whose main role is to confer substrate specificity.
The anaphase-promoting complex or cyclosome (APC/C) comprises a multisubunit E3, which likely belongs to the latter class. Targets of APC/C include securin proteins (Pds1p in budding yeast and Cut2p in fission yeast), whose destruction is required for the separation of sister chromatids at anaphase (Cohen-Fix et al., 1996; Funabiki et al., 1996; Ciosk et al., 1998), and mitotic cyclins, such as Clb2p, which must be destroyed before mitotic exit (Glotzer et al., 1991; Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995). APC activity must, therefore, be tightly regulated to ensure that the timing and order of mitotic events are strictly maintained. The work of several groups has shown that APC activity is regulated at multiple levels, including APC subunit phosphorylation (King et al., 1995; Lahav-Baratz et al., 1995; Peters et al., 1996; Kotani et al., 1998; Shirayama et al., 1998; Kotani et al., 1999), association with substrate-selective WD40 repeat activator proteins such as Cdc20p and Hct1p/Cdh1p (Schwab et al., 1997; Visintin et al., 1997), and the spindle assembly checkpoint, which ensures that duplicated chromosomes are properly aligned and attached to spindles before being separated at anaphase (Li et al., 1997; He et al., 1997; Hwang et al., 1998; Kim et al., 1998; Fang et al., 1998).
While much has been learned about the regulation of APC activity, little is known about the biochemical roles played by its individual subunits. The budding yeast complex is perhaps the best characterized and comprises at least 12 subunits (Peters, 1999). Cdc16p/APC6, Cdc23p/APC8, Cdc27p/APC3, and APC7 were identified as tetratricopeptide repeat (TPR)- containing proteins, which function in the nucleus (Sikorski et al., 1990; Lamb et al., 1994; Lamb et al., 1995). Cdc16p, Cdc23p, and Cdc27p have been shown to associate in vivo and may form the stable core of the assembly (Lamb et al., 1994). Like the Skp1/Cdc53/cullin/F box (SCF) E3 complex, which functions in the G1 to S transition, the APC contains a cullin-like subunit, Apc2p (Yu et al., 1998; Zachariae et al., 1998; Kramer et al., 1998) and a small, RING-H2 finger subunit, Apc11p (Zachariae et al., 1998; Ohta et al., 1999; Tan et al., 1999; Kamura et al., 1999a; Seol et al., 1999). Together, the cullin and RING-H2 subunits have been proposed to form the catalytic core of APC and SCF complexes (Seol et al., 1999). RING finger proteins have been implicated in diverse processes (Freemont, 1993), and several members of this family, including BRCA1, c-Cbl, and Bmi-1, are known tumor suppressors or proto-oncoproteins (Langdon et al., 1989; van Lohuizen et al., 1991; Miki et al., 1994). The RING-H2 domain, defined as C1XXC2X(12–35)C3XH1XXH2XXC4X(8–39)C5XXC6, consists of a cluster of cysteine and histidine residues that chelate two atoms of zinc. The metal-stabilized “cross-brace” motif so formed is thought to provide a scaffold for intermolecular interactions, and indeed several RING finger family members have been found in large, multiprotein complexes (Borden and Freemont, 1996).
Recently, RING finger proteins have come to prominence for their role in ubiquitin-mediated protein degradation. The c-Cbl RING finger was recently shown to mediate E2-dependent ubiquitination and has been proposed to mediate the down-regulation of activated growth factor receptors by promoting their ubiquitin-mediated proteolysis (Joazeiro et al., 1999; Waterman et al., 1999; Lee et al., 1999; Yokouchi et al., 1999; Levkowitz et al., 1999). Lorick and coworkers demonstrated that several RING finger family members, including AO7, NF-X1, and BRCA1, can bind directly to E2s and mediate E2-dependent ubiquitin transfer (Lorick et al., 1999). We investigated whether the budding yeast RING-H2 finger protein Apc11 plays a direct role in APC-mediated ubiquitination.
MATERIALS AND METHODS
Yeast Strains Used in this Study
W303: MATa/α ura3–52/ura3–52 lys2–801/lys2–801 ade2- 101/ade2–101 leu2–3, -112/leu2–3, -112 trp1-Δ901/trp1-Δ901 his3–300, -Δ200/his3–300,-Δ 200
YAP140: MATa ura3–52, lys2–801, ade2–101, leu2-Δ1, trp1-Δ63, his3-Δ200, apc11::HIS3 pAP33 (pRS316-APC11)
YAP160: MATa ura3–52, lys2–801, ade2–101, trp1-Δ63, his3-Δ200, apc11::HIS3, leu2-Δ1::APC11;LEU2
YAP201: MATa ura3–52, lys2–801, ade2–101, trp1-Δ63, his3-Δ200, apc11::HIS3, leu2-Δ1::apc11–13;LEU2
YAP219: MATa ura3–52, lys2–801, ade2–101, trp1-Δ63, his3-Δ200, apc11::HIS3, leu2-Δ1::apc11–22;LEU2
YPH501: MATa/α ura3–52/ura3–52 lys2–801/lys2–801 ade2–101/ade2–101 leu2-Δ1/leu2-Δ1 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-D200
All of the strains used in this study, with the exception of W303, were in the S288c background.
DNA Constructs and Mutagenesis
A genomic fragment of APC11, including 317 bp of sequence upstream of the start codon and 320 bp downstream of the stop codon, was cloned into centromeric plasmid pRS314 using ClaI and NotI to create pAP28. APC11 was PCR amplified from pAP28 using the primers (5′-GGAATTCTAAAAGTTAAAATAAACGAAGTGCACAG-3′) and (5′-GAGCTCGAGTCGTAACAAAAAGTCTTCGTCCAGG-3′) and cloned using EcoRI and XhoI into pGEX-KG to create GST-APC11, and into pHis8–3 (Jez et al., 2000) to create His-APC11. pRS314-APC11-3HA constructs were constructed by ligating an ∼ 300 bp Tth111I/HpaI fragment from integration construct pWZV84, encoding 3 direct repeats of the hemagglutinin (HA) epitope tag in-frame with the Apc11p C-terminus, into pAP28 with the Tth111I/HpaI fragment removed. Mutagenesis of pAP28, GST-APC11, and pRS314-APC11-3HA was carried out using the QuikChange Site Directed Mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The sequence of each mutant was confirmed by automated sequencing (Applied Biosystems Inc., Foster City, CA).
