Abstract
A Mini-RNA from potato spindle tuber viroid (PSTVd) was constructed specifically for cleavage and ligation to circles in vitro. It contains the C-domain with the so-called central conserved region (CCR) of PSTVd with a 17 nt duplication in the upper strand and hairpin structures at the left and rights ends of the secondary structure. The CCR was previously shown to be essential for processing of in vitro transcripts. When folded under conditions which favor formation of a kinetically controlled conformation and incubated in a potato nuclear extract, the Mini-RNA is cleaved correctly at the 5′- and the 3′-end and ligated to a circle. Thus, the CCR obviously contains all structural and functional requirements for correct processing and therefore may be regarded as ‘processing domain’ of PSTVd. Using the Mini-RNA as a model substrate, the structural and functional relevance of its conserved non-canonical motifs GAAA tetraloop, loop E and G:U wobble base pair were studied by mutational analysis. It was found that (i) the conserved GAAA tetraloop is essential for processing by favoring the kinetically controlled conformation, (ii) a G:U wobble base pair at the 5′-cleavage site contributes to its correct recognition and (iii) an unpaired nucleotide in loop E, which is different from the corresponding nucleotide in the conserved loop E motif, is essential for ligation of the 5′- with the 3′-end. Hence all three structural motifs are functional elements for processing in a potato nuclear extract.
INTRODUCTION
Viroids are autonomously replicating subviral plant pathogens consisting of a non-encapsidated, circular, single-stranded RNA of 250–600 nt depending on the viroid species. There is no evidence for any viroid translation product. Thus viroid replication and pathogenesis rely on interactions with the host enzyme system. Obviously viroids contain specific RNA structural motifs which are recognized by host enzymes to carry out reactions like transcription, cleavage, and ligation (reviewed in 1–4).
The largest family of viroids are the Pospiviroidae, and its type strain potato spindle tuber viroid (PSTVd) has been discovered first (5) and studied in most detail (1–4,6). They adopt an unbranched, rod-like secondary structure with a high degree of intramolecular base pairing (7,8). Their replication occurs via an asymmetric rolling circle mechanism (9): the (by definition) (+)-strand circular RNA is transcribed into oligomeric, linear (–)-strand intermediates, which in turn are transcribed into oligomeric, linear (+)-strand intermediates. Both directions of transcription are carried out by the DNA-dependent RNA-polymerase II (10,11). Finally, the (+)-strand intermediates have to be processed to mature circles, i.e. to be cleaved to exact monomeric length and ligated. Whereas all three members of the family Avsunviroidae (ASBVd, PLMVd, CChMVd) can form hammerhead structures for self-cleavage (12), it has been shown for PSTVd that enzymes are necessary for cleavage and ligation (13).
Viroids of the genus Pospiviroid contain a C-domain and within this domain a central conserved region (CCR), i.e. the central part of the rod-like structure is formed by base pairs with high GC content of the upper CCR (UCCR) and the lower CCR (LCCR) (for exact definitions see 1,2). Linear in vitro transcripts with a short terminal duplication within the UCCR are highly infectious (14–17). This led to the conclusion that processing proceeds within this region, allowing effective synthesis of mature viroid circles (18–21). PSTVd constructs of this kind (‘more-than-unit-length transcripts’) can adopt quite different conformations (22), whereof only one is a substrate for processing in a potato nuclear extract (23). The active conformation is thermodynamically suboptimal but can be favored by rapid renaturation (‘snap cooling’). It can be regarded as a model for a metastable conformation formed by sequential folding of the RNA during transcription in vivo.
All conformations of the linear more-than-unit-length transcripts share most of the unperturbed, rod-like secondary structure of the mature circle. The duplication, however, introduces local structure differences within the CCR which obviously determine the substrate activity for correct processing. In fact, the cleavage and ligation sites were identified within the UCCR close to an internal loop with high sequence similarity to the loop E of the eucaryotic 5S rRNA (24). A homologous internal structure of the loops has been postulated since both are able to be crosslinked specifically by UV light (25,26). Actually, a highly conserved ‘loop E motif’ is apparent in quite different RNAs such as 16S and 23S rRNA, group I and II introns, RNase P and the hairpin ribozyme. The loop has been found to be highly structured by non-canonical base pairs and is involved in RNA–RNA as well as RNA–protein interactions (27). However, a series of studies, such as structure calculation, analysis by temperature-gradient gel electrophoresis (TGGE), modification by dimethylsulfate and UV crosslinking, established that the conformation active in processing does not contain the loop E, but can instead assume a branched structure consisting of six stem–loop structures arround a central loop (24,28). One of these stem–loop structures comprises the 5′-cleavage site which is located at a single G:U wobble base pair within the stem. The loop is a GAAA tetraloop (shown for the Mini-RNA in Fig. 3).
Figure 3.
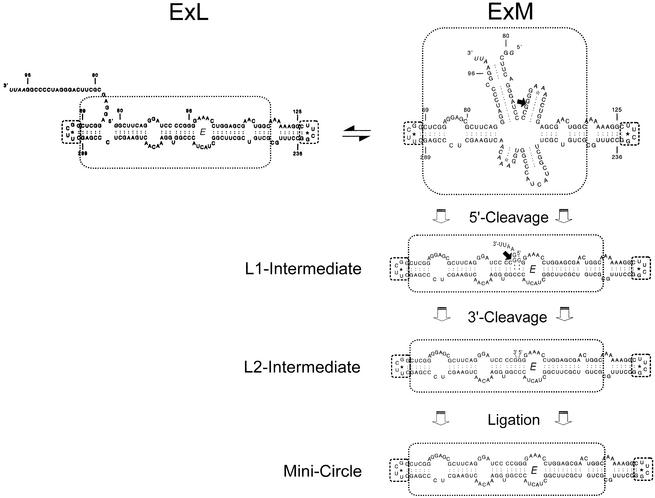
Mechanistic model for processing in vitro of the Mini-RNA analogous to the mechanistic model for the more-than-unit-length transcripts (Fig. 1) and model transcripts resulting thereof. The Mini-RNA forms either the thermodynamically favored conformatin ExL or the kinetically favored conformation ExM which is the substrate for the initial 5′-cleavage of processing (arrow). The consecutive step and intermediates correspond exactly to the steps and intermediates of the more-than-unit-length transcript in Figure 1.
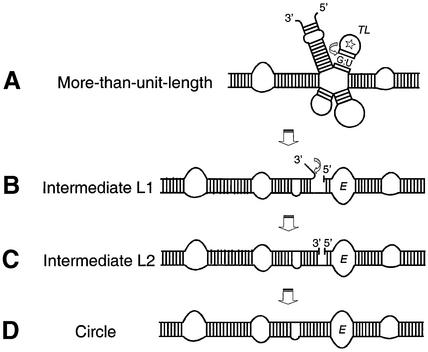
Detailed analysis of the more-than-unit-length transcripts and linear intermediates as substrates for processing led to a mechanistic model of processing (Fig. 1) (24): the metastable multi-helix conformation is substrate for initial 5′-cleavage, perhaps recognized at the GAAA tetraloop or the G:U wobble base pair by a host factor. First 5′-cleavage, followed by dissociation of the 5′-end, drives a structural rearrangement leading to the rod-like conformation containing loop E. Cleavage of the 3′-unpaired nucleotides results in a conformation of exact monomeric length with 3′- and 5′-ends in optimal juxtaposition, i.e. the 3′-end and the 5′-end are stacked onto each other coaxially and may readily be ligated by a host RNA ligase. Positioning of 3′- and 5′-ends essentially needs loop E formation which also might be involved in ligase recruiting. The segments of the RNA which are involved in the whole processing, in particular the specific RNA motifs like GAAA tetraloop, loop E and G:U wobble base pair, are located within the highly conserved CCR, implying that the processing mechanism is very similar for the whole PSTVd genus.
Figure 1.
