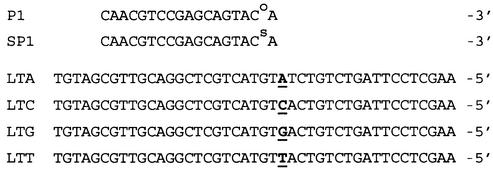Abstract
Model single base extension (SBE) genotyping reactions with individual deoxy-, dideoxy- and acyclonucleoside triphosphates are monitored by MALDI-TOF mass spectrometry. Three non-proofreading DNA polymerases display remarkably high misincorporation (up to 64% of correct incorporation) when extending primers with single substrates at saturating concentrations. Introduction of one phosphorothioate (PS) linkage into the primer 3′ terminus reduces misincorporation by these enzymes an average 1.4-fold (range 0- to 3.5-fold) versus correct incorporation. Combined use of 3′-PS primers with strongly proofreading DNA polymerases yields order of magnitude improvements in SBE fidelity over those produced by the equivalent non-proofreading enzymes. Errors are reduced to below MALDI-TOF detectable levels in almost all cases. The Sp diastereomer of the 3′-PS primer, which can be prepared in situ by incubation with proofreading polymerase, is stable to 3′-exonuclease activity over periods longer than 16 h. Products of correct extension by T7 DNAP are retained over 30–60 min during idling turnover at a dNTP concentration of 2.5 µM, indicating that the assay can be applied over a broad range of substrate concentrations. These results suggest that the use of PS primers and proofreading polymerases will offer a simple and cost-effective means to improve fidelity in a range of single-substrate SBE assay formats.
INTRODUCTION
Genotyping of single nucleotide polymorphisms (SNPs) has grown strongly in recent years, a major impetus for this growth coming from the search for disease-susceptibility genes (1) and from the promise of pharmacogenomics and personalised medicine for health care (2). Almost all SNP genotyping methods are based upon one or more of four fundamental molecular biological processes: hybridisation, polymerisation (nucleotidyl transfer), ligation and nucleolysis (3,4). Polymerisation is a particularly powerful tool, with applications ranging from full Sanger sequencing through limited pyrosequencing (5) to single base extension (SBE) or ‘minisequencing’ methods which identify a single allelic nucleotide immediately adjacent to a defined primer terminus (6). SBE has proven particularly attractive for its simplicity (the minimal implementation contains only three major added components: primer, polymerase and nucleoside triphosphate substrate) and for its adaptability to various detection formats. Typically, SBE is considered to offer about an order of magnitude better SNP discrimination than allele-specific hybridisation (7).
Most of the allele discrimination power of SBE resides in the polymerase active site. Generally accepted in vitro error rates for polymerase base insertion vary between 10–3 and 10–6 (8), suggesting that even low fidelity enzymes such as reverse transcriptase should be suitable for highly quantitative SBE genotyping. However, these low error rates have been measured for fully processive polymerisation under optimal conditions, whereas SBE is not truly processive and assay conditions are frequently suboptimal. For example, reactions may contain only one or two labelled terminator substrates at saturating levels, introducing the potential for ‘forced’ misincorporation. Although SBE genotyping studies do not usually report such data, up to 5% misincorporation of ddTTP and ddCTP has been described (9) or is apparent (10) in published assays. Anomalous allele frequencies that imply up to 30% misincorporation have been reported for homogeneous assays with the four ddNTPs (11) or four dye-labelled ddNTPs (12) and in limited-base extensions containing three ddNTPs and one dNTP (13). Inhomogeneous assays such as arrayed primer extension (APEX) and bead minisequencing present additional complications due to local concentration and steric effects, generally exhibiting significant variability (7,14,15) that can be at least partially attributed to misincorporation. Given these observations, current SBE methods generally do not lend themselves to the determination of allele frequencies in pooled or markedly heterogeneous samples, which is desirable for some applications. Moreover, while SBE is generally considered quite reliable for biallelic discrimination (6), there is benefit in further improving its robustness and increasing genotype-calling accuracy (14,16).
A direction towards improving the misincorporation properties of SBE has been provided by the primer-specific and mispair extension analysis (PSMEA) method (17,18), where the proofreading 3′→5′ exonuclease activity of Pfu DNA polymerase has been employed to provide highly sensitive discrimination between genotypes. Proofreading typically improves polymerase copy fidelity by one to two orders of magnitude (8), but direct application of proofreading polymerases to conventional SBE is not possible because the exonuclease activity of these enzymes causes extensive primer degradation (9,19). In principle, this degradation can be prevented through the use of primers with nuclease-resistant chain alterations (20), the simplest being the phosphorothioate (PS) modification, which is available from commercial oligonucleotide suppliers at modest cost. PS groups have long been known to inhibit nucleases, including the proofreading exonuclease of Escherichia coli DNA polymerase (DNAP) I (21), the phosphorothioate Sp diastereoisomer being more inhibitory than the Rp isomer (22). Phosphorothioate primers have previously been employed to expand the application of proofreading polymerases to PCR reactions, where primer degradation can result in erroneous amplification (23,24) and to multiply primed rolling circle amplification by Φ29 DNAP, where exonuclease-resistant primers greatly improve product yields (25).
In this report we examine the performance of PS-modifed primers and four representative proofreading polymerases in homogeneous single-substrate model SNP assays. Proof reading and non-proofreading variants of three polymerases (Klenow fragment and T7 DNAP in isothermal mode and Vent DNAP in thermocycling mode) are directly compared. For this proof-of-principle study, SBE products are detected by MALDI-TOF mass spectrometry to allow the simultaneous observation of all primer extension and degradation products. Suboptimal reaction conditions (high substrate concentrations) are employed to provide a stringent test of assay improvement for deoxy-, dideoxy- and acyclonucleotide substrate families under the sensitivity limitations of MALDI-TOF detection. The effects of primers and polymerases are separately quantitated to establish their relative contributions to SBE genotyping accuracy. The robustness of the single-substrate model assay to variation of polymerase and substrate concentrations is then evaluated as a function of reaction time.
MATERIALS AND METHODS
Oligonucleotides
The synthetic 18mer primers and 43mer templates shown in Figure 1 were purchased in desalted form from Sigma-Genosys or Genset Pacific. All oligonucleotides were purified by RP-HPLC on a 9.4 × 250 mm Zorbax ODS column with a 0–50% gradient of acetonitrile in 50 mM LiClO4. The major eluting peak volume was reduced in a vacuum concentrator (Eppendorf) prior to 10 vol of acetone being added for precipitation by centrifugation. Purified oligonucleotides were washed with acetone, dissolved in milli-Q water and desalted by spin chromatography (Micro Bio-Spin P-6; Bio-Rad). Oligonucleotides were quantitated by spectrophotometry using ε260 values provided by the supplier. Purity was checked by MALDI-TOF mass spectrometry.
Figure 1.
Primer and template sets. The corresponding phosphodiester and phosphorothioate groups of the P1 and SP1 primers are indicated by o and s, respectively. Template residues that direct primer extension are underlined in bold.
Polymerases and substrates
Wild-type and exo– Klenow fragment of E.coli DNAP I, T4 DNAP and T7 DNAP (MBI Fermentas) were purchased from Progen Industries. Sequenase 2.0 (USB) was purchased from AP Biotech. Wild-type and exo– Vent (Thermococcus litoralis) DNAPs were purchased from New England Biolabs. Exonuclease III (exo III) was from Promega. Deoxynucleotide triphosphates (Promega), dideoxynucleotide triphosphates (MBI) and acyclonucleotide triphosphates (NEB) were purchased commercially. Substrate concentrations were used as supplied by the manufacturer.
UV melting
Thermal denaturation experiments were performed on a Varian Cary Bio 100 spectrophotometer equipped with a thermal accessory. Primer/template pairs at 1 µM per strand were heated to 90°C in buffer (20 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 25 mM NaCl) and slow cooled before the experiment. Melting curves were analysed with Cary Thermal software, which includes derivative and van’t Hoff analysis modes.
Primer degradation
Individual primers or primer/template pairs were incubated with 1 U of appropriate polymerase in buffer. Primer remaining after incubation was quantitated relative to 100 pmol of a standard oligonucleotide (m/z 6010) added post-reaction.