For APC11Δ104–165, a cassette designed to introduce a stop codon after amino acid 103 in the APC11 open reading frame, followed by a His3MX6 module and APC11 3′ UTR flanking sequence, was PCR-amplified from pFA6a-3HA-His3MX6 using primers (5′- TGTCCGATGTGTAGGC-AAACTTTCCAGCTACAGAAGGGTTGAGGCG CGCCACTTCTAAA-3′) and (5′-GGAAATATAGCTAATTGTGATTTCTAAG TTTCTTTTTTAGAATTCGAGCT CGTTTAAAC-3′) (Longtine et al., 1998). The resulting product was transformed into YPH501. His+ transformants were isolated and screened for correct integration events by colony PCR using the primers (5′-GTGTTTGCTTGGTCATGGCAC-3′) and (5′-TGCAAGGATTGATAATGTAATAGG-3′). After integrants were sporulated on SPO medium, tetrads were dissected on YPD, grown at 25°C, and replica-plated to SC-His.
Generation of Temperature-sensitive APC11 Mutants and Complementation Assays
A 1.7 kb cassette containing APC11 under its own promoter and universal pRS vector sequence was PCR amplified from pAP28 with the primers (5′-CAAGTGTAGCGGTCACGC-3′) and (5′-CCCAATACGCAAACCGCC-3′), and cotransformed with BamHI-linearized pRS315 into an apc11::HIS3 shuffle strain (YAP140). Leu+ transformants were selected on SC-Leu at 25°C. To evict shuffle plasmid pAP33, transformants were replica-plated twice to complete media containing 1 mg/ml 5FOA, with 2 days of growth at 25°C between replicas. Surviving transformants were then replica-plated to SC-Leu plates and were grown at either 25°C or 37°C. Plasmid DNA was recovered from colonies that failed to grow at 37°C. The recovered plasmids were reshuffled into YAP140, and the resulting strains were tested for temperature sensitivity at 37°C, after eviction of pAP33 by multiple rounds of growth on 5FOA plates at 25°C. Sequence analysis of the recovered plasmids showed that the apc11–13 defect results from a single, nonconservative amino acid change (S10R). The apc11–22 mutant contains four nonconservative changes: F12S, K56R, E127K, and E128V, as well as single base changes in the 5′ and 3′ UTRs.
Cassettes containing mutant apc11 alleles were subcloned from the recovered plasmids into a LEU2 integrating vector (Δ-leu2 -LEU2) with XhoI and SpeI. The integrating constructs were then digested with NotI and transformed into YAP140. pAP33 was evicted from Leu+ transformants by two rounds of selection on 5FOA media, resulting in temperature-sensitive apc11 mutants integrated at the leu2 locus and marked with LEU2, covering a HIS3 deletion of the endogenous APC11 gene. The strains were then retested for temperature sensitivity at 37°C and amelioration of temperature sensitivity after introduction of a plasmid containing wild-type APC11 (pAP28).
For complementation assays, S. cerevisiae strain YAP201 was transformed using a standard lithium acetate procedure, streaked out to SC-Trp and incubated at 24°C. Single colonies were streaked out to SC-Trp and incubated at either the permissive (24°C) or restrictive (34°C) temperature.
Immunofluorescence and DAPI Staining
For immunofluorescence, cells were fixed in 37% formaldehyde and spheroplasted, before staining with 4, 6-diamidino-2-phenylindole (DAPI) or Yol 1–34 rat antitubulin antibody (Serotec, Oxford, UK) followed by fluorescein-conjugated goat antirat antibody (Cappel-ICN, Costa Mesa, CA). For flow cytometry, cells were fixed with 70% ethanol and 0.2 M Tris pH 7.5, according to standard protocols, and counted on a Becton-Dickinson FACSort (San Jose, CA).
Recombinant Proteins and GST Pull-Down Assays
Wild type or mutant GST-APC11 constructs were used to transform BL21/DE3 bacteria. Transformants were used to inoculate 50 ml cultures of TB/ampicillin, which were grown overnight at 37°C to stationary phase. A measure of 10 ml preculture was then used to inoculate 350 ml TB/ampicillin, plus 100 μM ZnSO4. The cultures were grown at 37°C to an OD600 of ∼ 0.6 before inducing with 0.4 mM IPTG for 3 h. Cell pellets were collected, resuspended in 9 ml lysis buffer (50 mM Tris-Cl pH 8.0, 120 mM NaCl, 1 mM DTT, plus 1 mM each PMSF and benzamidine, 20 μg/ml leupeptin, and 1.0 μg/ml aprotinin), and lysed by sonication. Triton X-100 was added to the lysates at a final concentration of 1% and left on ice for 20 min, before centrifugation at 10,000 rpm for 10 min (4°C). Cleared lysates were then bound to glutathione-agarose for 1.0 h, with rolling at 4°C. Beads were washed extensively with lysis buffer before eluting for 1.0 h in lysis buffer (pH 7.5) plus 20 mM glutathione. Eluted proteins were then dialyzed extensively against 20 mM Tris-Cl pH 8.0, 50 mM NaCl, 10% glycerol, and 1 mM DTT. GST pull-down assays were performed as described previously (Joazeiro et al., 1999). GST-Cbl RING finger was purified as described previously (Joazeiro et al., 1999).
His-tagged proteins were expressed in bacterial strain BL21/DE3, as described above. Cells were lysed by sonication in binding buffer (20 mM Tris-Cl pH 7.5, 100 mM NaCl, 10% glycerol, 10 μM ZnSO4) plus 1 mM imidazole. Cleared lysates were then bound to 1 ml bed volume Talon metal affinity resin (Clontech, Palo Alto, CA), washed with 10-bed volumes of binding buffer plus 10 mM imidazole, and eluted with 10-bed volumes of binding buffer plus 100 mM imidazole. Eluates were then concentrated in ubiquitination reaction buffer (see below) using Ultrafree-15 centrifugal filters (Millipore, Bedford, MA).
