Abstract
Site-directed mutagenesis is an invaluable tool for functional studies and genetic engineering. However, most current protocols require the target DNA to be cloned into a plasmid vector before mutagenesis can be performed, and none of them are effective for multiple-site mutagenesis. We now describe a method that allows mutagenesis on any DNA template (eg. cDNA, genomic DNA and plasmid DNA), and is highly efficient for multiple-site mutagenesis (up to 100%). The technology takes advantage of the requirement that, in order for DNA polymerases to elongate, it is crucial that the 3′ sequences of the primers match the template perfectly. When two outer mutagenic oligos are incorporated together with the desired mutagenic oligos into the newly synthesised mutant strand, they serve as anchors for PCR primers which have 3′ sequences matching the mutated nucleotides, thus amplifying the mutant strand only. The same principle can also be used for mutant screening.
INTRODUCTION
Site-directed mutagenesis (SDM) is widely used in molecular biology to study protein structure and functions. It is also used in genetic engineering to optimise enzyme activity and to generate genetically modified species. Various SDM protocols, both PCR and non-PCR based, have been described (1–5). However, none of these protocols is effective for multiple mutagenesis, with reported efficiencies for all methods being <30% for more than five mutation sites in a single round of reaction. Apart from the PCR-based methods, which can only mutate one site at a time, all other methods require the target DNA to be cloned into a plasmid vector before mutagenesis. Methods that involve virus DNA preparation also require f1 origin to be present in the plasmid.
In an attempt to overcome the limitations associated with current SDM protocols, we have developed the targeted amplification of mutant strand (TAMS; patent pending, more information is available at ozex.netfirms.com) technology. TAMS has proved to be efficient for multiple-site mutagenesis, and can be adapted to directly mutate cDNA, thus eliminating the need for sub-cloning prior to mutagenesis. The protocol takes advantage of the fact that primers with 3′ mismatches usually cannot amplify the target sequence (6,7). Therefore, if two anchor mutations are incorporated into the mutant DNA strand outside the desired mutations, they can be preferentially amplified by primers with 3′ sequence matching the mutant strand but not the wild-type strand. Since the desired mutations are in the mutant strand, the final PCR product will also contain the desired mutations.
MATERIALS AND METHODS
Single-stranded template preparation by linear PCR
A 3.7 kb linearised plasmid was amplified by Red-Taq polymerase (Sigma) with T7 primer only (5′-TAATAC GACTCACTATAGGG-3′) in a 50 µl reaction volume. The PCR conditions were 94°C for 2 min for 1 cycle; 94°C for 30 s, 56°C for 30 s, 72°C for 1 min for 30 cycles; and finally 1 cycle at 72°C for 5 min.
Mutant strand synthesis
Anti-sense outer mutation primers Anchor5′ and Anchor3′ (100 nM, respectively), inner mutation primers Mut1 and Mut2 (500 nM, respectively) were mixed in a total volume of 20 µl with 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 200 µM dNTP and 5 U T4 polynucleotide kinase (Promega). The tube was then incubated at 37°C. After 30 min, 2 µl of the single-stranded DNA (ssDNA) from step 1 was added, and the tube was left in a beaker containing 1 l of 75°C water and cooled over 30 min to room temperature. Mutant strand synthesis was then performed by adding 5 U of T4 DNA polymerase (Promega) and 2 U of T4 DNA ligase (Promega), and incubating at 37°C for 1 h.
Mutant strand amplification by PCR
The mutant strand (1 µl) was selectively amplified by Red-Taq with either 200 nM each of PCR primers PCR5′ and PCR3′, or 200 nM each of MutPCR1 and MutPCR2, in a final volume of 50 µl. PCR conditions were the same as those described in step 1. The wild-type ssDNA was amplified as control.
TA cloning and mutant screening
The PCR product (2 µl) was TA cloned into pGEM-T Easy vector (Promega) following the manufacturer’s protocol. Sixteen white colonies were selected and resuspended in 15 µl of LB media supplemented with 25 µg/l ampicillin, 1 µl of the mutants were amplified by Red-Taq with the primers MutPCR1 and MutPCR2 using the same PCR conditions described in step 1. The wild-type plasmid was used as control. In addition, four of the positive clones were sequenced to confirm the presence of the mutations.