Primer extension reactions
Each primer extension reaction contained either the P1 or SP1 primer (Fig. 1), one of the LT series templates (Fig. 1) and a single triphosphate substrate. The four templates each carried a different base at position n + 1 relative to the 3′ end of the primer. All primers were identical at other positions with the exception of LTA, which also differed at position n + 2 in order to avoid the intentional addition of two sequential dT residues with dTTP as substrate.
Primer extension reactions were initiated by polymerase addition. Reactions were performed in buffers supplied by the polymerase manufacturers: 50 mM Tris–HCl (pH 8.0 at 25°C), 5 mM MgCl2, 1 mM DTT for Klenow fragment species; 67 mM Tris–HCl (pH 8.8 at 25°C), 6.6 mM MgCl2, 1 mM DTT, 16.8 mM (NH4)2SO4 for T4 DNAP; 40 mM Tris–HCl (pH 7.5 at 25°C), 20 mM MgCl2, 50 mM NaCl for Sequenase 2.0; 40 mM Tris–HCl (pH 7.5 at 25°C), 10 mM MgCl2, 1 mM DTT for T7 DNAP; 20 mM Tris–HCl (pH 8.8 at 25°C), 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100 for Vent DNAP species.
The basic reaction mix for mesophilic polymerases contained 10 µM primer, 25 µM template and 1–2 U enzyme in a 10 µl volume. The basic reaction mix for thermophilic Vent polymerase contained 10 µM primer, 4 µM template and 1–2 U enzyme in a 10 µl volume. In trial experiments, reaction conditions for each combination of polymerase–substrate family (deoxy, dideoxy, acyclo) were established to accommodate the differing incorporation rates between these groups. Substrate concentrations and incubation times were varied so that incorporation of all correct substrates within each polymerase–nucleotide family utilised at least 50% of the available primer. Concentrations were then held at those values for the A, C, G and T members of that family. For the trial experiments, substrate concentrations were varied from 100 µM to 1 mM, incubation times from 15 min to 6 h and polymerase from 0.25 to 2 U per reaction. For all final reactions, nucleotide concentrations were kept at least four times above known Kd or Km values when available, with the enzyme concentration and incubation time varied accordingly. For reactions utilising extrinsic exo III, 0.1–10 U exonuclease were added to the normal polymerase reaction mixture. For time course experiments with limited substrate concentrations, 2.5 µM primer was combined with 10 µM LTG and 2.5, 10, 40 or 160 µM dCTP. Samples at each substrate concentration were incubated for 10, 30, 90 or 180 min with either 0.25 or 1 U enzyme in a 20 µl volume.
Mass spectrometry
Following incubation, all primer extension reactions were stopped by addition of EDTA to 20 mM, with 5 min heating at 95°C for mesophilic enzymes. Salts, detergents, proteins and other contaminants were removed with C18 Ziptips (Millipore) according to the manufacturer’s instructions. Samples were eluted onto a MALDI-TOF target (PerSeptive Biosystems) with a freshly prepared solution of nine parts 50 mg/ml 3-hydroxypicolinic acid in 50% acetonitrile/Milli-Q water with one part 50 mg/ml ammonium citrate in Milli-Q water. Mass data acquisition and analyses were performed with a Voyager-DE mass spectrometer (PerSeptive Biosystems).
For each data point, 100 shots were collected from various sites within the target spot and summed directly in software. A constant laser power setting (2020 arbitrary units) was employed for all experiments. Data were collected in linear negative ion mode, which in our hands yielded equivalent results to positive ion mode. Titration experiments with primer, extended primer and standard oligonucleotides indicated equivalent ionisation of these species within experimental error. Following data acquisition, noise reduction was performed by smoothing.
Voyager analysis software was used to determine peak areas for extension product distribution and quantitation. For experiments without an added internal standard, incorporation was determined according to E = Ae/(Ae + Ap) and converted into a percentage, where Ap = peak area for unextended primer and Ae = peak area(s) for extension product(s). Incorporation was determined as 2E for experiments in which a second aliquot of primer standard was added post-reaction. Linear response of peak area to amount of primer added was validated through construction of a standard curve. For each polymerase/primer pair, the average misincorporation ratio (I/Cavg) is expressed as the ratio of average incorrect to correct extension above the limit of detection. Standard deviations for repeated primer extension experiments were less than the limit of detection (5%).
RESULTS
Primer/template pairs
We investigated a wide range of SBE reactions using the set of synthetic primers and templates shown in Figure 1. When hybridised with primers, the LTA, LTC, LTG and LTT templates are designed to direct incorporation of a single T, G, C or A nucleotide, respectively. Two primers were used for comparative analysis: primer P1 contains a normal 3′-phosphodiester (PO) linkage, while the corresponding primer SP1 contains a single 3′-phosphorothioate (PS) linkage introduced for resistance to proofreading exonuclease activity. The use of a single primer–template set enables the direct comparison of misincorporation behaviour for several enzymes and three substrate classes.
Before introducing polymerases, UV melting experiments were performed to determine the extent of any primer/template destabilisation caused by the 3′ terminal PS group. The P1/LTA pair melted at a Tm of 65°C, with the corresponding SP1/LTA pair also melting at 65°C under the same conditions. As expected from the identical Tm values, van’t Hoff analysis yielded very similar energetic parameters for each of the primer/template pairs: ΔH = –123 kcal mol–1 and ΔS = –336 cal K–1 mol–1 for the P1/LTA pair, and ΔH = –118 kcal mol–1 and ΔS = –320 cal K–1 mol–1 for SP1/LTA.
Exonuclease activity
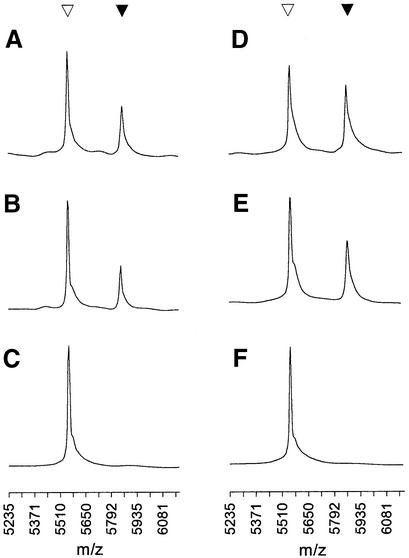
Proofreading polymerases have not previously been employed in SBE assays due to primer degradation by their 3′→5′ exonuclease activity (9,19). To qualitatively examine this issue, P1 and SP1 primers or P1/LTA and SP1/LTA primer/template pairs were incubated with proofreading polymerases and primer degradation monitored by MALDI-TOF mass spectrometry. Results for primer incubation with T4 DNAP, which has a strong exonuclease activity, are shown in Figure 2. As expected, T4 DNAP extensively hydrolyses the natural P1 primer; the intact P1 peak at m/z 5478 (Fig. 2A) is almost completely degraded to a mixture of shorter products in <10 min (Fig. 2B). The masses of these products correspond to the 5′ end of the progressively shortened P1 sequence, as expected. Less than 1 h is required to degrade the P1 primer to trinucleotide or smaller fragments (Fig. 2C). In contrast to the behaviour of P1, the 3′-phosphorothioate primer SP1 at m/z 5494 (Fig. 2D) remains largely intact after incubation with T4 DNAP for 1 h (Fig. 2E) or 16 h overnight (Fig. 2F). The exonuclease activities of T7 and Vent DNAPs show similar behaviour to T4 DNAP (data not shown). Vent and other thermophilic DNAPs are typically employed in cycled reactions with primer in substantial excess over template.
Figure 2.
Primer degradation by a proofreading polymerase with strong 3′→5′ exonuclease activity. Intact primer is indicated (inverted open triangle). (A–C) P1 primer incubated with T4 DNAP for 0, 0.17 and 1.0 h. (D–F) SP1 primer incubated with T4 DNAP for 0, 1.0 and 16.0 h.
