Abstract
The pharmacodynamics of moxifloxacin against Streptococcus pneumoniae and Pseudomonas aeruginosa were investigated in a pharmacokinetic infection model. Three strains of S. pneumoniae, moxifloxacin, and two strains of P. aeruginosa were used. Antibacterial effect and emergence of resistance were measured for both species over a 72-h period using an initial inoculum of about 108 CFU/ml. At equivalent area under the curve (AUC)/MIC ratios, S. pneumoniae was cleared from the model while P. aeruginosa was not. For S. pneumoniae, the area under the bacterial kill curve up to 72 h could be related to AUC/MIC ratio using an inhibitory maximum effect (Emax) model (concentration required for 50% Emax [EC50], 45 ± 22; r2, 0.97). For P. aeruginosa even at the highest AUC/MIC ratio (427), bacterial clearance was insufficient for the EC50 to be calculated. Emergence of resistance occurred with P. aeruginosa but not to any significant extent with S. pneumoniae. Emergence of resistance in P. aeruginosa as measured by population analysis profile (PAP-AUC) was dependent on drug exposure and time of exposure. In weighted least-squares regression analysis AUC/MIC ratio was predictive of PAP-AUC. When emergence of resistance was measured by the time for the colony counts on media containing antibiotic to increase by 2 logs, again AUC/MIC was the best predictor of emergence of resistance. However, for both experiments using S. pneumoniae and P. aeruginosa the correlation between all the pharmacodynamic parameters was high. These data indicate that for a given fluoroquinolone the magnitude of the AUC/MIC ratio for antibacterial effect is dependent on the bacterial species. Emergence of resistance is dependent on (i) species, (ii) duration of drug exposure, and (iii) drug exposure. A single AUC/MIC ratio magnitude is not adequate to predict antibacterial effect or emergence of resistance for all bacterial species.
In vitro pharmacokinetic models of infection are widely used to study antibacterial pharmacodynamics, especially within the fluoroquinolone drug class. It is known that area under the curve (AUC)/MIC is the main pharmacokinetic/pharmacodynamic index related to the antibacterial effects of fluoroquinolones in in vitro models. In 1996, Madaras-Kelly et al. (15) studied the pharmacodynamics of ofloxacin and ciprofloxacin against three strains of Pseudomonas aeruginosa. The area under the bacterial killing curve (AUBKC) was used as the measure of antibacterial effects. Multivariate regression analysis indicated that the AUC/MIC was related to the antibacterial effect and that this relationship was well described by a sigmoid maximum-effect (Emax) model, the AUC/MIC at which 50% of the isolates were inhibited (MIC50) being about 120 and the maximum antibacterial effect occurring at an AUC/MIC of 400 to 600. With similar techniques, the effects of levofloxacin, ofloxacin, and ciprofloxacin on Streptococcus pneumoniae were tested. Again, with an Emax model to relate the AUC/MIC and the antibacterial effect, the AUC/MIC50 was found to be 60 (K. J. Madaras-Kelly, T. Demasters, D. Soaves, and M. McLaughlin, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. A-27, p. 6, 1997). Subsequently, it was reported that AUC/MIC ratios of 44 to 49 resulted in the clearance of S. pneumoniae by levofloxacin, ciprofloxacin, or trovafloxacin in an in vitro model (9, 10). Levofloxacin AUC/MIC ratios of 30 to 55 were also shown to be effective against S. pneumoniae (8) in an in vitro system. In contrast, AUC/MIC ratios of 50 to 62 for ciprofloxacin, levofloxacin, or sparfloxacin did not result in clearance within 24 h in an infected fibrin clot model (6).
Other experiments with in vitro models have indicated that the AUC/MIC ratio for the clearance of S. pneumoniae in the first 24 h may be much higher than 50 (12). The reasons for these discrepant results are not clear but may be related to methodological variations in the models used, different strains used, or different choices of antibacterial effect measures. However, it has been suggested on the basis of these data that the magnitude of the AUC/MIC ratio required for an antibacterial effect is lower for S. pneumoniae than for P. aeruginosa, although this notion has not been established by direct comparison in the same in vitro pharmacokinetic model.
In addition, although the pharmacodynamics of the emergence of resistance in P. aeruginosa have been studied with a number of fluoroquinolones in in vitro models (1, 15), this type of study has not been done with S. pneumoniae.
The objective of this study was to perform a direct comparison of the pharmacodynamics of moxifloxacin against S. pneumoniae and P. aeruginosa to define the magnitude of the AUC/MIC ratio required for an antibacterial effect as well as the factors important in determining the emergence of resistance.
MATERIALS AND METHODS
Models.