Immunoprecipitation Assays
Immunoprecipitation reactions were carried out as described previously (Lamb et al., 1994). After being immunoprecipitated with anti-HA monoclonal antibody 12CA5, equal amounts of wild-type and mutant Apc11pHA3 proteins were loaded for SDS-PAGE. Western blots were subsequently probed with either anti-HA monoclonal antibody 12CA5, anti-Cdc16p polyclonal antibody JHU855 (Lamb et al., 1994), or anti-Cdc27p polyclonal antibody JHU729 (Lamb et al., 1994).
In Vitro Ubiquitination Reactions
In vitro ubiquitination assays were carried out as described previously (Joazeiro et al., 1999), using bacterially expressed E1 and Ubc4. Approximately 3 μg of wild-type or mutant GST-Apc11p or His-Apc11p were incubated with 50–500 nM His-E1, 0.5–5 μM His-Ubc4 or His-Cdc34p, 10 μM bovine ubiquitin or GST-ubiquitin, and 2 mM ATP in reaction buffer (50 mM Tris-Cl pH 7.5, 2.5 mM MgCl2, and 0.5 mM DTT). Approximately 1 μg His-Clb2p-HA or His-Clb2pΔDB-HA were added to reactions as indicated. After 90 min at room temperature, reactions were stopped with 2× SDS buffer containing 4% SDS and 5.8 M β-mercaptoethanol, separated by SDS-PAGE, and analyzed by immunoblotting with either anti-GST monoclonal antibody SC-138 (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-HA monoclonal antibody 12CA5.
RESULTS
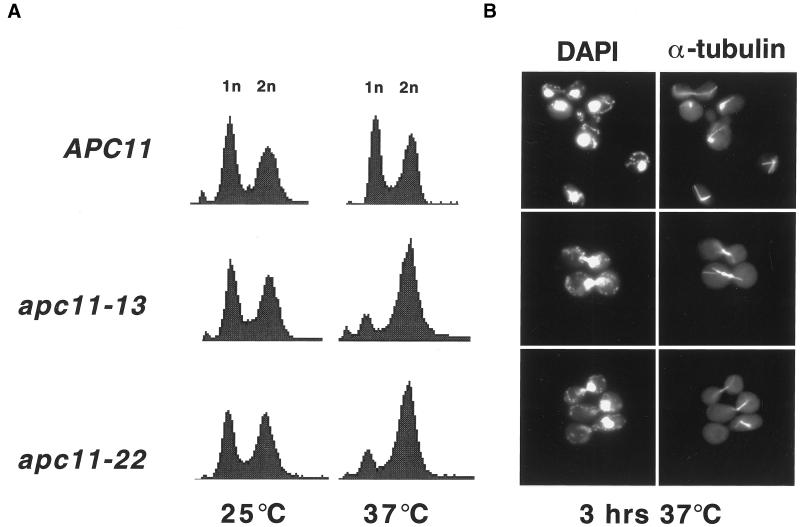
To investigate the role of the RING-H2 protein Apc11 in APC-mediated ubiquitination, we generated yeast strains bearing temperature-sensitive alleles of APC11 (see MATERIALS AND METHODS). Temperature sensitive strains apc11–13 and apc11–22 were grown to mid-log phase at 25°C, shifted to 37°C for 3 h, and analyzed by flow cytometry. At the restrictive temperature, apc11–13 and apc11–22 cells arrested with predominantly 2n DNA content, compared with an APC11 wild-type strain (Figure 1A). Immunofluorescence staining demonstrated that, for both mutants, >70% of the cells arrested with large buds, short mitotic spindles, and DAPI-staining masses at the bud-neck (Figure 1B). Using a strain that is temperature sensitive due to the addition of a 9 Myc tag to Apc11p, Zachariae and coworkers also reported that disrupting Apc11p function results in a mitotic arrest (Zachariae et al., 1998). The apc11–13 and apc11–22 arrest phenotypes are thus similar to those seen in other apc mutants (Irniger et al., 1995; Yu et al., 1998, Zachariae et al., 1998) and suggest that the defects result from a failure to degrade APC substrates.
Figure 1.
Apc11 Mutants arrest in G2/M as large-budded cells with short mitotic spindles. (A) Yeast strains bearing wild-type (APC11) or temperature-sensitive (apc11–13 and apc11–22) alleles of APC11 were grown to midlog phase at 25°C, shifted to 37°C for 3 h, and analyzed for DNA content by fluorescence-activated cell sorting (FACS). (B) Wild-type and apc11 ts cells incubated at 37°C for 3 h were analyzed by DAPI staining (left panels) or by immunofluorescence staining for mitotic spindles with antitubulin antibody (right panels).
The Apc11p RING-H2 Finger Is Essential for Cell Viability
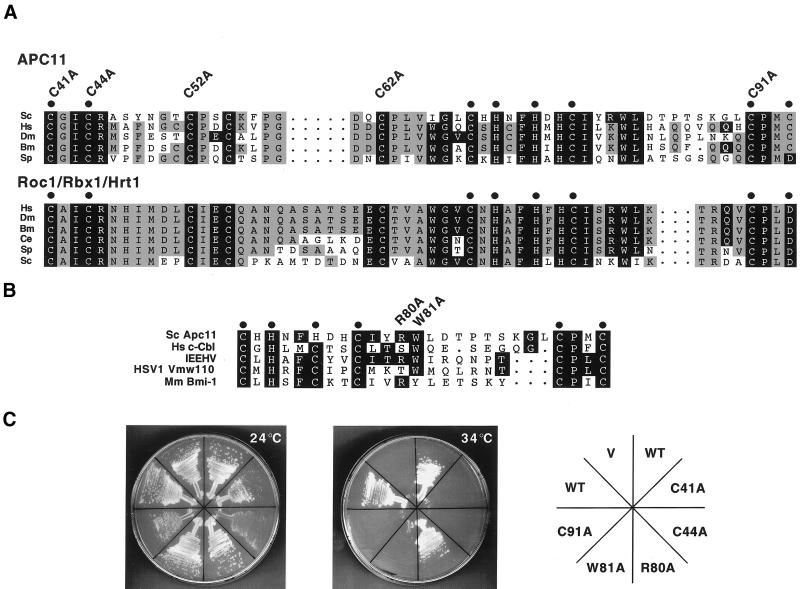
We went on to perform basic structure-function analyses of Apc11p utilizing the apc11–13 temperature sensitive strain. Roc1/Rbx1/Hrt1 and APC11 proteins from diverse species possess closely related RING-H2 finger domains (Figure 2A). To test the importance of the RING-H2 domain in Apc11p, we mutated residues predicted to participate in metal binding and the formation of the cross-brace motif (Figure 2A, C41A, C44A, C91A). By mutating cysteines to alanines, we expected to disrupt metal binding while introducing minimal structural changes. Each mutant was expressed from a centromeric plasmid under the control of its own promoter to test for its ability to complement the apc11–13 ts defect. As expected, wild-type APC11 expressed from the centromeric vector fully complemented the ts defect, while the vector alone did not (Figure 2C). Cells transformed with the C41A, C44A, or C91A mutants grew noticeably slower than their counterparts at the permissive temperature (Figure 2C, left panel) and failed to complement the apc11–13 ts defect at the restrictive temperature (Figure 2C, middle panel). Thus, the Apc11p RING-H2 finger performs a crucial function that is essential for cell viability.