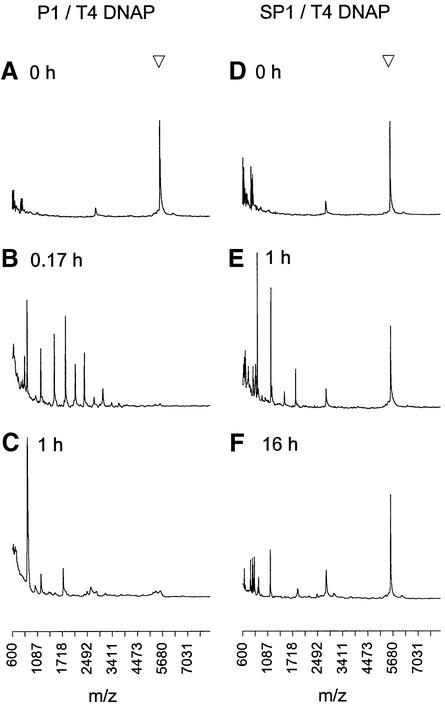
While a large amount of intact SP1 primer clearly remains after long-term incubation with T4 DNAP and other proofreading polymerases, there is some degradation apparent in the appearance of new peaks, mostly at m/z < 1200, corresponding to products smaller than tetramers (Fig. 2E and F). To investigate further, we performed quantitative degradation experiments with two proofreading polymerases. As shown in Figure 3, exo+ Klenow fragment (KF) causes minimal degradation of SP1 over 16 h, consistent with efficient inhibition of its relatively weak 3′→5′ exonuclease by both the Rp and Sp phosphorothioate diastereomers (21,22,26). In contrast, the strong exonuclease activity of T4 DNAP degrades primer SP1 to a level near 50% within 1 h, followed by minimal further degradation out to 16 h incubation (Fig. 3). This biphasic behaviour by T4 DNAP is consistent with a mechanism of 3′→5′ exonuclease inhibition where the Sp isomer is much more stable than the Rp isomer to degradation, as reported in a structural study of Klenow fragment (22). The Rp isomer therefore only partially protects the 3′ primer terminus against a strong proofreading exonuclease, while the Sp isomer is essentially fully protective. The net effect of the PS modification is to produce an ‘all-or-nothing’ population of essentially stable PS primers and very short non-priming degradation products; any slow degradation of the PS primer terminus (Figs 2E and F and 3) is followed by rapid degradation of the remaining PO groups (Fig. 2B). Primer stability is somewhat improved by the presence of template (data not shown), but the all-or-nothing population distribution of PS primer remains (see below).
Figure 3.
Time course of SP1 primer incubation with exo+ Klenow fragment (open triangle) and T4 DNAP (filled circle). Percentage intact primer is reported for triplicate samples relative to an internal standard oligonucleotide added post-reaction.
Primer extension
To establish the utility of proofreading SBE reactions, we compared incorporation of deoxy-, dideoxy- and acyclonucleotide substrates for PO primer/non-proofreading, PS primer/non-proofreading and PS primer/proofreading pairings. Because most polymerases incorporate these three substrate classes with widely differing efficiencies, it was first necessary to establish experimental conditions suitable for comparison by MALDI-TOF mass spectrometry. Because MALDI-TOF detection is usually insensitive to mutant or misincorporation frequencies <5% (27), experimental conditions that result in relatively high misincorporation are preferred for informative comparisons. Suitable conditions were established in trial experiments where primer and template concentrations were held constant while polymerase and substrate concentrations were varied until correct primer extension was observed at levels above 50% of available primer. The percentage of available primer extended under various conditions is reported in Tables 1–3.
Table 1. Primer extension with dNTP substrates.
| Template |
Substrate |
Primer extension by dNTP (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymerase | KF– | KF– | KF+ | T4+ | T7– | T7+ | V– | V– | V+ | |
| Primer | P1a | SP1a | SP1a | SP1a | P1a | SP1a | P1b | SP1b | SP1b | |
| LTA | dATP | 69 | 53 | 17 | 6 | 23 | – | 13 | 12 | – |
| dCTP | 38 | 23 | – | – | 7 | – | – | – | – | |
| dGTP | – | – | – | – | – | – | – | – | – | |
| dTTP | 94 | 88 | 86 | 79 | 81 | 63 | 100 | 91 | 81 | |
| LTC | dATP | 52 | 43 | 9 | – | 15 | – | 26 | 12 | – |
| dCTP | – | – | – | – | – | – | – | – | – | |
| dGTP | 100 | 100 | 100 | 100 | 93 | 81 | 100 | 85 | 81 | |
| dTTP | – | – | – | – | – | – | 64 | 32 | 13 | |
| LTG | dATP | 19 | 15 | – | – | – | – | – | – | – |
| dCTP | 100 | 100 | 100 | 100 | 100 | 88 | 100 | 81 | 75 | |
| dGTP | 48 | 35 | 11 | – | 16 | – | 17 | 15 | – | |
| dTTP | 24 | 18 | 11 | – | 28 | – | 37 | 16 | – | |
| LTT | dATP | 92 | 90 | 86 | 73 | 82 | 67 | 100 | 84 | 76 |
| dCTP | 20 | 17 | 6 | – | – | – | 19 | 9 | – | |
| dGTP | 74 | 67 | 21 | 8 | 13 | – | 25 | 17 | 7 | |
| |
dTTP |
– |
– |
– |
– |
– |
– |
38 |
11 |
– |
| Incorrect average | 29 | 23 | 6 | 1 | 9 | 0 | 20 | 10 | 2 | |
| Correct average | 97 | 95 | 93 | 88 | 89 | 75 | 100 | 85 | 78 | |
| I/Cavg (%) | 30 | 24 | 7 | 1 | 10 | 0 | 20 | 12 | 2 | |
Exonuclease-deficient (–) and proofreading (+) variants of Klenow fragment (KF), T4 DNAP (T4), T7 DNAP (T7) and Vent DNAP (V) are indicated. Correct nucleotide incorporation for each template is shown in bold. – entries indicate values below the limit of detection (<5%).
a10 µM primer, 25 µM template, 200 µM substrate, 1 U polymerase, 60 min at 37°C.
b10 µM primer, 4 µM template, 200 µM substrate, 1 U polymerase, 25 cycles of 30 s at 85°C, 1 min at 53°C, 1 min at 63°C.
Table 3. Primer extension with acyNTP substrates.
| Template |
Substrate |
Primer extension by acyNTP (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymerase | KF– | KF– | KF+ | T4+ | T7– | T7+ | V– | V– | V+ | |
| Primer | P1a | SP1a | SP1a | SP1b | P1b | SP1b | P1c | SP1c | SP1c | |
| LTA | acyATP | 10 | 10 | – | – | – | – | – | – | – |
| acyCTP | – | – | – | – | – | – | – | – | – | |
| acyGTP | – | – | – | – | – | – | 8 | – | – | |
| acyTTP | 64 | 56 | 59 | 78 | 81 | 82 | 74 | 64 | 62 | |
| LTC | acyATP | – | – | – | – | – | – | 9 | – | – |
| acyCTP | 14 | 9 | – | – | – | – | – | – | – | |
| acyGTP | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| acyTTP | – | – | – | – | – | – | 29 | 20 | – | |
| LTG | acyATP | – | – | – | – | – | – | – | – | – |
| acyCTP | 100 | 100 | 100 | 100 | 100 | 93 | 100 | 100 | 100 | |
| acyGTP | 12 | 11 | – | – | 39 | – | 45 | 28 | – | |
| acyTTP | – | – | – | – | – | – | 42 | 26 | – | |
| LTT | acyATP | 100 | 100 | 100 | 92 | 93 | 86 | 79 | 68 | 69 |
| acyCTP | 10 | – | – | – | – | – | 42 | 34 | – | |
| acyGTP | 15 | 12 | – | – | – | – | 23 | 10 | – | |
| |
acyTTP |
– |
– |
– |
– |
– |
– |
37 |
34 |
– |
| Incorrect average | 5 | 4 | 0 | 0 | 3 | 0 | 20 | 13 | 0 | |
| Correct average | 91 | 89 | 90 | 93 | 94 | 90 | 88 | 83 | 83 | |
| I/Cavg (%) | 6 | 4 | 0 | 0 | 3 | 0 | 22 | 15 | 0 | |
Polymerase identities are the same as those listed for Tables 1 and 2. – entries indicate data below the limit of detection (<5%).
a10 µM primer, 25 µM template, 500 µM substrate, 2 U polymerase, 4 h at 37°C.
b10 µM primer, 25 µM template, 500 µM substrate, 2 U polymerase, 6 h at 37°C.
c10 µM primer, 4 µM template, 200 µM substrate, 2 U polymerase, 35 cycles of 30 s at 85°C, 1 min at 53°C, 1 min at 63°C.