A New Brunswick (Hatfield, Hertfordshire, England) Bioflo 1000 in vitro pharmacokinetic model was used to simulate the serum drug concentrations associated with the oral administration of moxifloxacin. The apparatus, which has been described before, consists of a single central chamber connected to a dosing chamber, which is in turn attached to a reservoir containing broth. The central chamber is connected to a collecting vessel for overflow (12). The contents of the dosing chamber and the central chamber were diluted with broth by using a peristaltic pump (Ismatec; Bennett & Co., Weston-super-Mare, England) at a flow rate of 66 ml/h. The temperature was maintained at 37°C, and the broth in the dosing and central chambers was agitated by a magnetic stirrer.
Media.
75% Brain heart infusion broth was used in the experiments with S. pneumoniae. For P. aeruginosa, 2% Iso-Sensitest broth (Oxoid, Basingstoke, England) was used. Magnesium chloride (1%; BDH, Poole, Dorset, England) was incorporated into nutrient agar plates (Merck, Dorset, England). For experiments with S. pneumoniae, 5% whole horse blood (TCS Microbiology, Buckingham, England) was also added. No magnesium chloride was used in the antibiotic-containing plates in studies of the emergence of resistance.
Strains.
Strains of S. pneumoniae and P. aeruginosa for which MICs were similar were selected. The S. pneumoniae strains were SMH 11622, SMH 12716, and SMH 12648; the P. aeruginosa strains were SMH 21356 and SMH 20574. Strain SMH 21356 was provided by J. Andrews, Department of Microbiology, City NHS Trust, Birmingham, United Kingdom; other strains were isolated from cases of clinical infection.
Antibiotic.
Moxifloxacin was obtained from Bayer AG, Leverkusen, Germany. Stock solutions were prepared according to British Society of Antimicrobial Chemotherapy guidelines (2) and stored at −70°C.
MICs.
MICs were determined with a standard broth dilution method according to British Society of Antimicrobial Chemotherapy guidelines (2).
Pharmacokinetics.
The following simulated moxifloxacin doses were used: 200 mg every 24 h, 400 mg every 24 h, 400 mg every 12 h, and 800 mg every 24 h for 72 h. The maximum concentrations (Cmax) after day 1 were 1.25 mg/liter for 200 mg, 2.50 mg/liter for 400 mg, and 5.0 mg/liter for 800 mg. The simulated half-life was 12.5 h, and the mean target AUCs from 0 to 24 h (AUC0-24) were as follows: for the 200-mg regimen, 28 mg/liter · h; for the 400-mg regimen, 55 mg/liter · h; and for the 800-mg regimen, 107 mg/liter · h.
Antibacterial effect.
For all experiments, 100 μl of an overnight broth culture of either the S. pneumoniae or the P. aeruginosa test strain was inoculated into the central culture chamber (volume, 360 ml), and the model was run for 18 h to allow the organism growth to reach steady state at a density of about 107 to 108 CFU/ml. Moxifloxacin was then added to the dosing chamber, and samples were taken from the central chamber throughout the 72-h period at 0, 1, 2, 3, 4, 5, 6, 7, 12, 24, 25, 26, 28, 29, 30, 31, 36, 48, 55, 60, and 72 h for the determination of viable counts. Bacteria were quantified by using a Spiral Plater (Don Whitley Spiral Systems, Shipley, West Yorkshire, England); the minimum detection level was 102 CFU/ml. Additional aliquots were also stored at −70°C for the measurement of moxifloxacin by using a bioassay described previously (3)
Emergence of resistance.
Drug resistance was assessed before moxifloxacin exposure (time zero) and 12, 24, 36, 48, 60, and 72 h after exposure. Samples were plated on agar containing no moxifloxacin and containing antibiotic at 1, 2, 4, 8, and 16 times the MIC in order to quantify the fluoroquinolone-resistant subpopulation.
All pharmacokinetic simulations to determine antibacterial effect and emergence of resistance were performed at least in triplicate.
Pharmacodynamics, measurement of antibacterial effect, measurement of emergence of resistance, and statistical analysis.
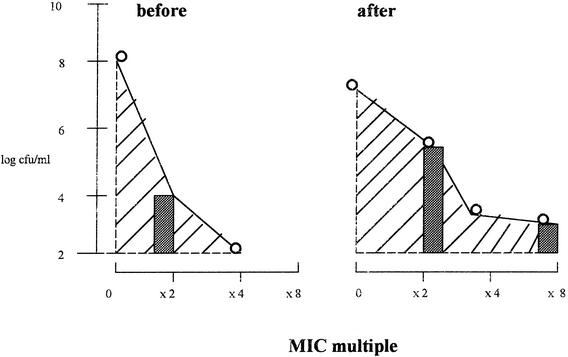
The antibacterial effect was assessed by calculating the log change in viable counts between time zero and 24 h (Δ24), 48 h (Δ48), and 72 h (Δ72). The maximum reduction in counts (Δmax) was also recorded, as was the time taken for the inoculum to fall to 99.9% its value at time zero (T99.9). In addition, the AUBKC (measured as log CFU per milliliter · h) was calculated by using the log linear trapezoidal rule for the period from 0 to 72 h (AUBKC0-72). For drug resistance studies, the measures of resistance were changes in the bacterial counts on plates containing antibiotic and the AUC for the population analysis profile (PAP). The latter value was calculated by using the log linear trapezoidal rule at time zero and at 24, 48, and 72 h (Fig. 1).