RESULTS
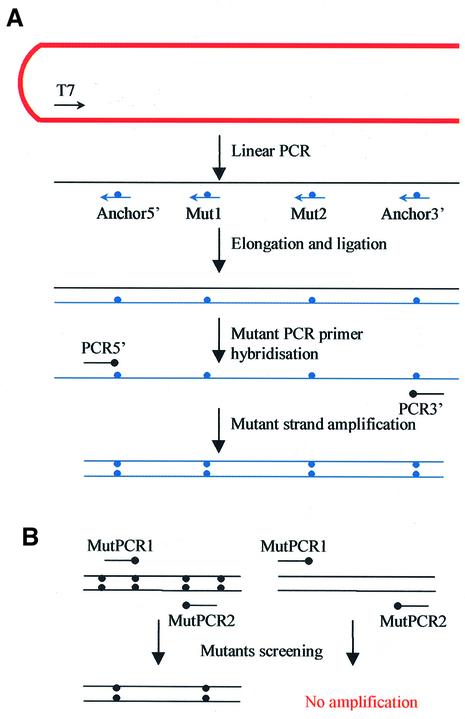
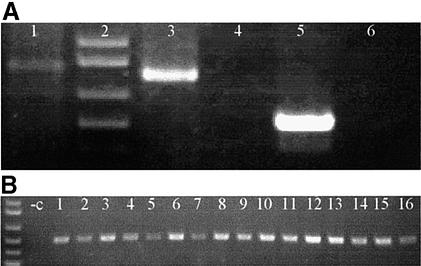
The sequences of primers that were used in this experiment are shown in Table 1, and the mechanism of the mutagenesis method and the positions of the primers are depicted in Figure 1. To determine the efficiency of this method, the TAMS technology was used to mutate a wild-type vector. Mutant strand selection was accomplished in three steps. (i) Single-stranded templates were synthesised by linear PCR extending from the T7 primer, resulting in a template 840 bp in length (Fig. 2A, lane 1). (ii) Four mutagenic oligos were phosphorylated with T4 polynucleotide kinase, and hybridised to the template. The phosphorylated primers were extended with T4 DNA polymerase, which lacks 5′→3′ exonuclease activity, therefore would not displace the hybridised primers, thus would produce a nicked mutant strand. The nicks were then ligated by T4 DNA ligase to form a continuous strand. (iii) The mutant strands were preferentially amplified by 3′ mismatched primers. As DNA polymerase would only extend the 3′ end of perfectly matched primers, only the mutant strand, which had the mutations incorporated, thus matched the 3′ end of the PCR primers, could be amplified (Fig. 2A).
Table 1. Nucleotide sequence of the primers used, with the mutation points shown in lower case.
| Name | Sequence |
|---|---|
| Anchor5′ | 5′-CGAATTCCCGgtaCCGCCA-3′ |
| Anchor3′ | 5′-TGCAGGCGGCCGCGAgTcgACTAGT-3′ |
| Mut1 | 5′-TAAGAAGATCaCTATGCCC-3′ |
| Mut2 | 5′-GCATGCTACTaAGCTTTCA-3′ |
| PCR5′ | 5′-CCGGCCGCCATGGCGGtac-3′ |
| PCR3′ | 5′-TGCAGGCGGCCGCGAgTcg-3′ |
| MutPCR1 | 5′-GGCAACTCAAGGGCATAGt-3′ |
| MutPCR2 | 5′-GCTCTGAGCATGCTACTa-3′ |
Figure 1.
Schematic diagrams of the TAMS technology. (A) ssDNA template was generated by linear PCR on the linearised vector. Approximate positions of the primers are shown with arrows, blunt arrowheads mean sequence matching mutant strand. Large dots indicate mutated nucleotides. (B) Same mechanism can be used to screen mutations from bacteria colonies.
Figure 2.