Deoxynucleotide incorporation
We first examined SBE reactions for each of the four deoxynucleoside triphosphates opposite four different templates to cover all 16 possible substrate–template interactions. These experiments were performed first on the basis that all DNAPs incorporate their natural dNTP subtrates with high efficiency. Results for extension of the P1 and SP1 primers by KF, T4, T7 and Vent DNAPs are summarised in Tables 1 and S1 (Supplementary Material). For the mesophilic polymerases, template was present at 2.5 times the concentration of primer, with near saturating 200 µM substrate levels. The behaviour of KF, which displays the greatest misincorporation, will now be described in detail for illustration. As shown for the P1/LTT primer/template pair, two major classes of error are observed upon extension by exo– KF (Fig. 4A and E). Misincorporation of dG opposite the T of the LTT template occurs at the remarkably high level of 74% (Fig. 4A), the worst misincorporation result for any experimental combination (Table 1). The average incorrect/correct extension ratio above the limit of detection (I/Cavg) for the complete P1/exo– KF/dNTP set is 30%, including those combinations without measurable misincorporation (Table 1).
Figure 4.
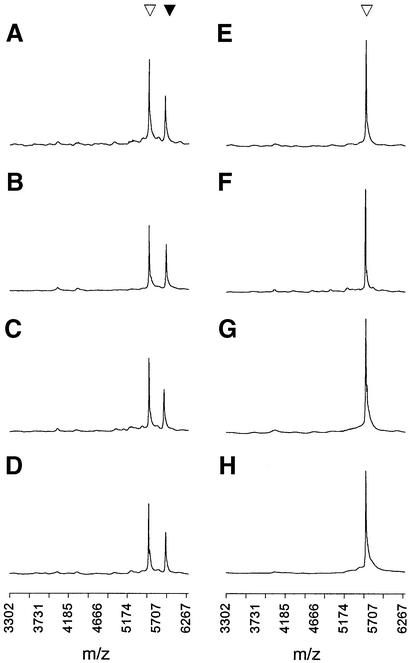
Primer extension with deoxynucleotide triphosphates. The positions of unextended (inverted open triangle), singly extended (inverted black triangle) and doubly extended primers (inverted grey triangle) are indicated. All spectra contain one additional equivalent of primer as a post-reaction standard. (A–D) Incorporation of dGTP for the following primer/template/polymerase combinations. (A) P1/LTT/exo– KF. (B) SP1/LTT/exo– KF. (C) SP1/LTT/exo+ KF. (D) SP1/LTC/exo+ KF. (E–H) Incorporation of dATP for the following primer/template/polymerase combinations. (E) P1/LTT/exo– KF. (F) SP1/LTT/exo– KF. (G) SP1/LTT/exo+ KF. (H) SP1/LTT/T4 DNAP. All spectra include a signal from primer added post-reaction as an internal standard.
Replacement of P1 by the 3′-PS SP1 primer produces a small but significant reduction in misincorporation by exo– KF. Misincorporation of dG declines from 74 to 67% for LTT/dGTP (Fig. 4B) and the overall I/Cavg for KF with dNTP substrates drops from 30 to 24% (Table 1). The SP1 phosphorothioate group also causes average correct incorporation levels to drop slightly, but much less significantly (Table 1). On top of this mild PS primer effect, combination of proofreading exo+ KF with the SP1 primer causes a strong reduction in misincorporation of dG opposite LTT to 21% (Fig. 4C), with the overall I/Cavg for exo+ KF decreasing to 7%. Two previously significant misincorporations drop to below the limit of detection (Table 1). As expected, correct SP1 extension by exo+ KF produces clean results (Fig. 4D) at high yields (Table 1). While the relative benefit of proofreading is apparent in the examples of Figure 4, it is clear that KF exonuclease activity is not adequate to entirely eliminate misincorporation under the present conditions.
To determine the effect of stronger proofreading activity, we examined the exo+ T4 DNAP/SP1 pair for all substrates. As indicated in Table 1, T4 DNAP reduces all misincorporations to near or below the limit of detection, with an I/Cavg of 1%. A similar improvement in assay performance is observed upon comparison of the Sequenase 2.0/P1 pair with the proofreading T7 DNAP/SP1 pair. Sequenase 2.0 (T7–) intrinsically makes fewer errors than KF (I/Cavg = 10 versus 30%), but all errors for the proofreading T7 DNAP/SP1 pair are further reduced to below the limit of detection. The absence of additional peaks in Figure 5 shows that this improvement is obtained without any detectable degradation–extension events. For conditions favouring both correct substrate incorporation (Fig. 5A–D) and misincorporation (Fig. 5E–H), only clean products are observable in the mass window where any aberrant products such as those formed by the extension of partially degraded primer would be expected to appear. This indicates that any unprotected primers are quickly degraded to minimal polynucleotides that cannot give rise to more than transient extension products. In agreement with the results obtained for these mesophilic enzymes, thermocycling of thermophilic Vent DNAP variants run with excess primer yields similar results (Table 1): a significant improvement in misincorporation upon replacing P1 primer (I/Cavg = 20%) by SP1 primer (I/Cavg = 12%) and a major improvement upon introducing proofreading activity (I/Cavg = 2%).
Figure 5.
Extension of SP1 primer by proofreading T7 DNAP. The positions of unextended (inverted open triangle) and extended (inverted black triangle) primers are indicated. All spectra contain one additional equivalent of primer as a post-reaction internal standard. (A–D) Correct primer–substrate pairs. (A) LTT + dATP. (B) LTA + dTTP. (C) LTG + dCTP. (D) LTC + dGTP. (E–H) Incorrect primer–substrate pairs. (E) LTG + dATP. (F) LTC + dTTP. (G) LTA + dCTP. (H) LTA + dGTP.
Improvements in SBE fidelity are maintained upon changes in buffer conditions. Replacement of magnesium by manganese in thermocycled Vent DNAP reactions results in a large increase in misincorporation, reaching an extreme of 93% for addition of dTTP opposite G (Table S2). The I/Cavg value for the exo– Vent/SP1 pairing (64%) drops significantly in the exo+ Vent/SP1 system (19%), but is still high in comparison to the behaviour in normal buffer, suggesting that manganese may affect the exonuclease active site as well as the polymerase site.
Deoxynucleotide substrates display a novel second type of error, where extension with the correct nucleotide is followed by further extension with mispairing (Fig. 4E–G and Table S1). As expected, highly efficient double extension (Δm/z 626) of the P1 primer by exo– KF (Fig. 4E) is reduced significantly by substitution of SP1 primer (Fig. 4F) and more substantially in the proofreading exo+ KF/SP1 system, so that a single correct dA extension (Δm/z 313) predominates (Fig. 4G). The strong proofreading activity of T4 DNAP reduces double incorporation to below detectable levels (Fig. 4H and Table S1). In contrast to KF, Sequenase 2.0 displays no significant double incorporation (Table S1).
Isothermal reactions with terminator substrates
Many SBE experiments use dideoxynucleotide (ddNTP) terminators to ensure that only the base immediately adjacent to the primer terminus is extended. Values and trends for extension of the P1 and SP1 primers by ddNTPs (Table 2) tend to mirror the behaviour of dNTPs (Table 1), although not perfectly. The relatively low fidelity of the exo– KF/P1 pair (I/Cavg = 46%) is improved first by substitution with SP1 primer (I/Cavg = 31%) and further by substitution with proofreading activity (I/Cavg = 9%). Better performance is observed for the other mesophilic proofreading DNAPs, the T4 DNAP/SP1 and T7 DNAP/SP1 pairs displaying no or almost no observable misincorporation. This is a substantial improvement over their exo– DNAP/P1 counterparts (Table 2). Very high ddNTP concentrations were employed in these experiments to ensure substrate saturation.