Figure 2.
Mutational analysis of the RING-H2 finger protein Apc11p. (A) Alignment of APC11 and Roc1/Rbx1/Hrt1 RING-H2 finger domains from various species, including Homo sapiens (Hs), Drosophila melanogaster (Dm), Bombyx mori (Bm), Caenorhabditis elegans (Ce), Schizosaccharomyces pombe (Sp), and Saccharomyces cerevisiae (Sc). Residues conserved between both protein families are shaded black, while those conserved only among APC11 or Roc1/Rbx1/Hrt1 family members are shaded gray. Cysteines and histidines predicted to mediate zinc binding are denoted by dots. Apc11p cysteine residues 41, 44, 52, 62, and 91 were individually mutated to alanine, and mutants were subsequently tested by complementation analysis, as described in C. (B) Alignment of the Saccharomyces cerevisiae Apc11p RING-H2 domain with RING domains from c-Cbl, the ENX fragment of the immediate early equine herpes virus protein (IEEHV), herpes simplex virus 1 protein Vmw110, and Bmi-1. Tryptophan 81 of Apc11p corresponds to c-Cbl tryptophan 408, which is essential for full E3 activity in vitro (Joazeiro et al., 1999). R80 and W81 were individually mutated to alanine, and the mutants tested for complementation of the apc11–13 ts allele. (C) Complementation analysis was performed by transforming the apc11–13 temperature sensitive strain (YAP201) with centromeric plasmids expressing either wild-type Apc11p (WT) or the mutant versions described in A and B (C41A, C44A, R80A, W81A, C91A). Empty vector (V) was employed as a negative control. Transformed cells were allowed to grow at the permissive temperature of 24°C before being restreaked to selective medium at either 24°C (left) or the restrictive temperature of 34°C (center). (D) Schematic depiction of APC11 proteins from various species. The RING-H2 domain is indicated by a hatched box. S. cerevisiae Apc11p contains a C-terminal extension (residues 104–165, shaded black) not found in other family members. (E) Diploid cells harboring one copy of a truncated version of APC11 (APC11Δ104–165) were sporulated, and the tetrads analyzed for viability on YPD (left panel). Spore colonies were then replica-plated to SC-His to confirm 2:2 segregation of wild-type APC11 (His−) and APC11Δ104–165 (His+) (right panel).
Certain RING finger family members, including APC11, c-Cbl, and the immediate early equine herpes virus (IEEHV) protein, contain arginine and/or tryptophan residues at identical positions C-terminal to the third metal-chelating pair (Figure 2B, see also Joazeiro et al., 1999). In terms of length and primary sequence, this region is perhaps the most diverse among RING finger family members and has been proposed to confer some level of functional specificity to these proteins (Freemont, 1993; Borden et al., 1995). The 1H-NMR solution structure of IEEHV revealed that an amphipathic alpha helix forms in this region (Barlow et al., 1994), and residues in the analogous region of the IEEHV homolog Vmw110 were later shown to be crucial for its function (Everett et al., 1995). Based on sequence conservation, we mutated Apc11p R80 or W81 to alanine and again tested the mutants for the ability to complement the apc11–13 ts allele. While the R80A mutant could fully complement the ts defect, the W81A mutant was nonfunctional in this assay (Figure 2C). Interestingly, mutating the corresponding residue (W408) in the c-Cbl RING finger lead to a reduction in its E3 activity (Joazeiro et al., 1999), suggesting that the tryptophan at this position plays a crucial role in the function of at least a small subset of RING finger proteins.
Saccharomyces cerevisiae Apc11p is unique among known members of the APC11 family, in that it contains a C-terminal extension (residues 104–165, Figure 2D). This region is somewhat acidic, but does not exhibit significant homology to other known proteins. Because yeast strains expressing Apc11p with large, C-terminal epitope tags are temperature sensitive (Zachariae et al., 1998), we decided to test whether this region is required for Apc11p function in vivo. To this end we constructed diploid strains in which a stop codon was introduced after amino acid 103 in one copy of the APC11 gene (see MATERIALS AND METHODS). Tetrad analysis performed on three independent isolates yielded 4 viable spores in 35 out of 36 tetrads examined (12 tetrads per isolate). In 34 of the 35 4-spored tetrads, spores segregated 2:2 for both His+:His− and MATa:MATα (Figure 2e, and our unpublished results). Strains derived from APC11Δ104–165 spores showed no temperature sensitivity when compared with APC11 spores. These data clearly indicate that the C-terminal tail comprising amino acids 104–165 is not essential for Apc11p function.