Mechanistic model for viroid processing in vitro according to Baumstark et al. (24). The structures represent schematically the CCR from the more-than-unit-length transcripts. (A) The multi-helix conformation is substrate for initial 5′-cleavage at the G:U wobble base pair (arrow) within the short stem of the GAAA tetraloop hairpin (TL, marked with a star). (B) Dissociation of the 5′-end drives a structural rearrangement leading to the rod-like conformation containing loop E (denoted E). Cleavage of the 3′-unpaired nucleotides (arrow) results in a conformation of exact monomeric length with the 3′- and 5′-end in juxtaposition stabilized by coaxial stacking (C). (D) Ligation of 5′- and 3′-end results in mature circular progeny.
In this work a Mini-RNA has been constructed as a substrate for processing in vitro which contains almost only the C-domain with the identical 17 nt duplication used by Baumstark et al. (23,24). If the mechanism proposed earlier is right, the same processing reaction should occur also within the Mini-RNA. This indeed was found and site directed mutagenesis within the structural motifs tetraloop, wobble pair and loop E allowed us to analyze their relevance for structure formation and function. Whereas the GAAA tetraloop turned out to be essential for kinetically driven structure formation, the G:U wobble base pair and the loop E seem to be involved in recognition by host factors.
MATERIALS AND METHODS
DNA oligonucleotides and plasmids
The Mini-RNA cDNA was synthesized by ligation of three oligonucleotides, A, B, and C. Oligonucleotide A contains the BglII restriction site and the T7 RNA polymerase promotor sequence for in vitro transcription of pRH701 (21) followed by the PSTVd sequence G80-C109 (5′-AAACTGCAGATCTA ATTAATACGACTCACTATAG G80-C109-3′); sequence and numbering refers to Gross et al. (7). Oligonucleotides B and C contain the subsequent PSTVd sequence in the Mini-RNA cDNA up to G96 so that G80-G96 are present at the 5′- and the 3′-ends (see Fig. 2). However, the right and the left arms of the rod-like secondary structure of PSTVd, i.e. A126-C235 and A290-359/1-G68, respectively, are substituted by CTTCGG sequences in the Mini-RNA cDNA. The CTTCGG sequences in their RNA version form stable UUCG-tetraloops closed by an extra G:C base pair within the resulting in vitro transcripts replacing the deleted ‘arms’ (see Fig. 2). In detail, oligo B is: 5′-G110-G125 CTTCGG C236-G268-3′; oligo C contains an EcoRI restriction site at the 3′-end: 5′-G269-G289 CTTCGG C69-G96 GAATTCCTAGC-3′. To synthesize mutant Mini-RNA variants, oligonucleotides A and B containing point mutations were used [A(R+): A100G; A(A4): G98A; A(GC): T104C; A(AU): G95A; A(UU): G95T; A(AC): G95A; B(LE): C259G]. To synthesize cDNAs for the in vitro transcription of the L1-intermediates, modified oligonucleotides A with a 16 nt deletion (G80-G96; A-L1) were used. To synthesize mutated L1-intermediates, variants of the deleted oligonucleotide A-L1 containing identical point mutations to the mutated oligonucleotides A were used. For the ligation of the (+)-strand oligonucleotides A, B and C (or their variants) they were hybridized with (–)-strand oligonucleotides covering the ligation sites (A/B: complementary to A122-C93; B/C: complementary to G287-C53). For PCR amplification of the Mini-RNA cDNA the primers T7 [(+) strand, identical to the 5′-end of oligonucleotide A up to G80] and Eco [(–) strand, complementary to the 3′-end of oligonucleotide C from C93 plus an additional GGTC at the 3′-end optimizing the annealing temperature] were used. Subsequently the Mini-RNA cDNA was cloned into the plasmid pRH701 (21).
Figure 2.
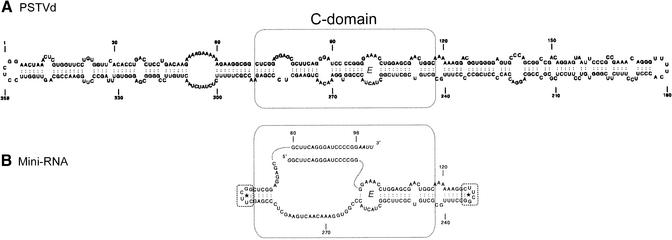
Mini-RNA for processing in vitro. (A) Rod-like conformation of PSTVd and numbering of sequence according to Gross et al. (7). The C-domain which contains the CCR is highlighted by a dotted box. (B) Schematic representation of the 148 nt Mini-RNA containing almost only the CCR of PSTVd (dotted box) with the 17 nt duplication (G80-G96) identical to the more-than-unit-length transcripts. Nucleotides A126-C235 (‘right arm’) and A290-359/1-G68 (‘left arm’) of PSTVd were substituted by extraordinary stable UUCG tetraloops closed by an additional G-C base pair (dashed box) to maintain the rod-like conformation. Theoretical secondary structure analyses were carried out by using the program LinAll (29).
Secondary structure calculations
The calculation of RNA secondary structures and structural transitions was carried out using the algorithm LinALL (29) with additional implementation of thermodynamic parameters for tetraloop formation (30–32). RT–PCR primers were chosen according to calculations using a program developed by Steger (33).
Synthesis of a viroid-specific Mini-RNA cDNA and its mutants
The Mini-RNA cDNA for in vitro transcription was synthesized by enzymatic ligation of (+)-strand DNA oligonucleotides (A, B, C; see above), amplification as a double-stranded DNA by PCR and cloning in pRH701. By using appropriately modified oligonucleotides, point mutations and end deletions were introduced resulting in mutant Mini-RNAs and the L1- and L2-intermediates, respectively. For ligation the oligonucleotides B and C (and their modified variants) were 5′-phosphorylated by polynucleotide kinase (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s protocols. Oligonucleotides A, B and C then were hybridized with (–)-strand oligonucleotides complementary to the ligation sites (A/B, B/C), thereby linking together the (+)-strand oligonucleotides to be ligated. For this, 1 pmol of each of the oligonucleotides together was heated for 5 min at 90°C and slowly cooled to room temperature overnight in 10 µl of 3 mM MgCl2, 6 mM Tris–HCl pH 8.0. Following adjustment of the buffer to 1 mM dithiothreitol and 1 mM ATP in 50 µl, 5 U T4 DNA ligase (Roche Diagnostics) was added followed by incubation at room temperature for 6 h. After extraction of the DNA by phenol–chloroform and ethanol precipitation, the pellet was dissolved in 100 µl TE buffer whereof 5 µl were used as template for PCR amplification. The PCR was carried out in 100 µl buffer containing 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl pH 9.0, 0.01% Triton X-100, 4 mM each of the dNTPs, 100 pmol each of primers T7 and Eco and 2.5 U Taq-polymerase (Promega, USA). Thirty amplification cycles were run (94, 50 and 72°C for 60 s each) with an extra 3 min denaturation at 94°C at the beginning and a 10 min final elongation at 72°C. PCR products were purified and desalted with a commercial kit (QIAquick PCR Purification Kit, Qiagen, Hilden, Germany). Typically, 1–2 µg of PCR product was finally eluted in 50 µl of TE buffer. The PCR products were digested with the endonucleases BglII and EcoRI using commercially available standard enzymes and protocols, and cloned in pRH701.
Enzymatic reactions
All standard enzymes for modifications of nucleic acids (e.g. restriction enzymes, ligases, phosphatases, etc.) were used according to the manufacturer’s instructions unless described in detail within one of the methods herein. T7 RNA polymerase for in vitro transcription (21,23) was purified in house according to the literature (34). Transcripts were labeled by addition of [α-32P]-UTP to the transcription mixture; labeled transcripts were separated on and subsequently eluted from a high resolution denaturing gel as described by Krupp (35) for removal of non-full-length transcripts.