Table 2. Primer extension with ddNTP substrates.
| Template |
Substrate |
Primer extension by ddNTP (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymerase | KF– | KF– | KF+ | T4+ | T7– | T7+ | V– | V– | V+ | |
| Primer | P1a | SP1a | SP1a | SP1b | P1b | SP1b | P1c | SP1c | SP1c | |
| LTA | ddATP | 36 | 27 | – | – | – | – | – | – | – |
| ddCTP | 53 | 32 | 8 | – | 11 | – | – | – | – | |
| ddGTP | 26 | 17 | – | – | – | – | 17 | 10 | – | |
| ddTTP | 76 | 73 | 73 | 57 | 62 | 67 | 91 | 82 | 72 | |
| LTC | ddATP | 25 | 19 | – | – | 9 | – | – | – | – |
| ddCTP | 23 | 13 | – | – | 10 | – | – | – | – | |
| ddGTP | 100 | 100 | 100 | 76 | 100 | 83 | 100 | 100 | 92 | |
| ddTTP | 21 | 13 | – | – | – | – | 35 | 28 | – | |
| LTG | ddATP | 19 | 16 | – | – | – | – | 36 | 26 | 9 |
| ddCTP | 100 | 100 | 88 | 72 | 83 | 80 | 100 | 100 | 100 | |
| ddGTP | 50 | 26 | – | – | 11 | – | 27 | 19 | – | |
| ddTTP | 19 | 16 | – | – | – | – | 23 | 12 | – | |
| LTT | ddATP | 87 | 79 | 73 | 77 | 89 | 86 | 79 | 70 | 63 |
| ddCTP | 79 | 51 | 32 | – | 22 | – | 32 | 22 | – | |
| ddGTP | 100 | 75 | 41 | – | 18 | – | 9 | – | – | |
| |
ddTTP |
49 |
26 |
9 |
– |
12 |
– |
25 |
14 |
– |
| Incorrect average | 42 | 28 | 8 | 0 | 8 | 0 | 17 | 11 | 1 | |
| Correct average | 91 | 88 | 84 | 71 | 84 | 79 | 93 | 88 | 82 | |
| I/Cavg (%) | 46 | 31 | 9 | 0 | 9 | 0 | 18 | 12 | 1 | |
Exonuclease-deficient (–) and proofreading (+) variants of Klenow fragment (KF), T4 DNAP (T4), T7 DNAP (T7) and Vent DNAP (V) are indicated. Correct nucleotide incorporation appears in bold. – entries indicate values below the limit of detection (<5%).
a10 µM primer, 25 µM template, 1 mM substrate, 2 U polymerase, 4 h at 37°C.
b10 µM primer, 25 µM template, 1 mM substrate, 2 U polymerase, 6 h at 37°C.
c10 µM primer, 4 µM template, 1 mM substrate, 2 U polymerase, 35 cycles of 30 s at 85°C, 1 min at 53°C, 1 min at 63°C.
In acyclonucleotide terminators (acyNTPs) the furanose sugar moiety is replaced by a 2-hydroxyethoxymethyl group. These species are coming into more widespread use (28). Table 3 summarises the results of acyNTP incorporation experiments. For these substrates, misincorporation by exo– KF occurs at much lower levels than for the corresponding dNTP or ddNTP substrates. All three isothermal proofreading systems display misincorporation levels below the limit of detection, the best performance for any substrate family.
Thermocycled reactions with terminator substrates
One of the most commonly employed polymerases for thermocycled reactions utilising terminator nucleotides is ThermoSequenase. However, being a Taq DNAP derivative, no exo+ variant exists for comparison with the natural exo– form. Vent DNAP, which is available in both exo+ and exo– forms, was instead chosen to examine SBE performed by thermocycling with terminator nucleotides. Like the mesophilic polymerases, non-proofreading Vent DNAP misincorporates dideoxynucleotides (Table 2) into the P1 and SP1 primers in a pattern similar to that for the deoxynucleotides (Table 1). The performance of the exo+ Vent under thermocycling is comparable to the strongly proofreading T4 and exo+ T7 DNAPs used in isothermal mode, with misincorporation often reduced markedly in comparison to the exo– form (Fig. 6A–C). I/Cavg values drop predictably on proceeding from the exo– Vent/P1 (18%) to exo– Vent/SP1 (12%) to exo+ Vent/SP1 (1%) pairings. With exo– Vent DNAP, in contrast to the mesophilic polymerases, acyclonucleotides are misincorporated at equivalent or slightly higher levels than deoxy- or dideoxynucleotides (Table 3). However, the exo+ Vent/SP1 pairing strikingly outperforms the exo– variant (Fig. 6E and F), with misincorporation undetectable in any of the assays performed.
Figure 6.
SBE reactions of Vent DNAP variants with dideoxy- and acyclonucleotide terminators. The positions of unextended (inverted open triangle) and extended (inverted black triangle) primers are indicated. (A–C) Primer extension for LTC/ddTTP. (A) P1/exo– Vent. (B) SP1/exo– Vent. (C) SP1/exo+ Vent. (D–F) Primer extension for LTT/acyCTP. (D) P1/exo– Vent. (E) SP1/exo– Vent. (F) SP1/exo+ Vent.
Extrinsic exonuclease
In addition to the application of intrinsic 3′→5′ exonuclease activity, we also examined whether proofreading could be performed by an added nuclease. These experiments were carried out with exo– KF, which displays relatively poor misincorporation behaviour, SP1 primer and the widely available E.coli exo III. In general, increasing amounts of added exo III activity were found to diminish errors. For the strong misincorporation of dGTP opposite the LTT primer (see Fig. 4A), 5 U of exo III were sufficient to reduce the error to a low level (Table S3). The same observation held for double addition of dGTP opposite LTC. These improvements were associated with minimal effects on correct extension reactions with both dNTP and ddNTP substrates.
Concentration dependence of idling turnover
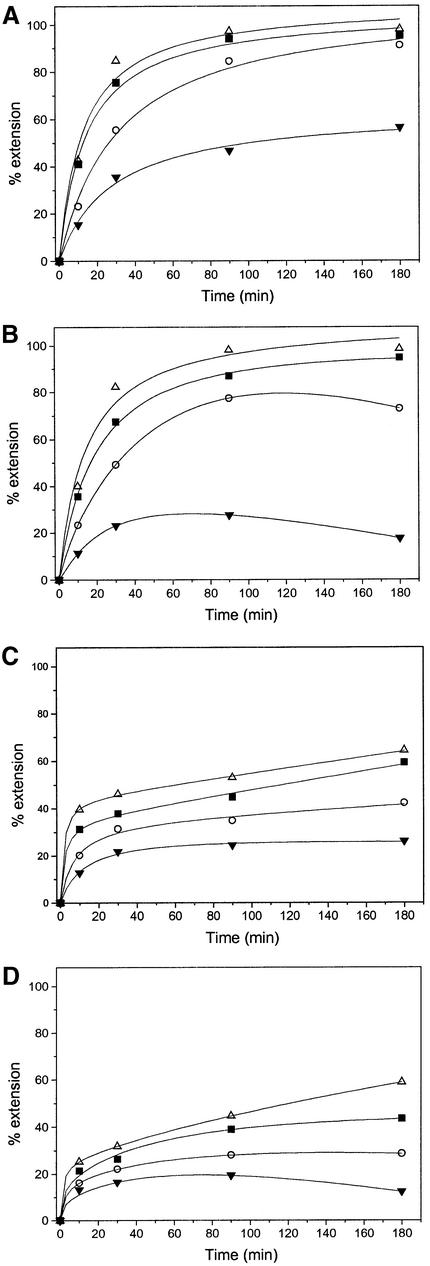
Proofreading polymerases are capable of repetitive extension/cleavage reactions known as ‘idling turnover’ (26). Depending upon the ratio of exonuclease activity to polymerase activity, there is the potential for correctly added nucleotides to be excised from the primer terminus, reducing correct product yield. This process results in the net conversion of triphosphate substrate into monophosphate. With the exonuclease activity of T7 DNAP for example being close to that of the polymerase activity (29), it is important to examine whether high yields of correct extension products can be maintained over a range of experimental conditions, including low substrate concentrations.