FIG. 1.
Theoretical plots of measures of drug resistance. Hatched areas indicate the AUC for the PAP before and after exposure; gray bars indicate the log CFU per milliliter at two or eight times the MIC before and after exposure.
The variability in the measurement of emergence of resistance was assessed by calculating the coefficient of variation (CV) for each measure and then using the following equation: percent CV = (1 standard deviation/mean) × 100. For pharmacodynamic analysis, the AUC/MIC, Cmax/MIC, and time above the MIC (T>MIC) were calculated for the 72-h observation period. The AUBKC0-72 was compared to the AUC/MIC, Cmax/MIC, and T>MIC by using an inhibitory Emax model with Win Nonlin Software (Scientific Consulting). The correlation between the pharmacodynamic variables was assessed by using Spearman's rank correlation coefficient. For emergence of resistance, the AUC for the PAP was compared to the AUC/MIC, Cmax/MIC, and T>MIC by using an inhibitory Emax model as described above. Subsequently, weighted least-squares regression analysis was used to examine the combined effects of the AUC/MIC, Cmax/MIC, and T>MIC on the AUC for the PAP for both S. pneumoniae and P. aeruginosa.
Cox proportional-hazard regression analysis was used to assess whether the AUC/MIC, Cmax/MIC, or T>MIC was predictive of the time taken for the bacterial population counts on antibiotic-containing plates to increase by 2 log units.
RESULTS
MICs.
The moxifloxacin MICs for the S. pneumoniae strains were as follows: strain SMH 11622, 0.25 mg/liter; strain 12716, 1.0 mg/liter; and strain 12648, 3.6 mg/liter. The MICs for the P. aeruginosa strains were as follows: strain 21356, 0.25 mg/liter; and strain 20574, 1.0 mg/liter.
Pharmacokinetic curves and pharmacodynamic parameters.
There was good agreement between target and achieved moxifloxacin concentrations in the model (data not shown). The target pharmacodynamic parameters (at 24 h) for S. pneumoniae were as follows: AUC/MIC, 15 to 427; Cmax/MIC, 0.9 to 26.0; and T>MIC, 7 to 100%. The target pharmacodynamic parameters (at 24 h) for P. aeruginosa were as follows: AUC/MIC, 107 to 427; Cmax/MIC, 4.7 to 26.0; and T>MIC, all ≥99%. The correlation between the pharmacodynamic parameters was high: for the S. pneumoniae experiments, Spearman r values were 0.9820 for AUC/MIC ∼ Cmax/MIC, 0.9751 for AUC/MIC ∼ T>MIC, and 0.9452 for Cmax/MIC ∼ T>MIC; for the P. aeruginosa experiments, the Spearman r value was 0.971 for AUC/MIC ∼ Cmax/MIC.
Antibacterial effect.
The antibacterial effect measures for S. pneumoniae and P. aeruginosa are shown in Tables 1 and 2. For S. pneumoniae strain 11622, the MIC of 0.25 mg/liter in the simulation with 400 mg every 24 h had a poorer antibacterial effect than either the simulation with 400 mg every 12 h or the simulation with 800 mg every 24 h, as indicated by a smaller Δ24, a longer T99.9, and a larger AUBKC0-72. However, Δmax values were similar. The antibacterial effects of 400 mg every 12 h and 800 mg every 24 h were similar for this strain and strain 12716 (MIC, 1.0 mg/liter). There was a minimal antibacterial effect against strain 12648 (MIC, 3.6 mg/liter) with any of the simulations; Δ24, Δ48, Δ72, and Δmax values indicated a small reduction or no reduction in bacterial counts, therefore producing a large AUBKC0-72 and a T99.9 of >72 h.
TABLE 1.