Gel electrophoresis
TGGE (36) was carried out as described previously for application to a similar analysis (23). The gels contained 12% (w/v) polyacrylamide, 0.4% (w/v) bisacrylamide, 0.1% (v/v) TEMED, 0.2× TBE and 0.04% ammonium peroxidisulfate (APS); electrophoresis was run for 45 min at 500 V.
For analysis of in vitro processing products and UV crosslinking, the RNA was separated on high resolution denaturing gels typically used for sequencing (Electrophoresis Apparatus Model S2, Invitrogen, Gaithersburg, USA). The gels contained 8 M urea, 8% polyacrylamide, 0.27% bisacrylamide, 0.1% (v/v) TEMED, 0.2× TBE and 0.04% APS; electrophoresis was run for 1.5 h applying 100 W, resulting in a temperature of 60°C. The samples (∼5 µl) had been denatured for 3 min at 95°C and cooled rapidly on ice before loading onto the gel. The gels were exposed to X-ray film (Kodak Xomat AR).
Preparation of nucleic extract for processing in vitro
The processing reaction was performed in a nuclear extract derived from the host plant potato. Preparation of nuclei and the nuclear extract from a suspension cell culture was carried out as described previously (23,24).
Processing reaction in vitro
Standard processing reactions of the Mini-RNA were performed according to the protocol established for ‘more-than-unit-length’ transcripts (23,24) with minor modifications: radioactively labeled, gel-eluted Mini-RNA in vitro transcripts (∼105 c.p.m. per assay) were pre-incubated for ExM and ExL structure formation (see below) and incubated with nuclear extract at 30°C for the times indicated (0–90 min). An aliquot of 10 µl nuclear extract in OS buffer [20 mM HEPES–KOH pH 7.9, 50 mM KOAc, 10 mM Mg(OAc)2, 5 mM EDTA, 12 mM β-mercaptoethanol, 25% glycerol] and 5 µl of 100 mM HEPES–KOH pH 7.9, 10 mM DTT, and 4 µl of 100 mM MgCl2 was added to H2O to a final volume of 50 µl. The reaction was stopped by addition of 150 µl 27 mM EDTA, 0.5% SDS and transition on ice immediately followed by extraction by phenol and phenol–chloroform. Subsequently the RNA was precipitated by ethanol, dissolved in formamid-urea loading solution and analyzed on high resolution denaturing gels (see above).
UV crosslinking and assay
UV crosslinking of Mini-RNA in vitro transcripts for loop E formation analysis was carried out as described previously for ‘more-than-unit-length’ transcripts with minor modifications (24). Radioactively labeled, gel-eluted Mini-RNA in vitro transcripts (∼105 c.p.m. per assay in ∼5 µl in TE buffer) was pre-incubated for ExM and ExL structure formation (see below) and on ice UV-irradiated at 254 nm wavelength (Stratalinker, Model 1800, Stratagene) for the times indicated (0–20 min). The samples were eluted with 1 vol of loading solution and analyzed on high resolution denaturing gels (see above) afterwards.
For verification of the crosslinked nucleotides being the well established loop E crosslink (24,25) a primer extension analysis of the crosslinked transcripts was applied. Therefore, a crosslink assay using 5 µg of non-radioactively labeled in vitro transcripts was carried out: after pre-incubation for ExL structure formation the RNA was UV-irradiated in 5× 150 µl aliquots for 40 min, precipitated by ethanol and separated on a preparative denaturing 10% polyacrylamide gel. The RNA then was stained with ethidium bromide and the crosslinked transcripts were eluted by overnight incubation of the gel slice in 300 µl 300 mM NaOAc, 1 mM EDTA, 1% SDS at 4°C. Following ethanol precipitation and dialysis against TE buffer ∼20 ng per assay of the RNA was annealed to ∼105 c.p.m. radioactively labeled primer AF-10 (complementary to C259-C236) for identification of G98, and OS-3 (complementary to G95-G75) for identification of U260. The primer extension reaction occurred as described previously (24) using the Superscript II RNase H(–) Kit (Gibco BRL) with the exception that incubation at 42°C turned out to be favorable over incubation at 48°C as published for longer transcripts.
Pre-incubation of the Mini-RNA for secondary structure formation
Secondary structure formation by defined pre-incubation procedures as established for the ‘more-than-unit-length’ transcripts (23,24) was transferred to the Mini-RNA: fast renaturation at –15°C in low ionic strength TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA; ‘low salt snap’) resulted in ExM-structure formation, whereas slow renaturation in high ionic strength, i.e. 45 min incubation at 40°C followed by slow cooling to room temperature overnight in 500 mM NaCl, 4 M urea, 1 mM cacodylate, 0.1 mM EDTA (‘high salt slow’) resulted in ExL-structure formation. Analysis of secondary structure formation was done by TGGE and UV crosslinking.
RESULTS
Construction of the Mini-RNA
A Mini-RNA of 148 nt containing almost only the CCR of PSTVd was designed with the aim to restore the processing characteristics of the more-than-unit-length transcripts TB110 (23) but to exclude as much as possible from the rest of the PSTVd molecule. It is shown in Figure 2. It comprises the 5′-end (GG80-G125) and the 3′-end (C69-G96AAUU) of TB110, i.e. containing the identical 17 nt terminal duplication G80-G96, and the corresponding lower strand of the rod-like secondary structure C236-G289. The left and right ‘arms’, A126-C235 and A290-359/1-G68, are substituted by 5′-CUUCGG-3′ segments forming extrastable UUCG tetraloops closed by C:G base pairs as artificial termini. A cDNA was synthesized by ligating appropriate oligonucleotides, PCR amplification and cloning in an in vitro transcription vector (see Materials and Methods). The construct of the Mini-RNA was optimized by secondary structure calculation with the program LinAll (29).
Analysis of structure and processing activity
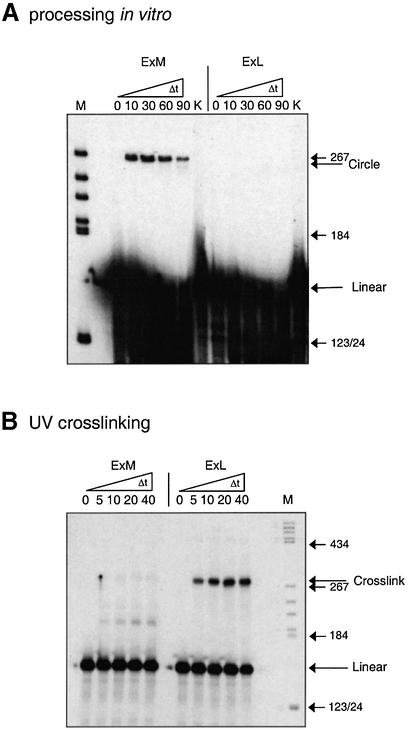
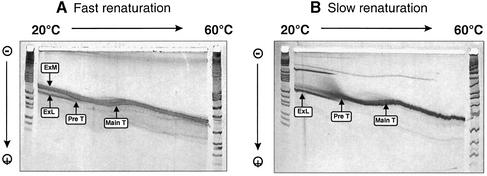
By analogy to the more-than-unit-length transcripts TB110 the thermodynamically optimal conformation of the Mini-RNA is expected to be the so-called ExL (extended left) conformation (Fig. 3). It looks similar to the native rod-like conformation of the mature circle with the 3′-segment of the duplication protruding unpaired. For TB110 the structure had been characterized by its ability to form the typical crosslink within loop E by UV irradiation and by its inactivity as template for correct processing in a potato nuclear extract (23). After pre-incubation conditions for kinetically favored structures the metastable ExM (extended middle) conformation was formed in TB110. It was processed to the mature circle in vitro and cannot be crosslinked, i.e. does not contain loop E. In correspondence with the experiments on the full-size transcript TB110, Mini-RNA in vitro transcripts were pre-incubated for formation of either the ExL (thermodynamic) or the ExM (kinetic) conformation (see Materials and Methods) and analyzed by UV crosslinking and processing in vitro (Fig. 4). Analyses of radioactively labeled transcripts on a sequencing gel after 0–40 min UV irradiation showed a retarded band for the thermodynamically pre-incubated transcripts only, in accordance with formation of ExL. Primer extension analyses of crosslinked transcripts eluted from the gel identified the crosslink site within loop E (data not shown). When the transcripts in the ExL or ExM conformation were incubated in the potato nuclear extract a retarded band was obtained only for the ExM conformation showing that the transcript in the ExM conformation was processed to a circular product. RT–PCR sequencing of the processed product, i.e. the retarded band, revealed a perfect minicircle with respect to the postulated processing mechanism (data not shown) (see Fig. 3). Thus the results of Figure 4 demonstrate that the Mini-RNA and the more-than-unit-length transcript TB110 behave identically concerning structure formation and processing activity. Conformations ExM and ExL are also distinguishable by TGGE (see Fig 5). The interpretation of these transition curves was given in detail for TB110 by Baumstark and Riesner (23). At 20°C, the two conformations of the Mini-RNA transcripts can be well separated with the branched ExM conformation being more retarded than the rod-like ExL conformation. In a pre-transition ExL shifts to ExM which denatures into the unpaired conformation within the main transition.