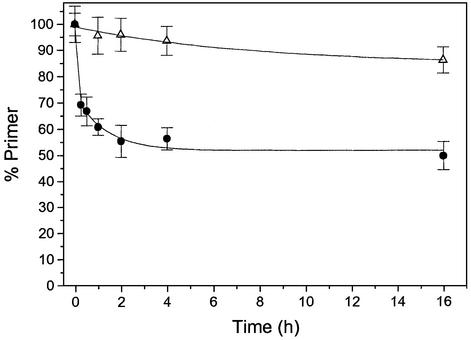
To establish baseline behaviour in the absence of proofreading, we first monitored primer extension by Sequenase 2.0 (1 U) in the presence of excess SP1/LTG primer/template (2.5 µM/10 µM) and the correct dCTP substrate at concentrations between 2.5 and 160 µM (Fig. 7A). Equivalent incorporation is observed for substrate concentrations ≥40 µM, with somewhat lower incorporation at 10 µM. By comparison, lowering the substrate concentration to 2.5 µM results in a substantial drop in the rate and level of extension at all time points (Fig. 7A).
Figure 7.
Primer extension under limiting substrate concentrations. LTG template with dCTP: 2.5 µM (inverted black triangle), 10 µM (open circle), 40 µM (filled square) and 160 µM (open triangle). (A) 1 U Sequenase 2.0. (B) 1 U exo+ T7 DNAP. (C) 0.25 U Sequenase 2.0. (D) 0.25 U exo+ T7 DNAP.
For the corresponding experiment with proofreading T7 DNAP (Fig. 7B), the net level of incorporation is only marginally lower than that for the exo– variant at substrate concentrations ≥40 µM, without any significant difference in extension after 90 min. However, while behaviour out to 30–60 min incubation is consistent under all conditions tested, a substrate concentration of 10 µM results in a slight decline in net extension at protracted time points, an effect which is more pronounced at 2.5 µM substrate concentration (Fig. 7B). This behaviour can be attributed to idling turnover under strong substrate limitation. A similar pattern is observed for reactions with further limitation of enzyme (Fig. 7C and D), where 0.25 U T7 DNAP causes net primer extension to decline somewhat following long incubation at low substrate concentrations (Fig. 7D).
DISCUSSION
MALDI-TOF detection
As a tool to first establish the utility of proofreading in SBE genotyping reactions, MALDI-TOF mass spectrometry has a major advantage: it permits the full range of primer extension products (extended, unextended and nuclease-degraded) to be detected directly and unambiguously. For this and other reasons, MALDI-TOF has a prominent position as a genotyping detection method (4,27,30–32). It has, however, the disadvantage of relatively poor signal-to-noise, with a generally accepted limit of quantitation ∼5% (27), necessitating the use of conditions that actively promote misincorporation in order to examine the issue of SBE fidelity effectively. These ‘forcing’ conditions, where a single substrate is employed at a concentration several times the Km for its incorporation, provide a sufficient dynamic range for improvements in extension fidelity to be assessed. The non-proofreading SBE data of Tables 1–3 therefore represent worst case misincorporation contributed solely by the polymerase active site. Because proofreading relies upon an intramolecular relationship between the polymerase and 3′→5′ exonuclease active sites, we expect the relative improvements in SBE fidelity reported here to be broadly applicable to a range of reaction conditions and detection formats.
Phosphorothioate resistance
Primer stabilisation is a key requirement to enable the application of proofreading to SBE genotyping. Inhibition of the E.coli DNAP I 3′→5′ exonuclease activity by PS linkages is well known (21). The Rp diastereomer is reported to inhibit exonucleolysis by Klenow fragment some 15- to 100-fold while the Sp isomer inhibits over 600-fold (22,26). A structural explanation for this isomer preference has been provided in terms of steric crowding of the KF exonuclease active site by the relatively bulky sulfur atom (22). It is clear from our results that a single Sp isomer at the 3′ primer terminus is also effectively resistant to the highly active proofreading exonucleases of other DNAPs. Since this inhibition is obtained for both a Pol A family member (T7 DNAP) and Pol B family members (T4 and Vent DNAPs), it is likely that this will hold for all proofreading DNAPs. In contrast, the Rp phosphorothioate isomer is only significantly resistant towards the relatively weak KF proofreading activity, but not to the other enzymes such as T4 DNAP (26). This differential behaviour of the Sp and Rp isomers may explain previous disagreement over the resistance of PS groups to Vent DNAP where either one (23) or three PS groups (24) were reported as necessary to protect the termini of PCR primers. The Sp phosphorothioate isomer also displays preferential resistance to snake venom phosphodiesterase and human serum exonuclease activities (33).
A significant attraction of 3′-PS primers for proofreading SBE genotyping assays is that they can be purchased from commercial oligonucleotide suppliers at modest cost; the modification currently adds ∼20% to the price of a 20mer primer. Because the cost of most SBE assays is dominated by the cost of polymerase (11), the additional expense of the PS primer is minor. Phosphorothioate primers are usually supplied as Rp/Sp racemic mixtures. In this study we have obtained good proofreading behaviour using racemic primers without further workup beyond normal purification (Fig. 5 and Tables 1–3). This is in part due to the long incubation times employed, causing Rp primers to be fully degraded by strong exonuclease activity. For short (<30 min) incubation times, the simple expedient of initiating extension by nucleoside triphosphate addition (i.e. preincubating primer with polymerase) ensures that only Sp primers participate. This reagent addition sequence is compatible with the PCR template workup commonly employed in homogeneous primer extension assays, where exonuclease I/shrimp alkaline phosphatase treatment degrades PCR primers and unincorporated dNTPs before addition of primer extension reagents (12). In cases where pure Sp primer is desired beforehand, the racemic mixture can sometimes be resolved by HPLC (34). Alternatively, stereospecific Sp phosphorothioate syntheses have been described (33,35,36), but are not generally available. A simple approach is to incubate racemic primer with a strongly proofreading polymerase or 3′-exonuclease prior to normal purification. Other backbone modifications can also produce the required proofreading exonuclease resistance. For example, phosphorodithioates (37,38) intrinsically contain a sulfur atom at the equivalent of the Sp position without the complication of stereoisomerism, although these species are also not commonly available. Nuclease-resistant modifications need not be limited to phosphodiester analogues. Inclusion of a single locked nucleic acid (LNA) residue is also sufficient to stabilise primers against the proofreading exonuclease activities of KF, T7, Vent and Pfu DNAPs (unpublished results).
Importantly for the general applicability of proofreading to SBE, we have not found it necessary to protect template strands against 3′→5′ exonuclease activity. Minor degradation of the template 3′ terminus has no effect upon the primer– template duplex, which both inhibits progression of exonuclease along the template and sequesters polymerase at the primer–template terminus by preferential binding (Kd typically <20 nM). Several other genotyping assays use phosphorothioates in alternative ways, including the GOOD assay (39,40) and pyrosequencing (5), which both employ α-thio-triphosphate substrates. Because polymerases only accept the Sp diastereomer and invert the configuration to Rp upon incorporation (41), the resulting extension product is not protective against strongly proofreading polymerases. Similarly, the modifications introduced into the primer terminus in the GOOD assay are directed towards phosphodiesterase resistance rather than proofreading exonuclease resistance. As we have shown here, use of appropriate primer modifications would permit proofreading to be employed in the GOOD primer extension reaction.
Misincorporation by non-proofreading polymerases
Complete kinetic parameters for primer extension by T7 DNAP (42) indicate that dissociation of the polymerase– primer–template ternary complex is the rate-determining step in our SBE assay, where polymerase concentrations are 10–80 nM. This is consistent with the relatively slow time courses observed and the small differences in correct nucleotide incorporation efficiencies between the A, C, G and T members of each substrate family. The misincorporation events for non-proofreading polymerases reach remarkably high values, up to 115% of correct incorporation for exo– KF (ddGTP/LTT), 64% for exo– Vent DNAP (dTTP/LTC) and 28% for Sequenase 2.0 (dTTP/LTG). High concentrations of single nucleoside triphosphates play a large part in producing such high level misincorporation (9). There are several SBE implementations where a single-substrate mode is employed, such as colourimetric detection of hapten-labelled products (19,43) and electronic detection of electrochemically labelled products (10,44,45). Most SBE reactions are conducted in ‘two-colour’ (i.e. two nucleotide triphosphates) or ‘four-colour’ modes where direct competition between substrates can occur. This competition will reduce misincorporation, but not eliminate it entirely. From our results it is clear that misincorporation can be a significant issue for a range of SBE genotyping implementations, and is likely to be at least partially responsible for anomalous results reported in the literature (7,11–16). In order to address this issue quantitatively, we are separately examining the effect of SBE proofreading with sensitive fluorescence detection of labelled substrates.