Antibacterial effect measures for each strain of S. pneumoniae with each moxifloxacin dose simulation
| Strain (MIC, mg/liter) | Dose regimen
|
Viable counts (log CFU/ml)
|
AUBKC0-72 (log CFU/ml · h) | T99.9 (h) | |||||
|---|---|---|---|---|---|---|---|---|---|
| mg | Interval between doses (h) | n | Δ24 | Δ48 | Δ72 | Δmax | |||
| 11622 (0.25) | 400 | 24 | 3 | −1.7 ± 1.1 | −5.4 ± 0.8 | −2.9 ± 0.2 | −5.7 ± 0.5 | 220 ± 8 | 30 ± 6 |
| 400 | 12 | 3 | −4.8 ± 0.5 | −4.2 ± 1.0 | −3.4 ± 1.2 | −5.2 ± 0.5 | 150 ± 24 | 16 ± 7 | |
| 800 | 24 | 3 | −4.6 ± 1.5 | −4.5 ± 1.2 | −5.4 ± 0.2 | −5.4 ± 0.2 | 124 ± 22 | 18 ± 12 | |
| 12716 (1.0) | 400 | 12 | 5 | −3.3 ± 1.8 | −5.7 ± 0.6 | −6.2 ± 0.7 | −6.2 ± 0.7 | 159 ± 44 | 19 ± 8 |
| 800 | 24 | 3 | −1.9 ± 1.1 | −6.3 ± 0.8 | −6.0 ± 1.2 | −6.7 ± 0.1 | 174 ± 28 | 29 ± 6 | |
| 12648 (3.6) | 400 | 24 | 3 | +0.6 ± 0.6 | +0.2 ± 0.4 | +0.6 ± 0.6 | −0.1 ± 0.1 | 456 ± 35 | >72 |
| 400 | 12 | 3 | +0.2 ± 0.7 | +0.3 ± 0.2 | +0.5 ± 0.4 | −0.6 ± 0.4 | 440 ± 24 | >72 | |
| 800 | 24 | 3 | −0.5 ± 0.5 | +0.2 ± 0.2 | −0.1 ± 0.3 | −0.9 ± 0.1 | 425 ± 22 | >72 | |
TABLE 2.
Antibacterial effect measures for each strain of P. aeruginosa with each moxifloxacin dose simulationa
| Strain (MIC, mg/liter) | Dose regimen
|
Viable counts (log CFU/ml)
|
AUBKC0-72 (log CFU/ml · h) | T99.9 (h) | |||||
|---|---|---|---|---|---|---|---|---|---|
| mg | Interval between doses (h) | n | Δ24 | Δ48 | Δ72 | Δmax | |||
| 21356 (0.25) | 200 | 24 | 3 | −0.1 ± 0.1 | −0.2 ± 0.2 | ND | −0.2 ± 0.2 | 417 ± 6 | >72 |
| 400 | 24 | 3 | −1.1 ± 0.6 | −0.7 ± 0.5 | −0.1 ± 0.2 | −1.8 ± 0.3 | 381 ± 29 | >72 | |
| 400 | 12 | 9 | −1.0 ± 0.5 | −2.5 ± 1.7 | −0.9 ± 1.0 | −3.2 ± 1.4 | 331 ± 52 | >72 | |
| 800 | 24 | 6 | −1.1 ± 0.6 | −1.3 ± 1.0 | −0.9 ± 0.9 | −4.0 ± 0.9 | 315 ± 39 | 30 ± 5 | |
| 20574 (1.0) | 400 | 12 | 7 | −2.8 ± 0.8 | −0.5 ± 0.4 | −0.2 ± 0.4 | −3.8 ± 0.7 | 321 ± 34 | 29 ± 21 |
| 800 | 24 | 4 | −2.8 ± 0.9 | −0.2 ± 0.6 | −0.1 ± 0.5 | −4.3 ± 0.5 | 336 ± 34 | 11 ± 10 | |
ND, not determined.
For P. aeruginosa strain 21356 (MIC, 0.25 mg/liter), the simulation with 200 mg every 24 h had a minimal effect. The antibacterial effect was greater with the simulation with 400 mg every 24 h and greatest with the simulations with 400 mg every 12 h and 800 mg every 24 h, which had the largest Δ72 and Δmax values and the smallest AUBKC0-72. As with S. pneumoniae, the antibacterial effects with 400 mg every 12 h and 800 mg every 24 h tended to be similar for P. aeruginosa strain 21356 (MIC, 0.25 mg/liter) and strain 20574 (MIC, 1.0 mg/liter).
A comparison of S. pneumoniae and P. aeruginosa strains for which MICs were similar, for example, strains 11622 and 21356 (MICs for both, 0.25 mg/liter) or strains 12716 and 20574 (MICs for both, 1.0 mg/liter), indicated that the antibacterial effect of moxifloxacin against P. aeruginosa was less marked than that against S. pneumoniae. For the strains for which the MIC was 0.25 mg/liter, Δmax values were smaller for P. aeruginosa than for S. pneumoniae, the AUBKC0-72 was larger, and the T99.9 was longer. The data were similar for strains for which the MIC was 1.0 mg/liter.