Figure 4.
Mini-RNA processing activity versus loop E formation in dependence upon the conformation of the Mini-RNA. Processing of the Mini-RNA, observed by minicircle formation, occurs only when pre-incubation for ExM formation was applied (A), whereas loop E formation, observed by UV crosslinking, only occurs when pre-incubation for ExL formation was applied (B). Pre-incubation, processing and UV crosslinking conditions were as described in Materials and Methods. K, control, 90 min mock incubation with buffer. M, marker, pBR322 cleaved with HaeIII.
Figure 5.
Secondary structure analysis of Mini-RNAs by TGGE. (A) Analysis after fast renaturation (‘low salt snap’) and (B) after slow renaturation (‘high salt slow’); for exact conditions see Materials and Methods. Fast renaturation leads predominantly to the ExM structure (ExM, arrow) which denatures in a single main transition at 40°C (Main T, arrow). A small amount of transcript forms the ExL conformation (ExL, arrow) which denatures in a pretransition at 30°C (Pre T, arrow). (B) Slow renaturation leads predominantly to the ExL conformation and only very little of the ExM conformation. Due to the slow renaturation process some bimolecular complexes appear which are characterized by high retardation and second order transitions resulting in the monomeric transcript. Interpretation of the faster migrating conformation as the extended structure ExL and of the slower migrating conformation as the bulky structure ExM was similar to that described previously (23).
Mutations in the structural motifs
The Mini-RNA obviously comprises all structural prerequisites for correct processing in vitro. As was discussed earlier (see also Fig. 3) a switch from a tetraloop to a loop E containing structure is essential for the mechanism, and the first cleavage occurs at a G:U wobble pair. Therefore, the structural and functional relevance of the three structural motifs mentioned were studied by site directed mutagenesis of the Mini-RNA, and analysis of the mutants was carried out with respect to their secondary structure by TGGE and crosslink analysis and to their template activity by the processing reaction in vitro.
Analysis of the mutants of the GAAA tetraloop and loop E
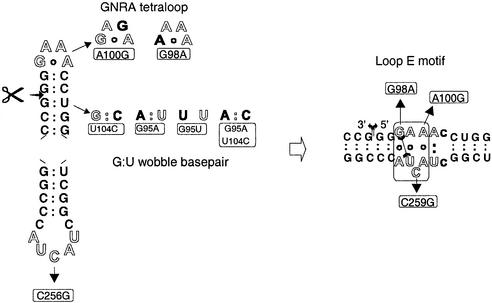
The mechanism of processing (see Fig. 3) shows that the identical sequence GAAA forms the tetraloop in the ExM conformation and the upper strand of loop E in the intermediates L1 and L2, respectively, as well as in the circular product. Thus, mutations in the tetraloop do also mutate loop E (see Fig. 6). In order to differentiate the effect of the mutations on the initial 5′-cleavage reaction in the ExM conformation from the effect on 3′-cleavage and ligation in loop E conformation, the linear intermediates L1 and L2 were synthesized and mutated as analogous Mini-RNAs as well. Independent analysis of processing activity of the ‘full-length’ Mini-RNA and the intermediates might allow us to identify the mechanistic steps influenced by the mutations. By single point mutations the GAAA tetraloop was substituted by a GAGA tetraloop, restoring the GNRA tetraloop consensus sequence and structure, or a ‘random’ AAAA loop without tetraloop features. The influence of these mutations on the structure of loop E is not known. Within the lower strand of loop E mutation C259G was introduced. Nucleotide C259 is the only nucleotide differing between loop E of PSTVd and the conserved loop E motif in 5S RNA and rRNA, thus restoring the general loop E motif. It should be mentioned that a natural mutation in this site (C259U) makes PSTVd (PSTVd-NT) replicating in the non-host plant Nicotiana tabacum (37). In the more-than-unit-length transcript TB110-NT the mutation did not influence structure formation and processing in the potato nuclear extract (O.Schrader, unpublished results).
Figure 6.
Mutations of the GNRA tetraloop, G:U wobble base pair and loop E motif in the Mini-RNA. The same nucleotides of the upper and the lower strand of the CCR (C93-G106 and U249-G263, respectively) form two small hairpins within the ExM conformation (left), and the loop E and its flanking helices within the ExL conformation (right). The L1- and L2-intermediates and the mature circle form also loop E. In the GAAA tetraloop single nucleotides were substituted leading to a GAGA tetraloop (A100G, designated in the text as R+) and an unstructured AAAA loop (G98A, designated in the text as A4). The single nucleotide mutations of the GNRA tetraloop necessarily also mutate the loop E motif (right). Mutation C259G is the only nucleotide of the conserved loop E motif of PSTVd which differs from and restores the known consensus loop E motif of other RNAs. The G:U wobble base pair at the 5′-cleavage site (marked by scissors) were substituted by G:C and A:U Watson–Crick base pairs (U104C, G95A), an A:C wobble base pair (G95A + U104C) and a U:U mismatch (G95U). In the ExL conformation, in the L1- and L2-intermediates and in the mature circle, the mutation U104C is located within the right flanking helix of the loop E motif, stabilizing it by substituting a G:U wobble base pair with a G:C Watson–Crick base pair. The mutations of G95 (A/U) are not present in the L1- and L2-intermediates and the circle because the mutated G95 is part of the 5′-end which is removed by the initial 5′-cleavage.
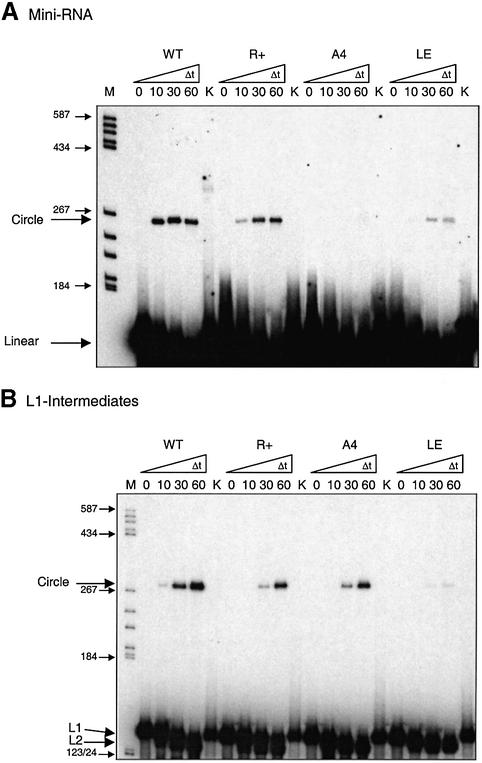
The processing activities of ‘full-length’ Mini-RNAs wild-type (GAAA tetraloop) and mutant R+ (GAGA tetraloop) are very similar whereas the mutant containing an AAAA loop (A4) does not show any processed, i.e. circularized product (Fig. 7A). The mutant containing the ‘restored’ loop E motif (LE) is processed only with very low efficiency. Processing of the L1 and L2 intermediates occurs with high efficiency for the wild-type and all mutants except LE. In Figure 7B L1 was used as substrate which is cleaved to L2 and circularized. All circularized processing products have been identified as correctly processed minicircles by elution from the gel and sequencing (data not shown). This clearly indicates that ‘restoration’ of the conserved loop E motif inhibits processing by interference with ligation. Whereas substitution of the GAAA tetraloop by a non-GNRA loop inhibits 5′-cleavage, the mutation does not interfere with 3′-cleavage and ligation when present in loop E.