Apc11p Binds Directly to Ubc4 In Vitro
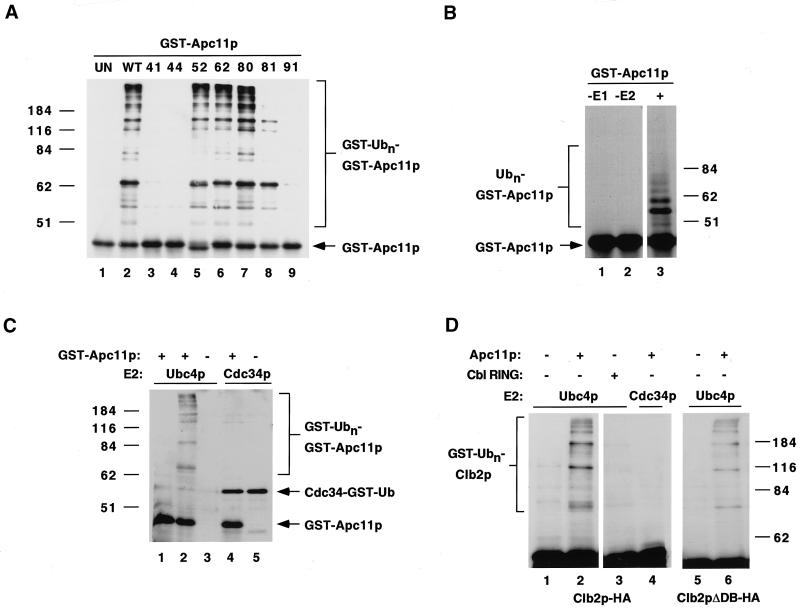
Several members of the HECT domain family of E3s had previously been shown to bind ubiquitin conjugating enzymes (Kumar et al., 1997). We and others have now shown that the RING finger domain can play a similar role in recruiting E2s to nonHECT E3s (Joazeiro et al., 1999; Yokouchi et al., 1999; Lorick et al., 1999). Because Ubc4 proteins can support APC-mediated ubiquitination in vitro (King et al., 1995; Yu et al., 1996; Charles et al., 1998), we decided to test whether Apc11p can bind directly to Ubc4. Employing a standard GST pull-down assay, we tested wild-type and mutant versions of GST-Apc11p for the ability to bind recombinant human Ubc4 (Figure 3). While little or no Ubc4 bound to GST alone (Figure 3, lane 2), GST-Apc11p bound significant amounts of Ubc4 (Figure 3, lane 3). GST-Apc11p failed to bind other purified, recombinant proteins added at equivalent concentrations (our unpublished results), suggesting that the interaction with Ubc4 was specific. The R80A mutant, which complemented the apc11–13 ts defect, bound Ubc4 at levels comparable to wild-type Apc11p (Figure 3, lane 8). However, mutating the adjacent tryptophan to alanine severely inhibited Ubc4 binding (W81A, Figure 3, lane 9). Mutating cysteine residues 52 or 62 also significantly reduced Ubc4 binding (Figure 3, lanes 6 and 7), and mutating predicted metal-binding cysteines reduced (C41A and C44A, Figure 3, lanes 4 and 5) or completely abolished (C91A, Figure 3, lane 10) Ubc4 binding, demonstrating that the integrity of the Apc11p RING-H2 finger is required for strong E2 binding.
Figure 3.
Apc11p interacts with Ubc4 in vitro. GST pull-down assays were employed to test for interactions between Apc11p and the ubiquitin conjugating enzyme (E2) Ubc4. Purified His-tagged human Ubc4 (L, 10% of the amount added to binding reactions) was incubated in the presence of GST alone (G), GST-Apc11p (WT), or various mutant versions (41, 44, 52, 62, 80, 81, and 91, as described in Figure 2) of Apc11p expressed as GST-fusion proteins. GST-fusions and interacting proteins were isolated using glutathione-agarose beads (see MATERIALS AND METHODS), separated by SDS-PAGE, and analyzed by immunoblotting for Ubc4 (upper panel). Blots were subsequently stained with coomassie blue (lower panel) to assess the levels of GST or GST-fusion proteins isolated (arrows). The asterisk indicates a contaminating protein present in all samples.
Because others have shown that strong E2 binding is not always required for the ubiquitin ligase activity of RING finger E3s (Lorick et al., 1999; Xie and Varshavsky, 1999), we also tested whether the mutant versions of Apc11p could still associate with the APC core complex. W303 cells were transformed with centromeric plasmids expressing wild-type or mutant versions of C-terminally 3xHA epitope-tagged Apc11p under the control of its own promoter. Apc11pHA3 proteins were immunoprecipitated from cell extracts using anti-HA antisera, separated by SDS-PAGE, and detected by immunoblotting (Figure 4, upper panel). The same blots were then probed for APC subunits Cdc16p (Figure 4, middle panel) or Cdc27p (Figure 4, lower panel). Perhaps surprisingly, all of the mutants were found to coimmunoprecipitate Cdc16p and Cdc27p as efficiently as wild-type Apc11p. Thus, while the RING-H2 finger mutants fail to bind wild-type levels of E2, they remain competent to assemble with other members of the APC, and this may in part account for their growth inhibitory effects at 24°C (Figure 2C, left panel).
Figure 4.
Wild-type and mutant versions of Apc11p assemble into the anaphase-promoting complex. W303 cells were transformed with centromeric plasmids expressing wild-type (WT) or mutant (41, 44, 80, 81, and 91, as described in Figure 2) versions of Apc11p bearing 3 HA epitope tags at their C-termini. Apc11pHA3 proteins were immunoprecipitated from cell extracts using monoclonal antibody 12CA5 and separated by SDS-PAGE. Gels were then blotted, cut into sections, and probed with either anti-HA monoclonal antibody (upper panel), anti-Cdc16 polyclonal antibody (middle panel), or anti-Cdc27 polyclonal antibody (lower panel). An arrow indicates immunoprecipitated Apc11pHA3 proteins, and the asterisk indicates the position of immunoglobulin light chain, which was detected in the 12CA5 immunoblot.