For S. pneumoniae, the AUBKC0-72 could be related to the AUC/MIC and the Cmax/MIC by using an inhibitory Emax model, were Emax represents the maximum effect, E0 represents the minimum effect, and EC50 represents the concentration required to obtain 50% the maximum effect. For both pharmacodynamic parameters, the relationship to AUBKC0-72 could be adequately described by the model, as judged by Akaike information criteria and plots of the fitted values. Parameters fitted by this model for the relationship of AUC/MIC (at 24 h) to AUBKC0-72 for S. pneumoniae were as follows: Emax, 456 ± 23 log CFU/μl · h (mean and standard deviation); E0, 163 ± 13 log CFU/μl · h; EC50, 45 ± 22; and r2, 0.9662. Parameters fitted by this model for the relationship of Cmax/MIC to AUBKC0-72 were as follows: Emax, 451 ± 19 log CFU/μl · h; E0, 164 ± 10 log CFU/μl · h; EC50, 2.3 ± 2.6; and r2, 0.9674. The data generated for P. aeruginosa indicated that even at the highest T>MIC, AUC/MIC, or Cmax/MIC values, bacterial clearance from the model was inadequate; therefore, the relationship between the pharmacodynamic parameters and AUBKC0-72 was not investigated further.
Emergence of resistance.
The emergence of resistance was studied by using three strains of S. pneumoniae (MICs, 0.25, 1.0, and 3.6 mg/liter) and two strains of P. aeruginosa (MICs, 0.25 and 1.0 mg/liter). Before exposure to moxifloxacin, few S. pneumoniae organisms grew on plates with the drug at the MIC (Table 3), indicating that the initial population was not very heterogeneous in its quinolone susceptibility. For S. pneumoniae strain 11622, no resistance emerged with the lowest dose simulation (400 mg every 24 h); with the higher dose simulations (400 mg every 12 h and 800 mg every 24 h), the organisms were cleared from the model. Similarly, S. pneumoniae strain 12716 (MIC, 1.0 mg/liter) was cleared from the model, and no resistance emerged. Strain 12648 (MIC, 3.6 mg/liter) was different in that the organisms were not cleared and the counts of bacteria growing on plates with the drug at the MIC increased from <2 log units to 3 to 5 log units with some simulations, although this pattern of resistance was also seen with strain 11622 (MIC, 0.25 mg/liter). No bacteria grew on plates with 8 or 16 times the MIC.
TABLE 3.
Emergence of resistance to moxifloxacin in three strains of S. pneumoniaea
| Strain (MIC, mg/liter) | Moxifloxacin concn in recovery medium (×) | Log CFU/liter at the following time (h) with the indicated dose regimen:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 24
|
48
|
72
|
||||||||
| 400 every 24 h | 400 every 12 h | 800 every 24 h | 400 every 24 h | 400 every 12 h | 800 every 24 h | 400 every 24 h | 400 every 12 h | 800 every 24 h | |||
| 11622 (0.25) | 0 | 7.8 ± 0.6 | 6.6 ± 0.9 | <2 | 2.9 ± 1.4 | 5.8 | 3.0 ± 1.1 | 2.8 ± 1.1 | 5.2 ± 0.2 | <2 | <2 |
| 1 | 4.6 ± 2.1 | <2 | <2 | <2 | 3.5 ± 1.4 | <2 | 2.7 ± 0.9 | 5.2 ± 0.1 | <2 | <2 | |
| 2 | <2 | <2 | <2 | <2 | 2.8 ± 0.9 | <2 | <2 | <2 | <2 | <2 | |
| 4 | <2 | <2 | <2 | <2 | 2.6 ± 0.6 | <2 | <2 | <2 | <2 | <2 | |
| 12716 (1.0) | 0 | 8.6 ± 0.2 | ND | 5.1 ± 2.0 | 6.9 ± 1.1 | ND | 2.5 ± 0.4 | <2 | ND | <2 | <2 |
| 1 | <2 | ND | <2 | <2 | ND | <2 | <2 | ND | <2 | <2 | |
| 2 | <2 | ND | <2 | <2 | ND | <2 | <2 | ND | <2 | <2 | |
| 4 | <2 | ND | <2 | <2 | ND | <2 | <2 | ND | <2 | <2 | |
| 12648 (3.6) | 0 | 7.9 ± 0.4 | 8.1 ± 0.1 | 8.2 ± 0.1 | 7.6 ± 0.7 | 8.2 ± 0.6 | 8.3 ± 0.1 | 8.2 ± 0.1 | 5.2 | 2.5 | 3.2 ± 1.7 |
| 1 | <2 | 5.1 ± 0.3 | 4.9 ± 0.3 | <2 | 4.9 ± 0.4 | 3.0 ± 0.9 | 5.1 ± 0.1 | <2 | <2 | <2 | |
| 2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | |
| 4 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | <2 | |
See Table 2, footnote a. Doses are given in milligrams.