The general fidelity of the non-proofreading polymerases follows the order: exo– KF < exo– Vent < Sequenase 2.0 (Tables 1–3). The overall average incorrect/correct incorporation values (I/Cavg) obtained here for the three substrate classes with each of these enzymes are 27% for exo– KF (deoxy 30%, dideoxy 46%, acyclo 6%), 20% for exo– Vent (20%, 18%, 22%) and 7% for Sequenase 2.0 (10%, 9%, 3%). Comparing substrate classes, the general fidelity with the non-proofreading polymerases as measured by I/Cavg values follows the order dideoxy (24%) < deoxy (20%) < acyclo (10%), although these values are skewed by the use of two Pol A polymerases and only one Pol B polymerase. It is clear from Table 3 that exo– Vent DNAP misincorporates acyclo derivatives much more efficiently than the other enzymes, which may be a consequence of its relatively high acyclo preference (28). When SBE misincorporation is examined across all substrate classes, we observe the non-proofreading enzymes to have somewhat different base preferences, the most notable being the high propensity of exo– Vent to misincorporate ‘T’ opposite C (64% for dTTP, 35% for ddTTP, 29% for acyTTP), a reaction of no significance for Sequenase 2.0 (Tables 1–3). In fact, Vent DNAP displays a general tendency to misincorporate ‘T’ opposite any template.
PS primer effect
Use of 3′-PS primers with non-proofreading polymerases consistently reduces misincorporation an average 1.4-fold (range 0- to 3.5-fold) across all substrate families (Tables 1–3 and Fig. 4). The level of correct extension also typically declines upon substitution of PS primers, but only by an average of 5% (range 2–15%). These global effects may be rationalised in terms of reduced primer–template stability. It has been demonstrated that DNA duplexes containing PS groups have lower melting temperatures, each introduction of a PS linkage decreasing the melting temperature by 0.4–0.6°C (37) with only a small isomer dependence (33). While we could not observe a significant difference between the melting temperatures of duplexes containing the P1 and SP1 primers, a small destabilisation is likely to be present. The selective effect upon misincorporation is consistent with the presence of primer–polymerase contacts that contribute to the energetics of nucleotide discrimination (8). A 1.4-fold reduction in misincorporation is a useful improvement that is directly applicable to existing non-proofreading SBE methods.
Proofreading
Use of proofreading polymerases and PS primers substantially improves SBE fidelity. T4, T7 and Vent DNAPs reduce misincorporation to near or below the limit of detection across all substrate classes, yielding an approximate order of magnitude improvement (Tables 1–3). The weaker proofreading activity of KF produces a significant decline in misincorporation, but is generally not as effective. These findings are in accord with expectations, where many studies indicate that proofreading increases processive polymerisation fidelity by a few-fold to over 100-fold (8). We expect this order of magnitude improvement to be beneficial for a range of SBE implementations. This is especially true for solid phase formats, where factors such as high local primer concentrations may result in complications such as transient mispriming. Analysis of solid phase SBE reactions is underway.
An extrinsic exonuclease can also be used to add proofreading activity to an exo– polymerase. We observe misincorporation to be reduced to a much greater extent than correct nucleotide incorporation upon addition of exo III, indicating that the extrinsic exonuclease preferentially detects the more frayed primer–template terminus.
Assay robustness
We have examined the substrate concentration dependence of the proofreading SBE assay with T7 DNAP, an enzyme for which a complete kinetic analysis of the reaction pathway has been determined. While most of the data for this paper were acquired at substrate concentrations >100 µM, the results of Figure 7 show that proofreading polymerases can also be employed at much lower concentrations without significant signal degradation. Idling turnover by T7 DNAP causes minimal loss of correct signal intensity for a dCTP concentration of 2.5 µM (one-seventh Km) out to incubation times of 30–60 min. This maintenance of signal is important for SBE implementations using expensive labelled substrates or where substrate incorporation is essentially driven to completion. The homogeneous FP-TDI assay for example employs fluorescent terminators at concentrations near 10 nM (12,46). Consideration of the detailed kinetic rates reported (29,42,47) for transport of matched and mismatched primer termini to the exonuclease active site of T7 DNAP (kp→x = 0.2 versus 2.3 s–1) and for dissociation of this enzyme from matched and mismatched primer termini (koff = 0.2 versus 0.4 s–1) suggest that its proofreading activity should improve SBE fidelity 6- to 12-fold at all substrate concentrations. Preliminary results with labelled substrates and fluorescence detection suggest that this expectation will be fulfilled. We are examining the behaviour of T7 DNAP and other proofreading polymerases with different kinetic parameters to determine their useful application ranges.
CONCLUSIONS
We arrive at three major conclusions from this study of single-substrate SBE reactions. (i) Relative destabilisation of the primer–template terminus improves non-proofreading SBE assay fidelity to a small but significant extent. (ii) Use of nuclease-resistant primers and proofreading polymerases improves SBE fidelity by approximately an order of magnitude. (iii) The Sp phosphorothioate diastereomer is responsible for resistance to strongly proofreading polymerases. We expect that the approach presented here will be directly transferable to two- and four-substrate reactions, offering a simple and cost-effective means to improve the fidelity of a range of SBE genotyping formats.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENT
We thank David Andrews for encouraging us to pursue the study of genotyping methods.
REFERENCES
- 1.Weaver T.A. (2000) High-throughput SNP discovery and typing for genome-wide genetic analysis. In New Technologies for the Life Sciences: A Trends Guide. Elsevier, Oxford, UK, pp. 36–42.