The Apc11p RING-H2 Finger Mediates E2-Dependent Ubiquitination
Because Apc11p could clearly bind Ubc4 in vitro, we next tested whether it could also mediate E2-dependent ubiquitin transfer. Purified GST-Apc11p fusion proteins were again employed, and served as both potential E3s and ubiquitination substrates (see Lorick et al., 1999). Roughly equivalent amounts of wild-type or mutant GST-Apc11p proteins were incubated in the presence of purified, recombinant E1, human Ubc4, GST-ubiquitin, and ATP. After 90 min, the reactions were stopped, separated by SDS-PAGE, and analyzed by immunoblotting with an anti-GST monoclonal antibody. As shown in Figure 5, GST-Apc11p was able to mediate high levels of ubiquitin transfer to proteins in the reaction mixture (Figure 5A, lane 2). An ascending ladder of bands corresponding to proteins that are mono- or poly-ubiquitinated can be observed and most likely represents ubiquitin-modified GST-Apc11p itself, as reactions carried out with nonGST-tagged ubiquitin also yielded ladders that could be detected by anti-GST antibody (Figure 5B, lane 3). Ubiquitination mediated by Apc11p was clearly dependent on the presence of both E1 and E2 (Figure 5B, lanes 1 and 2). While this reaction could also be mediated by budding yeast Ubc4p (Figure 5C, lane 2), Cdc34p, an E2 utilized by the SCF complex, failed to support Apc11p-mediated ubiquitination (Figure 5C, lane 4). Both purified His-tagged Apc11p and Apc11p purified away from the GST moiety by thrombin cleavage were also capable of mediating E1- and E2-dependent ubiquitination (Figure 5D, and our unpublished results), ruling out the possibility that the observed effects were an artifact of using GST-fusion proteins.
Figure 5.
The Apc11p RING-H2 finger mediates E1- and E2-dependent ubiquitination. (A) Wild-type (WT) or mutant (41, 44, 52, 62, 80, 81, and 91, as described in Figure 2) versions of Apc11p were tested for the ability to mediate polyubiquitination in vitro. Equivalent amounts of purified GST-Apc11p fusion proteins were incubated with purified E1, Ubc4 (E2), and GST-ubiquitin in reaction buffer containing ATP. After 90 min, reactions were stopped with 2x SDS gel loading buffer, separated by SDS-PAGE, and analyzed by immunoblotting with anti-GST monoclonal antibody. Lane 1 shows a reaction with GST-Apc11p which was stopped with SDS buffer before the addition of E1, E2, GST-ubiquitin, and reaction buffer (UN, unreacted). GST-ubiquitin-GST-Apc11p conjugates are indicated at right. Molecular weight markers, in kiloDaltons, are indicated at left. (B) Ubiquitination reactions were carried out with GST-Apc11p as described in A, using ubiquitin in place of GST-ubiquitin. Lanes 1 and 2 show reactions carried out in the absence of E1 or E2, respectively. Lane 3 shows a reaction carried out with the full complement (+) of components. GST-Apc11p and its ubiquitin-conjugated derivatives were detected by immunoblotting with anti-GST monoclonal antibody. Ubiquitin-GST-Apc11p conjugates are indicated at left. (C) Reactions were carried out as in A, with GST-Apc11p fusion proteins incubated in the presence of GST-ubiquitin, E1, and equivalent concentrations of either Ubc4p or Cdc34p. GST-fusion proteins were detected by immunoblotting with anti-GST monoclonal antibody. Autoubiquitinated Cdc34p is indicated by an arrow. (D) Ubiquitination reactions were carried out by incubating GST-ubiquitin and His-tagged Clb2p-HA or Clb2p lacking the destruction box (Clb2pΔDB-HA) in the presence of either His-tagged Apc11p or the c-Cbl RING finger purified as a GST-fusion protein. Reactions contained either Ubc4p or Cdc34p as indicated. Clb2 proteins and their ubiquitin-conjugated derivatives were detected by immunoblotting with anti-HA monoclonal antibody. GST-ubiquitin-Clb2p-HA conjugates are indicated at left.
We next tested the ability of the Apc11p mutants to mediate ubiquitination. As anticipated from its ability to bind Ubc4 and to complement the apc11–13 ts defect, the R80A mutant promoted ubiquitination to levels equivalent to wild-type Apc11p (Figure 5A, lane 7). However, the W81A mutant, which exhibited reduced Ubc4 binding (Figure 3, lane 9), also demonstrated a reduced ability to mediate ubiquitin transfer (Figure 5A, lane 8). A similar decrease in E3 activity was observed for the c-Cbl RING mutant W408A (Joazeiro et al., 1999). While the C52A and C62A mutants also bound reduced levels of Ubc4 (Figure 3, lanes 6 and 7), they were capable of mediating wild-type levels of ubiquitination in this assay (Figure 5A, lanes 5 and 6). However, Apc11p bearing mutations in putative metal-binding cysteines (C41, C44, or C91) showed little or no activity in this assay (Figure 5A, lanes 3, 4, and 9), demonstrating that the RING-H2 finger is absolutely required for its E3 activity. Although the C41A and C44A mutants bound slightly greater levels of Ubc4 than the C52A, C62A, or W81A mutants (Figure 3), their lack of activity in this assay indicates that simple E2 recruitment is not sufficient to induce the ubiquitination we observed (see DISCUSSION).
To test whether Apc11p was capable of mediating E2-dependent ubiquitin transfer to substrates other than itself or GST, we incubated purified His-tagged Apc11p with purified recombinant Clb2p, a known APC substrate. His-Apc11p mediated the polyubiquitination of Clb2p in the presence of Ubc4p, but not Cdc34p (Figure 5D, compare lanes 2 and 4). Clb2p was not modified when incubated with equivalent levels of purified c-Cbl RING finger (Figure 5D, lane 3), demonstrating that this effect was specific to Apc11p. His-Apc11p was also able to mediate the polyubiquitination of Clb2p lacking its N-terminal destruction box (Figure 5D, lane 6). These data indicate that, while Apc11p probably does not confer specificity toward destruction box-containing substrates, it is capable of mediating ubiquitin transfer from E2 to proteins other than itself, at least in vitro.
DISCUSSION
We have demonstrated that the RING-H2 protein Apc11p binds directly to the ubiquitin-conjugating enzyme (E2) Ubc4 and mediates Ubc4-dependent polyubiquitination of protein substrates in vitro. While Ubc4 binding was, in some cases, only reduced by mutations in putative metal-binding cysteines (Figure 3, C41A and C44A), substrate ubiquitination was wholly dependent on the integrity of the Apc11p RING-H2 finger. Intriguingly, certain mutations in nonmetal-binding residues reduced Ubc4 binding even further, yet left the protein capable of mediating protein ubiquitination (W81A and cysteine mutants C52A and C62A, see Figures 3 and 5A). Thus, while the RING-H2 finger of Apc11p is perhaps not absolutely required for E2 binding, it appears to be essential for mediating the E2-dependent transfer of ubiquitin to substrates. Indeed, for several other RING finger E3s, there is no strict correlation between the affinity of E2 binding and the ability to mediate ubiquitin transfer (Lorick et al., 1999; Xie and Varshavsky, 1999). Ubr1p, the N-end rule pathway E3 in budding yeast, was shown to bind Ubc2p through a domain distinct from its RING finger (Xie and Varshavsky, 1999). However, the integrity of the Ubr1p RING finger is strictly required for the degradation of N-end rule substrates. Thus, the RING finger is probably more than a simple E2-recruiting module, and it plays a crucial role in mediating ubiquitin transfer from E2s to substrate proteins.