In contrast to the S. pneumoniae strains, both strains of P. aeruginosa, 21356 and 20574, were heterogeneous; a substantial minority of cells within the population were able to grow on plates with two, four, and eight times the MIC, even before exposure to moxifloxacin. For strain 21356 (MIC, 0.25 mg/liter), the simulations with 200 mg every 24 h and 400 mg every 24 h resulted in an increase in the proportion of the population able to grow on plates with 4, 8, and 16 times the MIC, and this trend increased with the time of exposure (Table 4). No significant bacterial clearance occurred in the model. Strain 20574 (MIC, 1.0 mg/liter) showed a similar pattern with 400 mg every 12 h and 800 mg every 24 h (Table 4). In contrast, the more susceptible strain (MIC, 0.25 mg/liter) showed no convincing increase in the proportion of the population able to grow on the antibiotic-containing plates until perhaps 72 h of exposure to 400 mg every 12 h and 800 mg every 24 h.
TABLE 4.
Emergence of resistance to moxifloxacin in two strains of P. aeruginosaa
| Strain (MIC, mg/liter) | Moxifloxacin concn in recovery medium (×) | Log CFU/liter at the following time (h) with the indicated dose regimen:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 24
|
48
|
72
|
|||||||||||
| 200 every 24 h | 400 every 24 h | 400 every 12 h | 800 every 24 h | 200 every 24 h | 400 every 24 h | 400 every 12 h | 800 every 24 h | 200 every 24 h | 400 every 24 h | 400 every 12 h | 800 every 24 h | |||
| 21356 (0.25) | 0 | 8.5 ± 0.2 | 8.3 ± 0.1 | 7.6 ± 0.6 | 7.5 ± 0.6 | 7.5 ± 0.5 | 8.2 ± 0.1 | 7.5 ± 0.5 | 5.3 ± 1.7 | 7.3 ± 0.8 | 8.3 ± 0.1 | 8.2 ± 0.1 | 7.6 ± 0.9 | 7.5 ± 0.9 |
| 1 | 8.1 ± 0.2 | 8.2 ± 0.1 | 7.4 ± 0.8 | 7.5 ± 0.8 | 7.4 ± 0.5 | 8.2 ± 0.1 | 7.4 ± 0.5 | 5.2 ± 1.9 | 7.0 ± 1.0 | 8.2 ± 0.1 | 8.2 ± 0.1 | 6.9 ± 1.1 | ND | |
| 2 | 6.5 ± 1.7 | 8.1 ± 0.1 | 7.5 ± 0.7 | 3.8 ± 0.2 | 5.8 ± 0.9 | 8.2 ± 0.1 | 7.4 ± 0.5 | 4.9 ± 2.1 | 4.6 ± 1.1 | 8.2 ± 0.1 | 8.2 ± 0.1 | 6.4 ± 1.6 | ND | |
| 4 | 4.7 ± 1.5 | 8.0 ± 0.1 | 7.3 ± 0.8 | 2.6 ± 0.5 | 3.5 ± 1.0 | 8.1 ± 0.1 | 7.3 ± 0.6 | 2.9 ± 1.1 | 2.8 ± 1.1 | 8.0 ± 0.1 | 8.2 ± 0.1 | 4.0 ± 2.4 | 4.0 ± 0.4 | |
| 8 | 4.0 ± 1.5 | 7.7 ± 0.2 | 5.9 ± 1.6 | <2 | 2.9 ± 1.0 | 7.8 ± 0.1 | 7.2 ± 0.6 | <2 | <2 | 8.0 ± 0.1 | 8.2 ± 0.1 | 3.8 ± 3.1 | 3.6 ± 1.4 | |
| 16 | <2 | ND | 3.4 ± 1.6 | <2 | <2 | ND | 5.0 ± 0.6 | 3.8 ± 3.1 | <2 | ND | 8.3 ± 0.2 | 3.2 ± 1.8 | 3.5 ± 1.4 | |
| 20574 (1.0) | 0 | 8.6 ± 0.2 | ND | ND | 6.8 ± 0.8 | 8.2 | ND | ND | 7.3 ± 0.5 | 8.3 ± 0.3 | ND | ND | 8.3 | 8.0 ± 0.5 |
| 1 | 5.3 ± 0.3 | ND | ND | 6.0 ± 0.8 | 5.8 ± 1.0 | ND | ND | 7.2 ± 0.6 | 8.4 ± 0.4 | ND | ND | 8.1 | 8.3 ± 0.1 | |
| 2 | 4.4 ± 1.2 | ND | ND | 4.8 ± 0.2 | 4.7 ± 1.3 | ND | ND | 6.8 ± 0.2 | 8.2 ± 0.2 | ND | ND | 7.7 ± 0.9 | 8.2 ± 0.1 | |
| 4 | 3.8 ± 1.0 | ND | ND | 4.5 ± 0.1 | 4.7 ± 1.3 | ND | ND | 7.3 ± 0.7 | 8.2 | ND | ND | 8.2 ± 0.2 | 8.2 ± 0.2 | |
| 8 | 2.6 ± 0.6 | ND | ND | 4.1 ± 0.6 | 4.6 ± 1.1 | ND | ND | 6.3 ± 0.9 | 8.0 | ND | ND | 8.0 ± 0.4 | 7.4 ± 1.2 | |
| 16 | <2 | ND | ND | 3.5 ± 1.0 | 3.1 ± 0.1 | ND | ND | 6.6 ± 0.9 | 3.9 ± 2.0 | ND | ND | 7.7 ± 0.6 | 6.5 ± 1.3 | |
See Table 2, footnote a. Doses are given in milligrams.