PCR products checked on 1% agarose gel. (A) Single-stranded template was amplified by linear PCR (lane 1), after mutant strand synthesis, primers PCR5′ and PCR3′ amplified the mutant strand (lane 3) rather than the wild-type plasmid (lane 4); the same is true with primers MutPCR1 and MutPCR2 (lanes 5 and 6). (B) PCR product shown in lane 3 of (A) was cloned into pGEM-T Easy and bacteria transformed. The mutant primers PCRMut1 and PCRMut2 amplified all of the 16 white colonies selected (1–16), but not the wild-type vector (–c).
The PCR product shown in lane 3 of Fig. 2A was subsequently cloned into the pGEM-T Easy vector, and all 16 white colonies that were randomly selected for screening contained the desired mutations (Fig. 2B). Furthermore, four of the positive clones were sequenced, and all of them contained the desired mutations, although two of them also showed non-specific mutations (data not shown). These results suggest that the efficiency of the TAMS technology is up to 100%.
DISCUSSION
The TAMS protocol requires the use of more primers than most current mutagenesis methods, but this is more than compensated for by its simplicity, effectiveness and savings in other costs. The use of Taq polymerase may also introduce non-specific mutations during PCR amplification. However, since the mutagenesis efficiency of TAMS protocol is sufficiently high, usually at least one in four clones should prove to be the correct clone. If accuracy is of concern, a higher fidelity enzyme should be used. The 3′→5′ exonuclease activity of this type of enzymes, which may excise the 3′ mismatched nucleotides from the PCR primers, should not be a problem, as the shortened primers would have lower optimal annealing temperatures.
The reported efficiencies of current multiple-site mutagenesis protocols drop dramatically as the number of incorporated mutations increases. This is mainly because the annealing conditions do not allow mutagenic oligos to fully hybridise. For most protocols, the single point mutation efficiencies are ∼80%, which means the mutagenic oligos only anneal to 80% of the template at the given conditions. For this reason, the efficiency for five mutations would fall to ∼0.85 = 32.8%. To increase the chance for full hybridisation it is preferable that a ssDNA template is used, that enough annealing time and the full annealing temperature range for all mutagenic oligos are covered. The TAMS technology addresses these issues by first generating single-stranded template through simple linear PCR, then allowing extensive hybridisation of the mutagenic oligos by a gradual decrease of annealing temperature from 75°C to room temperature. Although only four mutagenic oligos were used in the present experiment, we believe that high hybridisation efficiency could be achieved even when more oligos are used. The fact that all 16 clones screened contained the desired mutations favours this assumption.
Since any ssDNA can be used as the template for the TAMS technology, it is possible to mutate first strand cDNA directly after removal of the RNA template with ribonuclease H. A separate experiment was performed to mutate the first strand cDNA of β-actin, apart from the two anchor mutations, three single point mutations, a 120 bp deletion and a 3 bp insertion were also introduced, restriction digestion of the resulting PCR product suggest that, depending on the primers used, the efficiency of TAMS can indeed be up to 100% (supporting data at http://ozex.netfirms.com/TS.html). Likewise, genomic DNA can also be mutated without cloning, as an asymmetrical PCR should generate enough single-stranded template.
In summary, the TAMS technology is a simple, efficient and cost effective method for both single and multiple-site mutagenesis, and a wide variety of DNA can be used as the templates.
REFERENCES
- 1.Ishii T.M., Zerr,P., Xia,X.M., Bond,C.T., Maylie,J. and Adelman,J.P. (1998) Site-directed mutagenesis. Methods Enzymol., 293, 53–71. [DOI] [PubMed] [Google Scholar]
- 2.Ling M.M. and Robinson,B.H. (1997) Approaches to DNA mutagenesis: an overview. Anal. Biochem., 254, 157–178. [DOI] [PubMed] [Google Scholar]
- 3.Braman J. (ed.) (2002) In Vitro Mutagenesis Protocols. 2nd Edn, Humana Press, Totowa, NJ.
- 4.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol., 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 5.Sawano A. and Miyawaki,A. (2000) Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res., 28, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha R.S., Zarbl,H., Keohavong,P. and Thilly,W.G. (1992) Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl., 2, 14–20. [DOI] [PubMed] [Google Scholar]
- 7.Huang M.M., Arnheim,N. and Goodman,M.F. (1992) Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res., 20, 4567–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]