Figure 7.
Processing activity of Mini-RNA mutants in the tetraloop and the loop E motif and their respective L1-intermediates. Conditions for processing are described in Materials and Methods. (A) Mini-RNA wild-type (WT) and mutants R+ (A100G), A4 (G98A) and LE (C259G, PSTVd loop E is substituted by the conserved loop E motif) as in Figure 6. (B) Corresponding L1-intermediate mutants.
Secondary structure analysis of the wild-type (Fig. 5) and mutant Mini-RNAs (data not shown) after pre-incubation for kinetically favored structures was carried out by TGGE. As expected from the processing activity, substitution of the GAAA tetraloop by GAGA tetraloop has no effect. In contrast, the mutant A4 forms the ExM conformation to very low degree only, most molecules form the ExL conformation. Furthermore, the small amount of ExM conformation is stable only below 25°C and most probably does not exist in the nuclear extract at 30°C. Thus, the correlation between absence of structure ExM and absence of processing activity is evident. The mutant containing the ‘restored’ 5S rRNA loop E forms both conformations, ExM and ExL, to almost equal extent. However, this lowered efficiency of forming the ExM conformation is not the main reason for inhibition of processing because processing of L1 and L2 intermediates is reduced as well (see above). Furthermore, the ExL conformation is stabilized. Whereas the ExL conformation of the wild-type and the R+ mutant are stable up to ∼30°C the ExL conformation of LE mutant is stable up to the main transition at ∼40°C. Obviously the restored loop E motif is more stable than the PSTVd loop E and therefore stabilizes the ExL conformation.
All mutants except R+ showed the typical loop E crosslink after pre-incubation for thermodynamically favored structure formation of the ‘full-length’ Mini-RNA (data not shown). The mutation A100G of R+ obviously abolishes loop E crosslinking by slight local structure rearrangement. However, this seems not to interfere with processing activity.
Analysis of mutants of the G:U wobble base pair
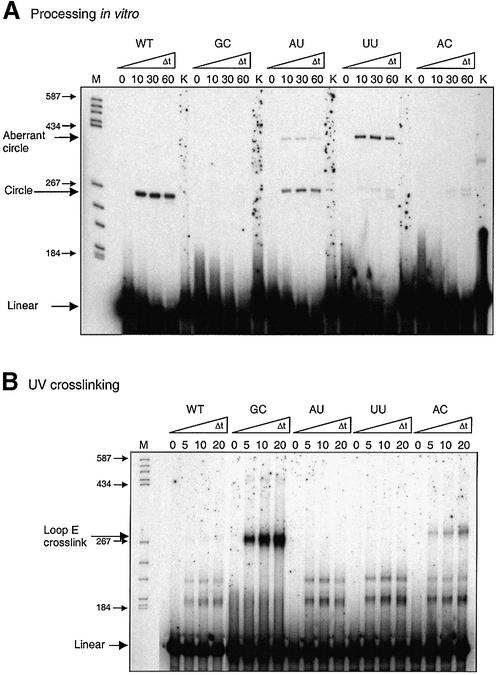
The properties of the mutants of the G:U wobble base pair are more difficult to understand; not only is formation of the correctly processed minicircle affected, but also aberrant minicircles, identified by a much more retarded band in the denaturing gel, were found with different intensities in different mutants (Fig. 8A). None of the mutants is processed correctly with an efficiency comparable to the wild-type. Substitution of the G:U base pair by a canonical Watson–Crick base pair resulted in complete suppression of processing for a G:C base pair and less effective but correct processing for an A:U base pair. The latter forms, in addition, a small amount of aberrant minicircles. U:U mismatch substitution almost totally abolishes correct processing but yields huge amounts of aberrant circles. A:C substitution also reduces strongly correct processing without aberrant minicircle production. Both minicircles have been eluted from the gel, amplified by RT–PCR and sequenced. Whereas the correct minicircles could be confirmed, the aberrant minicircles still contain the 17 nt duplication. Only the non-PSTVd nucleotides originating from the transcription vector were cut off (data not shown). Similar aberrant circles have been found as a side-product of in vitro processing of the more-than-unit-length transcripts TB110 (24).
Figure 8.
Processing activity versus loop E formation of the Mini-RNAs mutated in the G:U wobble base pair. Processing activity is observed by forming circles (A), and loop E formation by forming the specific crosslink (B). Incubation conditions were as in the experiment of Figure 4. Pre-incubation was chosen for favoring ExM structure. K, control, 90 min mock incubation with buffer. M, marker, pBR322 cleaved with HaeIII.
Crosslink analysis of the mutants after pre-incubation for kinetically favored structures clearly shows that the G:C mutant forms the crosslink typical for the loop E to high intensity extent, thus indicating the ExL conformation (Fig. 8B). One has to conclude from the absence of crosslinks in A:U and U:U mutants that these mutants form the ExM structure, as expected, and only the G:C mutant is dominated by the ExL structure. Consequently processing of the G:C mutant is inhibited because the right substrate conformation for the initial cleavage step is missing. Unexpectedly, the A:U and the U:U mutant, even after pre-incubation under thermodynamic equilibrium, do not show the crosslink, implying that the substrate conformation ExM, and not ExL, is the thermodynamically favored structure (data not shown). The A:C mutant does form some ExL conformation even after pre-incubation for kinetic structure, as can be seen from the low intensity loop E crosslink (Fig. 8A). These data are confirmed by TGGE secondary structure analysis in which ExM and ExL conformations can well be differentiated as was shown for the wild-type RNA in Figure 5. Whereas the G:C mutant predominantly forms the ExL conformation with very little ExM (with a markedly increased cooperativity of the main transition) all other three mutants form the ExM conformation (data not shown).
The reason for the drastic influence of G:U mutations on the secondary structure formation could be a priori the stabilization or destabilization of either the stem of the tetraloop hairpin in the ExM conformation or of the helices flanking loop E in the ExL conformation (see Fig. 6). In practice, the influence on ExL structure formation seems to dominate: mutation of G95 to A or U introduces a central mismatch in the helix left of the loopE which is not yet covalently linked at the G95/G96 position, thereby shifting the equilibrium to ExM conformation. Surprisingly the effect is seen even if a U:U mismatch is introduced in the stem in the ExM conformation. Mutation U104C results in a stable G:C base pair in the helix right of loop E, favoring the ExL formation over the ExM conformation, regardless of a stabilization of the GU to a GC substitution in the stem of ExM.
Thus, secondary structure formation does explain the abolished processing activity of the G:C mutant, but not easily the reduced activity of the A:U, U:U and A:C mutant. Considering formation of the ExM conformation the A:U pair and even the U:U mismatch should be at least an equivalent substrate for processing. The most plausible explanation for the inhibitory effect of these mutants therefore is a contribution of the original G:U wobble base pair to recognition of the 5′-cleavage site. This would also explain the induction of aberrant minicircles which are already ligated before the correct 5′-cleavage can occur.