Calculation of the AUC for the PAP helped to further quantify changes in the population profiles of the S. pneumoniae and P. aeruginosa strains. The average PAP AUC for the three strains of S. pneumoniae before drug exposure was 4.2 ± 2.1 (mean and standard deviation), and that for P. aeruginosa was 34.4 ± 16.9, reflecting the more heterogeneous nature of the Pseudomonas population (Table 5). None of the strains of S. pneumoniae showed a convincing trend in PAP AUCs. In contrast, for P. aeruginosa strain 21356 (MIC, 0.25 mg/liter) with the simulation with 400 mg every 24 h and strain 20574 (MIC, 1.0 mg/liter) with the simulations with 400 mg every 12 h and 800 mg every 24 h, the PAP AUCs increased as the period of exposure increased. This was not true for strain 21356 (MIC, 0.25 mg/liter) with the higher-dose simulations (400 mg every 12 h and 800 mg every 24 h). The emergence of resistance as measured by PAP AUCs was not different for the simulations with 400 mg every 12 h and 800 mg every 24 h (Table 5).
TABLE 5.
Emergence of resistance to moxifloxacin in S. pneumoniae and P. aeruginosa strains, as measured by changes in AUC for PAPa
| Strain (MIC, mg/liter) | Species | Dose simulation
|
AUC for PAP at the following times (h) postexposure:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| mg | Interval between doses (h) | 12 | 24 | 36 | 48 | 60 | 72 | ||
| 11622 (0.25) | S. pneumoniae | 400 | 24 | 3.1 ± 0.2 | 2.4 ± 0.5 | <1 | 4.7 ± 4.2 | ND | 9.7 ± 4.8 |
| 400 | 12 | 2.1 ± 1.4 | <1 | 2.9 ± 5.1 | <1 | <1 | <1 | ||
| 800 | 24 | 3.0 ± 2.2 | 1.2 ± 2.1 | <1 | 1.1 ± 1.4 | 1.5 ± 1.3 | 4.4 ± 8.8 | ||
| 12716 (1.0) | S. pneumoniae | 400 | 12 | 3.0 ± 0.1 | 1.1 ± 1.1 | <1 | <1 | <1 | <1 |
| 800 | 24 | 3.1 ± 0.3 | 2.4 ± 0.6 | <1 | <1 | <1 | <1 | ||
| 12648 (3.6) | S. pneumoniae | 400 | 24 | 5.8 ± 0.5 | 6.4 ± 0.5 | 6.2 ± 0.8 | 6.0 ± 0.6 | 6.4 ± 1.2 | 7.9 ± 2.7 |
| 400 | 12 | 3.8 ± 1.6 | 6.0 ± 0.3 | 4.1 ± 1.1 | 4.0 ± 0.8 | 5.3 | 3.6 | ||
| 800 | 24 | 3.3 ± 0.8 | 3.8 ± 1.7 | 6.2 ± 6.1 | 6.3 ± 0.1 | 5.7 ± 0.9 | 4.2 | ||
| 21356 (0.25) | P. aeruginosa | 200 | 24 | 62.7 ± 1.9 | 75.9 ± 3.0 | 79 ± 3.6 | 79.8 ± 3.5 | 81.3 ± 2.4 | 83.7 ± 0.6 |
| 400 | 24 | 59.1 ± 15.7 | 61.0 ± 18.5 | 75.7 ± 6.1 | 75.7 | ND | 99.6 ± 1.2 | ||
| 400 | 12 | 15.4 ± 2.5 | 13.8 ± 3.5 | 9.7 ± 1.9 | 13.8 ± 7.0 | 41 ± 32.5 | 34.1 ± 29 | ||
| 800 | 24 | 30.7 ± 8.3 | 23.1 ± 7.8 | 19.4 ± 8.9 | 15.9 ± 6.7 | 27 ± 11.5 | 28 ± 14.2 | ||
| 20574 (1.0) | P. aeruginosa | 400 | 12 | 24.7 ± 6.1 | 36.5 ± 6.7 | 59 ± 24.3 | 72.9 ± 9.3 | 93.7 ± 5.2 | 95.2 |
| 800 | 24 | 13.7 ± 7.1 | 32.2 ± 15.4 | 58.8 ± 19.6 | 74.7 ± 18.3 | 80.8 ± 15.7 | 87.7 ± 12.9 | ||
ND, not determined.