Ohta and coworkers have demonstrated that human Apc11 isolated from HeLa or transfected 293T cells is capable of mediating Ubc5-dependent polyubiquitination in vitro (Ohta et al., 1999). Because this work relied on the use of Apc11 immunocomplexes, the authors were unable to conclude definitively that Apc11 was sufficient to mediate polyubiquitination. In contrast, we have utilized an in vitro system which employs purified protein components to demonstrate that Apc11p alone is capable of mediating E1- and E2-dependent ubiquitin transfer. These results are especially intriguing in light of the fact that the close Apc11 relative Rbx1 was shown to require a cullin subunit, Cdc53, for full E3 activity (Seol et al., 1999).
Which E2s Are Utilized by the APC In Vivo?
While both human and budding yeast Ubc4 worked efficiently in our system to support Apc11p E3 activity (Figure 5), it remains unclear which E2s are utilized by the APC in vivo (see Page and Hieter, 1999, and Zachariae and Nasmyth, 1999 for reviews). Several groups have demonstrated that Ubc4 proteins can function in APC-mediated ubiquitination reactions (King et al., 1995; Yu et al., 1996; Charles et al., 1998), and it was recently demonstrated that the close Ubc4 relative Ubc5 can function in ubiquitination reactions mediated by human Apc11 immunocomplexes (Ohta et al., 1999). Interestingly, yeast strains deleted for UBC4 and UBC5 are fully viable (Arnason and Ellison, 1994), suggesting that the APC utilizes other E2s or, at least, is capable of doing so in the deletion strains. In fission yeast, the E2 UbcP4 is essential for the metaphase-to-anaphase transition and can rescue apc temperature-sensitive defects (Osaka et al., 1997). However, budding yeast deleted for the closest UbcP4 homolog, UBC11, are fully viable, as are strains lacking both UBC11 and UBC4 (Townsley and Ruderman, 1998). SCF complexes can utilize E2s other than Cdc34 (Yaron et al., 1998; Spencer et al., 1999), and it is quite possible that the APC employs multiple E2s for the ubiquitination of diverse substrates. Work ongoing in our laboratories is aimed at clarifying these issues.
The Role of Cullins in APC-mediated Ubiquitination
The combination of the SCF RING-H2 protein Hrt1/Roc1/Rbx1 and the cullin-like subunit Cdc53 potently stimulates Cdc34p autoubiquitination (Seol et al., 1999) and has been proposed to comprise a minimal E3 ubiquitin ligase activity (see Deshaies, 1999 for review). In contrast, the addition of purified cullin-like APC subunit Apc2p had no significant effect on Apc11p-mediated ubiquitination, and adding a purified C-terminal fragment of Apc2p (residues 471–853) containing the cullin homology domain (CHD) had a slight inhibitory effect (our unpublished results). We were also unable to observe a stimulation of Ubc4p autoubiquitination in the presence of Apc11p plus or minus Apc2p.
Because our assay employs a minimal system to monitor Apc11p E3 activity, the effects observed with Apc2p may not be representative of APC activity in vivo. Nevertheless, the results presented in Figure 5 demonstrate that Apc11p alone is capable of efficiently mediating ubiquitin transfer. While Apc2p was not required to effect ubiquitin transfer in vitro, it may function in a cellular context to tether Apc11p to APC core proteins, and to properly position the Apc11p RING-H2 finger with respect to its substrates. As has been suggested regarding SCF complexes (Deshaies, 1999), the APC is probably fully active only when the cullin and RING-H2 subunits meet substrate in the context of the full assembly.
Ohta and coworkers have employed coimmunoprecipitation experiments and two hybrid analyses in yeast to demonstrate interactions between human APC11 and mouse APC2, but they were unable to test interactions between yeast Apc11p and Apc2p (Ohta et al., 1999). While genetic evidence suggested that Apc11p and Apc2p are binding partners, we were unable to detect direct interactions between GST-Apc11p and a C-terminal fragment (residues 471–853) of Apc2p containing the cullin homology domain (our unpublished results). Roc1 binds the C-terminal 527 amino acids of CUL1 in a yeast two-hybrid system (Ohta et al., 1999), so perhaps more N-terminal regions of Apc2p are required for Apc11p binding. Alternatively, other APC subunits not present in our reactions may be involved in stabilizing Apc11p-Apc2p interactions within the complex. Finally, certain cullins are covalently modified by a single molecule of the ubiquitin relative Rub1p/Nedd8 (Osaka et al., 1998; Lammer et al., 1998; Liakopoulos et al., 1999; Wada et al., 1999), so perhaps Apc2p must be modified before binding and/or activating Apc11p. Intriguingly, Kamura and coworkers recently demonstrated a role for Rbx1 itself in mediating the modification of Cdc53/cullin by Rub1 (Kamura et al., 1999b), so it will be interesting to determine whether Apc11p plays a similar role in modifying Apc2p.
The RING Finger Family as Mediators of Ubiquitination
It is now clear that several members of the RING finger family bind directly to E2s and mediate E2-dependent ubiquitination. However, it remains to be seen whether this is a general function of the RING finger domain, or whether proteins have also employed this motif for distinct biochemical functions. The RING finger was originally proposed to function in DNA binding, and indeed, certain family members have been implicated in aspects of transcriptional regulation (see Freemont, 1993). The precise role of the RING finger in these processes is unclear, so it cannot be ruled out that these proteins also mediate protein ubiquitination.