DISCUSSION
A Mini-RNA of the CCR is substrate for processing in vitro
A Mini-RNA which contains little more than the CCR of PSTVd is a substrate for processing in a nuclear extract. This finding perfectly confirms the mechanistic model proposed by Baumstark et al. (24). According to this model, the CCR contains all structural features necessary for processing; thus it should be processable by the corresponding cellular enzymes in the same mode as the natural replication intermediates. The metastable conformation ExM was the substrate for initial 5′-cleavage, and the consecutive conformational shift to the rod-like conformation was interpreted as the driving force to 3′-cleavage and ligation. Exactly these conformational features and the conformational shift were reproduced in the Mini-RNA. It underlines the picture of the viroid as an RNA molecule which is built of independent structural and functional domains. The CCR thus may be regarded as the ‘processing domain’ of PSTVd and also of the whole genus of Pospiviroid due to the high conservation of the CCR. One should note that characterizing the CCR as ‘processing domain’ does not exclude processing at other sites of the molecule. Indeed Rakowski and Symons (38) reported that longer-than-unit-transcripts of citrus exocortis viroid are infectious when linearization is located in different parts of the molecule but the 3′- and 5′-ends can form double-stranded regions. In accordance with the results of the present work these results could be interpreted in terms of rare events of enzymatic cleavage, structural rearrangement and ligation. However, infectivity was not resolved into the different steps, and the authors favored restoration of the exact monomeric sequence by strand switching of the polymerase during transcription which mechanism would be quite different from the well defined steps of processing as described in the present work.
The genera Apscaviroid and Coleviroid also contain a CCR which might be involved in processing. In contrast, the family Avsunviroidae do not contain a CCR and form hammerhead structures for self-cleavage. Also in clear difference to PSTVd, which are replicating in the nucleus, these viroids are located in the chloroplast (39,40) and, therefore, are adapted to a different environment and corresponding host factors which might be recruited for replication.
Involvement of structural motifs in the processing mechanism
GAAA tetraloop. The GAAA tetraloop hairpin with the 5′-cleavage site at the G:U wobble pair is apparently a prerequisite for the processing activity of the ExM conformation; formation of the antagonistic loop E in the intermediates L1 and L2 drives 3′-cleavage and ligation. All three motifs are conserved within the CCR of the genus Pospiviroid (24). In this work they have been analyzed by the influence of point mutations on their structure and function.
Processing of the Mini-RNA in vitro tolerates a mutation of the GAAA tetraloop to a GAGA tetraloop. Both tetraloops belong to the type of GNRA tetraloops discussed in the literature as well defined and stable structures (41). Thus, a sequence specific interaction of the GAAA tetraloop does not appear essential for processing. Substitution of the GAAA tetraloop by the homoadenosin AAAA loop, however, completely abolished processing activity. It can be interpreted as loss of thermodynamic and structural features. The AAAA loop not only destabilizes the ExM conformation, as expected from its lower thermodynamic stability, but also inhibits the kinetically driven formation of the ExM conformation. It is known from the literature that small G:C-rich hairpins with small, favorable loop, i.e. tetraloops, form first on the hierarchical folding pathway of a higher order RNA structure (42–44). In the case of PSTVd, the stem region of the GAAA tetraloop stem, consisting of 80% G:C content, obviously functions as folding nucleus for the kinetically controlled formation of the conformation ExM which acts as substrate for processing. In the in vitro experiment kinetically favored structures are formed by ‘snap cooling’; the process might be regarded as a model for sequential folding of RNA during transcription in vivo. Co-transcriptional formation of the GNRA tetraloop hairpin presents the 5′-cleavage site and delays or prevents the stable rod-like structure within the CCR. Also for replication of PSTVd it was reported that sequential folding of metastable RNA structures is critical, namely the so-called hairpin II (HPII) in the (–)-strand replication intermediate (45–49). A functional role of co-transcriptionally formed and metastable structures was also considered in quite different systems such as folding of the Tetrahymena group I intron, translational regulation of bacterial mRNA and regulation of the Escherichia coli plasmid ColE1 copy number (50). The unusually high frequency of tetraloops in rRNA (51) might originate from a combination of tetraloop functions as folding nucleus, local high stability and importance in tertiary structure formation.
G:U wobble pair. Substitutions of the G:U pair at the 5′-cleavage site in the Mini-RNA were analyzed with respect to secondary structure formation and activity as a template for processing. The Watson–Crick A:U pair reduces correct processing efficiency and induces some aberrant circle production, but clearly maintains some correct processing. Substitution with a Watson–Crick G:C pair shifts the secondary structure to the inactive ExL conformation and therefore cannot be analyzed in terms of G:U recognition. Otherwise, substitution neither with an A:C nor with a U:U mismatch rescues significant amounts of correct circle production. Evidently, the G:U pair does contribute to correct processing by secondary structure formation, as seen for the G:C mutant, and in addition by recognition of the 5′-cleavage site within the conformation ExM; failure of recognition results in aberrant circles missing the correct 5′ cleavage.
Highly conserved G:U pairs are observed in various RNAs associated with diverse functions, e.g. as a major determinant for E.coli tRNAAla aminoacylation, regulation of the expression of the ribosomal protein S15 and at the cleavage sites of group I introns (5′) and the ribozyme part of the hepatitis delta virus RNA. The geometrical and dynamical characteristics of the wobble configuration as well as the pattern of hydrogen bonding donor and acceptor sites were discussed to promote specific recognition (52). For tRNAAla aminoacylation and group I intron 5′-cleavage the N2-exocyclic amino group of guanosine presented in the shallow groove was described as the major determinant (53,54). In those cases substitution of the G:U wobble pair by canonical Watson–Crick pair led to complete inactivation, whereas some group I intron activity was rescued by forming non-canonical pairs. Thus, the results in the literature on G:U recognition and the results of this work cannot be interpreted at present in a unique structural scheme.
Loop E. The functional relevance of the loop E motif in the processing mechanism was analyzed by the three mutations A100G (R+), G98A (A4) and C259G (LE). One has to keep in mind that A100G (R+) and G98A (A4) are mutations of the GAAA tetraloop as well (see above). Actually, the mutation A100G (R+) inhibits the typical loop E UV crosslink in the ExL conformation as well as in the intermediates, presumably by slightly local deformation of the non-canonical loop structure. The mutation G98A (A4), however, substituting the crosslinked nucleotide G98 itself, does not interfere with crosslink formation. Both mutations do not significantly inhibit processing by its influence on loop E formation as shown by efficient processing of the intermediates. In contrast, mutation C259G (LE) almost completely abolishes processing of the full-length Mini-RNA as well as of the intermediates. The result, that even processing of the L2 intermediate is inhibited, indicates that ligation is the affected mechanistic step (see Fig. 3). As outlined before, mutation of the same nucleotide to U enables PSTVd to replicate in the non-host plant N.tabacum (37), implying involvement in some kind of species-specific interaction.
Mutation of the analogous nucleotide in the loop E of 5S rRNA abolishes binding of transcription factor TFIIIA in vitro (55); the analogous nucleotide in the loop E motif of the so-called sarcin–ricin loop of 23S rRNA is protected from chemical modification by the elongation factors EF-G and EF-Tu (56) and is the recognition element for the ribotoxins α-sarcin (57) and restrictocin (58). Also theoretical and experimental structural studies (59) indicated that this nucleotide within the loop E motif apparently functions as a recognition site for RNA–protein interaction.