- 2.Phillips K.A., Veenstra,D.L., Oren,E., Lee,J.K. and Sadee,W. (2001) Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. J. Am. Med. Assoc., 286, 2270–2279. [DOI] [PubMed] [Google Scholar]
- 3.Kwok P.-Y. (2001) Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genomics Hum. Genet., 2, 235–258. [DOI] [PubMed] [Google Scholar]
- 4.Syvanen A.-C. (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nature Rev. Genet., 2, 930–942. [DOI] [PubMed] [Google Scholar]
- 5.Ronaghi M., Uhlen,M. and Nyren,P. (1998) A sequencing method based on real-time pyrophosphate. Science, 281, 363–365. [DOI] [PubMed] [Google Scholar]
- 6.Syvanen A.-C. (1999) From gels to chips: “minisequencing” primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum. Mutat., 13, 1–10. [DOI] [PubMed] [Google Scholar]
- 7.Pastinen T., Kurg,A., Metspalu,A., Peltonen,L. and Syvanen,A.-C. (1997) Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res., 7, 606–614. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel T.A. and Bebenek,K. (2000) DNA replication fidelity. Annu. Rev. Biochem., 69, 497–529. [DOI] [PubMed] [Google Scholar]
- 9.Haff L.A. and Smirnov,I.P. (1997) Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res., 7, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brazill S.A. and Kuhr,W.G. (2002) A single base extension technique for the analysis of known mutations utilizing capillary gel electrophoresis with electrochemical detection. Anal. Chem., 74, 3421–3428. [DOI] [PubMed] [Google Scholar]
- 11.Bray M.S., Boerwinkle,E. and Doris,P.A. (2001) High-throughput multiplex SNP genotyping with MALDI-TOF mass spectrometry: practice, problems and promise. Hum. Mutat., 17, 296–304. [DOI] [PubMed] [Google Scholar]
- 12.Hsu T.M., Chen,X., Duan,S., Miller,R.D. and Kwok,P.-Y. (2001) Universal SNP genotyping assay with fluorescence polarization detection. Biotechniques, 31, 560–570. [DOI] [PubMed] [Google Scholar]
- 13.Sun X., Ding,H., Hung,K. and Guo,B. (2000) A new MALDI-TOF based mini-sequencing assay for genotyping of SNPs. Nucleic Acids Res., 28, e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapero M.H., Leuther,K.K., Nguyen,A., Scott,M. and Jones,K.W. (2001) SNP genotyping by multiplexed solid-phase amplification and fluorescent minisequencing. Genome Res., 11, 1926–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindroos K., Liljedahl,U., Raitio,M. and Syvanen,A.-C. (2001) Minisequencing on oligonucleotide microarrays: comparison of immobilisation chemistries. Nucleic Acids Res., 29, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber M., Mundlein,A., Dornstauder,E., Schneeberger,C., Tempfer,C.B., Mueller,M.W. and Schmidt,W.M. (2002) Accessing single nucleotide polymorphisms in genomic DNA by direct multiplex polymerase chain reaction amplification on oligonucleotide microarrays. Anal. Biochem., 303, 25–33. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y.-W., Balaskas,E., Kessler,G., Issid,C., Scully,L.J., Murphy,D.G., Rinfret,A., Giulivi,A., Scalia,V. and Gill,P. (1998) Primer specific and mispair extension analysis (PSMEA) as a simple approach to fast genotyping. Nucleic Acids Res., 26, 5013–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y.-W., Balaskas,E., Furione,M., Yen,P.-H., Kessler,G., Scalia,V., Chui,L. and Sher,G. (2000) Comparison and application of a novel genotyping method, semiautomated primer-specific and mispair extension analysis and four other genotyping assays for detection of hepatitis C virus mixed-genotype infections. J. Clin. Microbiol., 38, 2807–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Syvanen A.-C., Aalto-Setala,K., Harju,L., Kontula,K. and Soderlund,H. (1990) A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics, 8, 684–692. [DOI] [PubMed] [Google Scholar]
- 20.Verma S. and Eckstein,F. (1998) Modified oligonucleotides: synthesis and strategy for users. Annu. Rev. Biochem., 67, 99–134. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel T.A., Eckstein,F., Mildvan,A.S., Koplitz,R.M. and Loeb,L.A. (1981) Deoxynucleoside [1-thio]triphosphates prevent proofreading during in vitro DNA synthesis. Proc. Natl Acad. Sci. USA, 78, 6734–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brautigam C.A. and Steitz,T.A. (1998) Structural principles for the inhibition of the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol., 277, 363–377. [DOI] [PubMed] [Google Scholar]
- 23.Skerra A. (1992) Phosphorothioate primers improve the amplification of DNA sequences by DNA polymerases with proofreading activity. Nucleic Acids Res., 20, 3551–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Noronha C.M. and Mullins,J.I. (1992) Amplimers with 3′-terminal phosphorothioate linkages resist degradation by Vent polymerase and reduce Taq polymerase mispriming. PCR Methods Appl., 2, 131–136. [DOI] [PubMed] [Google Scholar]
- 25.Dean F.B., Nelson,J.R., Giesler,T.L. and Lasken,R.S. (2001) Rapid amplification of plasmid and phage DNA using phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res., 11, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta A.P., Benkovic,P.A. and Benkovic,S.J. (1984) The effect of the 3′,5′ thiophosphoryl linkage on the exonuclease activities of T4 polymerase and the Klenow fragment. Nucleic Acids Res., 12, 5897–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross P.L., Hall,L.R. and Haff,L.A. (2000) Quantitative approach to single-nucleotide polymorphism analysis using MALDI-TOF mass spectrometry. Biotechniques, 29, 620–626, 628–629. [DOI] [PubMed] [Google Scholar]
- 28.Gardner A.F. and Jack,W.E. (2002) Acyclic and dideoxy terminator preferences denote divergent sugar recognition by archeon and Taq DNA polymerases. Nucleic Acids Res., 30, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donlin M.J., Patel,S.S. and Johnson,K.A. (1991) Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry, 30, 538–546. [DOI] [PubMed] [Google Scholar]
- 30.Juhasz P., Roskey,M.T., Smirnov,I.P., Haff,L.A., Vestal,M.L. and Martin,S. (1996) Applications of delayed extraction matrix-assisted laser desorption ionization time-of-flight mass spectrometry to oligonucleotide analysis. Anal. Chem., 68, 941–946. [DOI] [PubMed] [Google Scholar]
- 31.Lechner D., Lathrop,G.M. and Gut,I.G. (2001) Large-scale genotyping by mass spectrometry: experience, advances and obstacles. Curr. Opin. Chem. Biol., 6, 31–38. [DOI] [PubMed] [Google Scholar]
- 32.Buetow K.H., Edmonson,M., MacDonald,R., Clifford,R., Yip,P., Kelley,J., Little,D.P., Strausberg,R., Koester,H., Cantor,C.R. et al. (2001) High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl Acad. Sci. USA, 98, 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu D., Kandimalla,E.R., Roskey,A., Zhao,Q.Y., Chen,L.H., Chen,J.D. and Agrawal,S. (2000) Stereo-enriched phosphorothioate oligodeoxynucleotides: synthesis, biophysical and biological properties. Bioorg. Med. Chem., 8, 275–284. [DOI] [PubMed] [Google Scholar]
- 34.Stec W.J., Zon,G. and Uznanski,B. (1985) Reversed-phase high-performance liquid chromatographic separation of diasteromeric phosphorothioate analogues of oligodeoxyribonucleotides and other backbone-modified congeners of DNA. J. Chromatogr., 326, 263–280. [DOI] [PubMed] [Google Scholar]
- 35.Cosstick R. and Williams,D.M. (1987) An approach to the stereoselective synthesis of Sp-dinucleoside phosphorothioates using phosphotriester chemistry. Nucleic Acids Res., 10, 9921–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stec W.J., Grajkowski,A., Kobylanska,A., Karwowski,B., Koziolkiewicz,M., Misiura,K., Okruszek,A., Wilk,A., Guga,P. and Boczkowska,M. (1995) Diastereomers of nucleoside 3′-O-(2-thio-1,3,2-oxathia(selena)phospholanes) – building blocks for stereocontrolled synthesis of oligo(nucleoside phophorothioate)s. J. Am. Chem. Soc., 117, 12019–12029. [Google Scholar]
- 37.Bjergarde K. and Dahl,O. (1991) Solid phase synthesis of oligodeoxyribonucleoside phosphorodithioates from thiophosphoramidites. Nucleic Acids Res., 19, 5843–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall W.S. and Caruthers,M.H. (1993) Phosphorodithioate DNA as a potential therapeutic drug. Science, 259, 1564–1570. [DOI] [PubMed] [Google Scholar]
- 39.Sauer S., Lechner,D., Berlin,K., Lehrach,H., Escary,J.-L., Fox,N. and Gut,I.G. (2000) A novel procedure for efficient genotyping of single nucleotide polymorphisms. Nucleic Acids Res., 28, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauer S., Gelfand,D.H., Boussicault,F., Bauer,K., Reichert,F. and Gut,I.G. (2002) Facile method for automated genotyping of single nucleotide polymorphisms by mass spectrometry. Nucleic Acids Res., 30, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckstein F. (1985) Nucleoside phosphorothioates. Annu. Rev. Biochem., 54, 367–402. [DOI] [PubMed] [Google Scholar]
- 42.Patel S.S., Wong,I. and Johnson,K.A. (1991) Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry, 30, 511–525. [DOI] [PubMed] [Google Scholar]
- 43.Pecheniuk N.M., Marsh,N.A., Walsh,T.P. and Dale,J.L. (1997) Use of first nucleotide change technology to determine the frequency of factor V Leiden in a population of Australian blood donors. Blood Coagul. Fibrinolysis, 8, 491–495. [DOI] [PubMed] [Google Scholar]
- 44.Wlassoff W.A. and King,G.C. (2002) Ferrocene conjugates of dUTP for enzymatic redox labelling of DNA. Nucleic Acids Res., 30, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weizman H. and Tor,Y. (2002) Redox-active metal-containing nucleotides: synthesis, tunability and enzymatic incorporation into DNA. J. Am. Chem. Soc., 124, 1568–1569. [DOI] [PubMed] [Google Scholar]
- 46.Chen X., Levine,L. and Kwok,P.-Y. (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res., 9, 492–498. [PMC free article] [PubMed] [Google Scholar]
- 47.Wong I., Patel,S.S. and Johnson,K.A. (1991) An induced-fit mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry, 30, 526–537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.