We now know, at least for a subset of this family, that the RING finger functions as an E2 recruiting module, and it appears that a variety of strategies have been adopted for the targeting of RING substrates. The RING finger family is quite diverse, ranging from small proteins like Roc1/Rbx1/Hrt1 and Apc11, which consist almost solely of the RING domain, to large, modular proteins like c-Cbl, which bear several recognizable protein motifs in addition to the RING finger. In the case of SCF complexes, a cullin and the small RING-H2 protein Rbx1 bind to substrate-specificity determining F box proteins. Apc11p is most likely targeted to destruction box-containing substrates by other APC subunits and/or WD-40 activator proteins such as Cdc20p and Hct1p/Cdh1p. The c-Cbl proto-oncoprotein contains its own substrate-specificity determining motif—a modified phosphotyrosine-binding domain that binds to activated growth factor receptors. Other targeting strategies are likely to be revealed as work on RING finger proteins intensifies.
In light of our results, it will be interesting to determine whether other RING finger proteins require auxiliary factors, such as the cullins, for the full activation of their E3 activity. Our work, and that of others, indicates that RING-based E3s do not act as ubiquitin carriers that form thioester intermediates, but instead act as bridges between E2s and substrates that provide a favorable environment for the transfer of ubiquitin. Seol and coworkers have suggested that positively charged regions in the cullin and/or RING subunits could facilitate RING-mediated ubiquitin transfer (Seol et al., 1999), and it is conceivable that distinct domains in larger RING finger proteins play a similar role. Clearly, the precise mechanisms employed by the RING-based E3s in vivo remain to be elucidated.
ACKNOWLEDGMENTS
The authors thank W. Zachariae and K. Nasmyth for APC11 construct pWZV84, M. Nakao for Ubc4 constructs, F. Yamao for the His-E1 construct, and P. Kaiser and S. Reed for His-Clb2-HA constructs and purified His-Cdc34p. Thanks go to J. Meisenhelder, S. Simon, and H. Mondala for technical support. We also thank S. Forsburg and members of the Hunter laboratory for helpful discussions and advice. J.D.L. was supported by NIH training grant T32 CA09523, and is currently supported by American Cancer Society fellowship PF-99–228-01-CCG. C.A.P.J. is a postdoctoral fellow of the American Cancer Society, California Division. H.k.-H. is supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation, fellowship DRG-1531. The work of A.M.P. and P.H. was supported by NIH grant CA 16519. The work of J.D.L., C.A.P.J., H.k.-H., and T.H. was supported by NIH grants CA 14185 and 80100. T.H. is a Frank and Else Schilling American Cancer Society Professor.
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- CHD
cullin homology domain
- DAPI
4, 6-diamidino-2-phenylindole
- DTT
dithiothreitol
- FACS
fluorescence-activated cell sorting
- GST
glutathione-S-transferase
- HA
hemagglutinin
- HECT
homologous to E6AP C-terminus
- IEEHV
immediate early equine herpes virus
- PMSF
phenylmethylsulfonyl fluoride
- SC
synthetic complete
- SCF
Skp/Cdc53/cullin/F box
- TPR
tetratrico peptide repeat
- YPD
yeast extract peptone dextrose
REFERENCES
- Arnason T, Ellison MJ. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow PN, Luisi B, Milner A, Elliott M, Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994;237:201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- Borden KL, Boddy MN, Lally J, O'Reilly NJ, Martin S, Howe K, Solomon E, Freemont PS. The solution structure of the RING finger domain from the acute promyelocytic leukemia proto-oncoprotein PML. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL, Freemont PS. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/RING H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Everett R, O'Hare P, O'Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont PS. The RING finger. A novel protein sequence motif related to the zinc finger. Ann NY Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein Mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- Joazeiro CAP, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2- dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999a;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999b;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tugendreich S, Fujii M, Jorgensen PM, Watanabe N, Hoog C, Hieter P, Todokoro K. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tanaka H, Yasuda H, Todokoro K. Regulation of APC activity by phosphorylation and regulatory factors. J Cell Biol. 1999;146:791–800. doi: 10.1083/jcb.146.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kramer KM, Fesquet D, Johnson AL, Johnston LH. Budding yeast RSI1/APC2, a novel gene necessary for initiation of anaphase, encodes an APC subunit. EMBO J. 1998;17:498–506. doi: 10.1093/emboj/17.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kao WH, Howley PM. Physical interaction between specific E2 and HECT E3 enzymes determines functional cooperativity. J Biol Chem. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- Lahav-Baratz S, Sudakin V, Ruderman JV, Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc Natl Acad Sci USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Michaud WA, Sikorski RS, Hieter PA. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 1994;13:4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCF Cdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon WY, Hartley JW, Klinken SP, Ruscetti SK, Morse HC. v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early B-lineage lymphomas. Proc Natl Acad Sci USA. 1989;86:1168–1172. doi: 10.1073/pnas.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, Bowtell DD, Stanley ER. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 1999;18:3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc Natl Acad Sci USA. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Osaka F, Seino H, Seno T, Yamao F. A ubiquitin-conjugating enzyme in fission yeast that is essential for the onset of anaphase in mitosis. Mol Cell Biol. 1997;17:3388–3397. doi: 10.1128/mcb.17.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AM, Hieter PA. The anaphase-promoting complex: new subunits and regulators. Annu Rev Biochem. 1999;68:583–609. doi: 10.1146/annurev.biochem.68.1.583. [DOI] [PubMed] [Google Scholar]
- Peters JM, King RW, Hoog C, Kirschner MW. Identification of BIME as a subunit of the anaphase-promoting complex. Science. 1996;274:1199–1201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- Peters JM. Subunits and substrates of the anaphase-promoting complex. Exp Cell Res. 1999;248:339–349. doi: 10.1006/excr.1999.4443. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1–E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Shevchenko A, Deshaies RJ. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of S.C.F. define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boguski MS, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Ruderman JV. Functional analysis of the Saccharomyces cerevisiae UBC11 gene. Yeast. 1998;14:747–757. doi: 10.1002/(SICI)1097-0061(19980615)14:8<747::AID-YEA271>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in Eμ-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC- dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wada H, Yeh ET, Kamitani T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J Biol Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne WC, Zhang H, Yoshimura A, Baron R. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- Yu H, Peters JM, King RW, Page AM, Hieter P, Kirschner MW. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]