Binding of a host-specific ligase would best explain also the in vitro and in vivo mutation data with PSTVd. However, the mutated nucleotide is the only nucleotide differing between loop E of PSTVd and the highly conserved core of the loop E motifs mentioned above, and the mutation C259G which exactly restores the conserved sequence inhibits the ligation activity. Consequently, loop E in PSTVd has to have exceptional features as compared to the loop E structure in the other examples mentioned. The crucial nucleotide, the so-called bulged-G motif, is not involved in the non-canonical base pairs constituting the loop E structure but forms a base triple with one of them (60). An adenosine at this site does not form the base triple resulting in significant destabilization of the loop (61). The loop E motif in the hairpin ribozyme (loop B), containing a pyrimidine (U) at this site is also very unstable and does not form the base triple (62). Presumably, the wild-type PSTVd loop E also does not contain the base triple and is less stable than the ‘original’ loop E. In fact, restoration of the ‘original’ loop E stabilizes the loop E conformation, as has been observed in the TGGE analysis. One might argue that loop E in PSTVd-WT (C bulged out) and in PSTVd-NT (U bulged out) does not form a base triple at that site and therefore represents a more flexible structure. This flexibility may be essential for the interaction with a host ligase, which is certainly not optimized for recognition of the parasitic viroid RNA.
Sequence comparison between the PSTVd CCR and 5S rRNA of the host plant Solanum tuberosum (taken from the 5S ribosomal RNA database, http://rose.man.poznan.pl/5SData/) (63) reveals 70% homology between the loop E motifs plus the left and right hand closing helices (helix IV and V in 5S rRNA). Also loop D as a GNRA tetraloop and helix I of 5S rRNA are conserved in the CCR. Whereas most plant 5S rRNAs are not able to form an alternative conformation to loop E which would be homologous to the PSTVd conformation ExM, an ExM-like conformation was reported for the 5S rRNA of Xenopus laevis (64). A closer look at the non-homologous nucleotides between PSTVd CCR and the plant 5S rRNA reveals that exactly those nucleotides differ which enable PSTVd to form the ExM conformation (data not shown). One is tempted to speculate about a two-fold relationship between the ‘processing domain’ CCR of PSTVd and a domain of the host plant 5S rRNA: on one hand, a conservative relation with respect to sequence and structure is based on a common origin or co-evolution which is, on the other hand, modified by divergent functional requirements. PSTVd had to acquire the structural requirements of the processing mechanism, in particular the formation of a metastable initial substrate conformation. The CCR of the PSTVd subgroup might be envisaged as a host 5S rRNA domain picked up by the viroids and adapted to new functions, i.e. processing of viroid RNA.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Dr M. Wassenegger for providing us with plasmid pTB110-NT. Many thanks to Drs P. Klaff, M. Schmitz and G. Steger for numerous stimulating discussions, and to H. Gruber for help in preparing the manuscript. The work was supported by grants from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Riesner D. and Gross,H.J. (1985) Viroids. Annu. Rev. Biochem., 54, 531–564. [DOI] [PubMed] [Google Scholar]
- 2.Diener T.O. (ed.) (1987) The Viroids. Plenum Publishing Corporation. New York, NY.
- 3.Semancik J.S. (ed.) (1987) Viroids and Viroid-like Pathogens. CRC Press, Boca Raton, FL.
- 4.Symons R.H. (ed.) (1990) Viroids and related pathogenic RNAs. Semin. Virol., Vol. 1, Issue 2, Saunders Scientific Publications, Philadelphia, PA. [Google Scholar]
- 5.Diener T.O. (1971) Potato spindle tuber virus: a plant virus with properties of a free nucleic acid. III. Subcellular location of PSTV-RNA and the question of whether virions exist in extracts or in situ. Virology, 43, 75–82. [DOI] [PubMed] [Google Scholar]
- 6.Diener T.O. (1999) Viroids and the nature of viroid diseases. Arch. Virol. Suppl., 15, 203–220. [DOI] [PubMed] [Google Scholar]
- 7.Gross H.J., Domdey,H., Lossow,C., Jank,P., Raba,M., Alberty,H. and Sänger,H.L. (1978) Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature, 273, 203–208. [DOI] [PubMed] [Google Scholar]
- 8.Riesner D., Henco,K., Rokohl,U., Klotz,G., Kleinschmidt,A.K., Domdey,H., Jank,P., Gross,H.J. and Sänger,H.L. (1979) Structure and structure formation of viroids. J. Mol. Biol., 133, 85–115. [DOI] [PubMed] [Google Scholar]
- 9.Branch A.D. and Robertson,H.D. (1984) A replication cycle for viroids and other small infectious RNAs. Science, 223, 450–455. [DOI] [PubMed] [Google Scholar]
- 10.Mühlbach H.P. and Sänger,H.L. (1979) Viroid replication is inhibited by α-amanitin. Nature, 278, 185–188. [DOI] [PubMed] [Google Scholar]
- 11.Schindler I.-M. and Mühlbach,H.-P. (1992) Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci., 84, 221–229. [Google Scholar]
- 12.Flores R., Navarro,J.A., de la Pena,M., Navarro,B., Ambros,S. and Vera,A. (1999) Viroids with hammerhead ribozymes: some unique structural and functional aspects with respect to other members of the group. Biol. Chem., 380, 849–854. [DOI] [PubMed] [Google Scholar]
- 13.Tsagris M., Tabler,M. and Sänger,H.L. (1987) Oligomeric potato spindle tuber viroid (PSTV) RNA does not process autocatalytically under conditions where other RNAs do. Virology, 157, 227–231. [DOI] [PubMed] [Google Scholar]
- 14.Tabler M. and Sänger,H.L. (1984) Cloned single- and double-stranded DNA copies of potato spindle tuber viroid (PSTV) RNA and coinoculated subgenomic DNA fragments are infectious. EMBO J., 3, 3055–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meshi R., Ishikawa,M., Watanabe,Y., Yamaya,J., Okada,Y., Sano,T. and Shikata,E. (1985) The sequence necessary for the infectivity of hop stunt viroid cDNA clones. Mol. Gen. Genet., 200, 199–206. [Google Scholar]
- 16.Candresse T., Diener,T.O. and Owens,R.A. (1990) The role of the viroid central conserved region in cDNA infectivity. Virology, 175, 232–237. [DOI] [PubMed] [Google Scholar]
- 17.Rakowski A.G. and Symons,R.H. (1994) Infectivity of linear monomeric transcripts of citrus exocortis viroid: terminal sequence requirements for processing. Virology, 203, 328–335. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto J. and Machida,Y. (1985) The sequence in the potato spindle tuber viroid required for its cDNA to be infective: a putative processing site in viroid replication. J. Gen. Appl. Microbiol., 31, 551–561. [Google Scholar]
- 19.Diener T.O. (1986) Viroid processing: a model involving the central conserved region and hairpin I. Proc. Natl Acad. Sci. USA, 83, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steger G., Tabler,M., Brüggemann,W., Colpan,M., Klotz,G., Sänger,H.L. and Riesner,D. (1986) Structure of viroid replicative intermediates: physico-chemical studies on SP6 transcripts of cloned oligomeric potato spindle tuber viroid. Nucleic Acids Res., 14, 9613–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecker R., Wang,Z., Steger,G. and Riesner,D. (1988) Analysis of RNA structures by temperature-gradient gel electrophoresis: viroid replication and processing. Gene, 72, 59–74. [DOI] [PubMed] [Google Scholar]
- 22.Steger G., Baumstark,T., Mörchen,M., Tabler,M., Tsagris,M., Sänger,H.L. and Riesner,D. (1992) Structural requirements for viroid processing by RNase T1. J. Mol. Biol., 227, 719–737. [DOI] [PubMed] [Google Scholar]
- 23.Baumstark T. and Riesner,D. (1995) Only one of four possible secondary structures of the central conserved region of potato spindle tuber viroid is a substrate for processing in potato nuclear extract. Nucleic Acids Res., 23, 4246–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumstark T., Schröder,A.R.W. and Riesner,D. (1997) Viroid processing: a switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J., 16, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branch A.D., Benenfeld,B.J. and Robertson,H.D. (1985) Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc. Natl Acad. Sci. USA, 82, 6590–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul C.P., Levine,B.J., Robertsom,H.D. and Branch,A.D. (1992) Transcripts of the viroid central conserved region contain the local tertiary structural element found in full-length viroid. FEBS Lett., 305, 9–14. [DOI] [PubMed] [Google Scholar]
- 27.Leontis N.B. and Westhof,E. (1998) Conserved geometrical base-pairing patterns in RNA. Q. Rev. Biophys., 31, 399–455. [DOI] [PubMed] [Google Scholar]
- 28.Schröder A.R.W., Baumstark,T. and Riesner,D. (1998) Chemical mapping of co-existing RNA structures. Nucleic Acids Res., 26, 3449–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz M. and Steger,G. (1992) Base-pair probability profiles of RNA secondary structure. Comp. Appl. Biosci., 8, 389–399. [DOI] [PubMed] [Google Scholar]
- 30.Groebe D.R. and Uhlenbeck,O.C. (1988) Characterization of RNA hairpin loop stability. Nucleic Acids Res., 16, 11725–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antao V.P. and Tinoco,I.,Jr (1992) Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res., 20, 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antao V.P., Lai,S.Y. and Tinoco,I.,Jr (1991) A thermodynamic study of unusually stable RNA and DNA hairpin. Nucleic Acids Res., 19, 5901–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steger G. (1994) Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res., 22, 2760–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyatt I.R., Chastian,M. and Puglisi,I.D. (1991) Synthesis and purification of large amounts of RNA oligo. Biotechniques, 11, 764–769. [PubMed] [Google Scholar]
- 35.Krupp G. (1988) RNA synthesis: strategies for the use of bacteriophage RNA polymerases. Gene, 72, 75–89. [DOI] [PubMed] [Google Scholar]
- 36.Rosenbaum V. and Riesner,D. (1987) Temperature-gradient gel electrophoresis: thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys. Chem., 26, 235–246. [DOI] [PubMed] [Google Scholar]
- 37.Wassenegger M., Spieker,R.L., Thalmeir,S., Gast,F.-U., Riedel,L. and Sänger,H.L. (1996) A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology, 262, 191–197. [DOI] [PubMed] [Google Scholar]
- 38.Rakowski A.G. and Symons,R.H. (1994) Infectivity of linear monomeric transcripts of citrus exocortis viroid: terminal sequence requirements for processing. Virology, 203, 328–335. [DOI] [PubMed] [Google Scholar]
- 39.Bussiere F., Lehoux,J., Thompson,D.A., Skrzeczkowski,L.J. and Perreault,J. (1999) Subcellular localization and rolling circle replication of peach latent mosaic viroid: hallmarks of group A viroids. J. Virol., 73, 6353–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro J.A., Daros,J.A. and Flores,R. (1999) Complexes containing both polarity strands of avocado sunblotch viroid: indentification in chloroplasts and characterization. Virology, 253, 77–85. [DOI] [PubMed] [Google Scholar]
- 41.Varani G. (1995) Exceptionally stable nucleic acids hairpin. Annu. Rev. Biophys Biomol. Struct., 24, 379–404. [DOI] [PubMed] [Google Scholar]
- 42.Brion P. and Westhof,E. (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys Biomol. Struct., 26, 113–137. [DOI] [PubMed] [Google Scholar]
- 43.Tinoco I. Jr and Bustamante,C. (1999) How RNA folds. J. Mol. Biol., 293, 271–281. [DOI] [PubMed] [Google Scholar]
- 44.Woodson S.A. (2000) Recents insights into RNA folding mechanisms from catalytic RNA. Cell. Mol. Life Sci., 57, 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riesner D., Baumstark,T., Feng,Q., Klahn,T., Loss,P., Rosenbaum,V., Schmitz,M. and Steger,G. (1992) Physical basis and biological examples of metastable RNA structures. In Structural Tools for the Analysis of Protein-Nucleic Acids Complexes, Advances in Life Sci., Birkhäuser Verlag, Basel, Switzerland.
- 46.Schmitz M. and Steger,G. (1996) Description of RNA folding by “Simulated Annealing”. J. Mol. Biol., 255, 254–266. [DOI] [PubMed] [Google Scholar]
- 47.Gultyaev A.P., van Batenburg,F.H. and Pleij,C.W. (1998) Dynamic competition between alternative structures in viroid RNA simulated by an RNA folding algorithm. J. Mol. Biol., 276, 43–55. [DOI] [PubMed] [Google Scholar]
- 48.Repsilber D., Wiese,S., Rachen,M., Schröder,A.R.W., Riesner,D. and Steger,G. (1999) Formation of metastable RNA structures by sequential folding during transcription: time-resolved structural analysis of potato spindle tuber viroid (–)-strand RNA by temperature-gradient gel electrophoresis. RNA, 5, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schröder A.R.W. and Riesner,D. (2002) Detection and analysis of HP II, an essential metastable structural element in viroid replication intermediates. Nucleic Acids Res., 30, 3349–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klaff P., Riesner,D. and Steger,G. (1996) RNA structure and the regulation of gene expression. Plant Mol. Biol., 32, 89–106. [DOI] [PubMed] [Google Scholar]
- 51.Woese C.R., Winker,S. and Gutell,R.R. (1990) Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc. Natl Acad. Sci. USA, 87, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masquida B. and Westhof,E. (2000) On the wobble G:U and related pairs. RNA, 6, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musier-Forsyth K. and Schimmel,P. (1992) Functional contacts of a transfer RNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature, 357, 513. [DOI] [PubMed] [Google Scholar]
- 54.Strobel S.A. and Cech,T.R. (1995) Minor groove recognition of the conserved G:U pair at the Tetrahymena ribozyme reaction site. Science, 267, 675–679. [DOI] [PubMed] [Google Scholar]
- 55.Theunissen O., Ruidt,F. and Pieler,T. (1998) Structural determinants in 5S RNA and TFIIIA for 7S RNP formation. Eur. J. Biochem., 258, 758–767. [DOI] [PubMed] [Google Scholar]
- 56.Moazed D., Robertson,J.M. and Noller,H. (1988) Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature, 334, 362–364. [DOI] [PubMed] [Google Scholar]
- 57.Glück A. and Wool,I.G. (1996) Determination of the 28S ribosomal RNA identity element (G4319) for alpha-sarcin and the relationship of recognition to the selection of the catalytic site. J. Mol. Biol., 256, 838–848. [DOI] [PubMed] [Google Scholar]
- 58.Yang X. and Moffat,K. (1996) Insights into specifity of cleavage and mechanism of cell entry from the crystal structure of the highly specific Aspergillus ribotoxin, restrictocin. Structure, 4, 837–852. [DOI] [PubMed] [Google Scholar]
- 59.Chin K., Sharp,K.A., Honig,B. and Pyle,A.M. (1999) Calculating the electrostatic properties of RNA provides new insight into molecular interactions and function. Nature Struct. Biol., 6, 1055–1061. [DOI] [PubMed] [Google Scholar]
- 60.Corell C.C., Freeborn,B., Moore,P.B. and Steitz,T.A. (1997) Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell, 91, 705–712. [DOI] [PubMed] [Google Scholar]
- 61.Seggerson K. and Moore,P.B. (1998) Structure and stability of variants of the sarcin-ricin loop of 28S rRNA: NMR studies of the prokaryotic SRL and a functional mutant. RNA, 4, 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butcher S.E. and Burke,J.M. (1994) A photo-cross-linkable tertiary structure motif found in functionally distinct RNA molecules is essential for catalytic function of the hairpin ribozyme. Biochemistry, 33, 992–999. [DOI] [PubMed] [Google Scholar]
- 63.Erdmann V.A., Szymanski,M., Hochberg,A., de Groot,N. and Barciszewski,J. (2000) Non-coding, mRNA-like RNAs database Y2K. Nucleic Acids Res., 28, 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pieler T., Guddat,U., Oei,S.L. and Erdmann,V.A. (1986) Analysis of the RNA structural elements involved in the binding of the transcription factor III A from Xenopus laevis. Nucleic Acids Res., 14, 6313–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]