The variability of each measure of antibacterial effect was tested by calculating the percent CV, reported as the median (range), for changes in the log CFU per liter at 24, 48, and 72 h and the PAP AUC. These were 10 (0.2 to 31.1), 8.6 (0.6 to 50.8), 4 (0.1 to 81.1), and 13 (0.5 to 44.7), respectively, indicating that no measure was notably superior to the others. The mean percent CVs were all in the range of 12 to 15. In a subsequent analysis, the PAP AUC was taken as the primary end point for the emergence of resistance, as it seemed to be the measure which best encompassed all of the data.
Multiple regression analysis was used to assess the predictive influence of AUC/MIC and Cmax/MIC on PAP AUCs for P. aeruginosa and S. pneumoniae. For S. pneumoniae, all models explored included T>MIC but were inadequate, indicating that a regression model of this type was inappropriate for these data. For P. aeruginosa, there was evidence to suggest that AUC/MIC was associated with PAP AUC but that Cmax/MIC was not predictive of outcome. The model fitted the data as shown in Table 6. The model fit was assessed graphically and was satisfactory, with errors following an approximately normal distribution.
TABLE 6.
Regression model estimated with AUC for P. aeruginosa PAP as the measure of emergence of resistance
| Variable | Coefficient | SE | Pa | 95% confidence interval |
|---|---|---|---|---|
| AUC/MIC | −0.09628 | 0.0261 | >0.002 | −0.1515 to −0.04105 |
| Cmax/MIC | 1.6931 | 1.5442 | >0.289 | −1.5804 to 4.9667 |
| Constant | 115.292 | 7.7332 | 98.898 to 131.686 |
P values were determined with a t test.
In addition to PAP AUC, the emergence of resistance was measured by the time taken for the bacterial counts on the antibiotic-containing media to increase by 2 log units. Univariate analysis suggested that both AUC/MIC and Cmax/MIC were predictive of outcome (P < 0.0001); however, the parameters were highly confounded. AUC/MIC, Cmax/MIC, and antibiotic concentrations in the recovery media were fitted together in a multivariate model, and then each was omitted in turn to assess whether excluding that factor changed the model fit significantly. Two of the three factors, drug concentration and Cmax/MIC, were omitted from the model without a loss of fit. The final model therefore comprised one factor—AUC/MIC. The proportional-hazards assumption underpinning the model fit was assessed by using the “stphtest” procedure in Stata. The relative “risk” of achieving the 2-log-unit increase in counts, as estimated from the model, is given in Table 7. A 2-log-unit increase was less likely to be achieved for experiments with the highest AUC/MIC.
TABLE 7.
Relative risk of achieving a 2-log-unit increase in counts of resistant bacteria, as related to AUC/MIC
| Magnitude of AUC/MIC | Relative risk | 95% confidence interval | P |
|---|---|---|---|
| 110 | 1.00 | ||
| 221 | 0.84 | 0.40-1.74 | <0.0001 |
| 427 | 0.14 | 0.06-0.29 | <0.0001 |
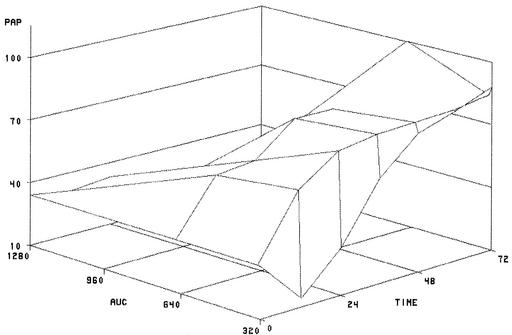
Surface plots were constructed to illustrate the relationship among AUC/MIC, time, and emergence of resistance (Fig. 2). These showed that the emergence of resistance, as measured by the PAP AUC, is more likely at low AUC/MIC ratios than at high ones and that the duration of drug exposure also determines the PAP AUC.
FIG. 2.
Surface plot of PAP data for time and AUC/MIC.
DISCUSSION
The strains of both S. pneumoniae and P. aeruginosa used in this study were not selected to be typical of their respective species in terms of moxifloxacin susceptibility. The S. pneumoniae strains for which MICs are 0.25 mg/liter and higher represent strains for which MICs are above the MIC90 of moxifloxacin, while the P. aeruginosa strains for which MICs are ≤1.0 mg/liter are relatively susceptible (13). The strains were selected to allow a direct comparison of the antibacterial effects of moxifloxacin on these species by using identical methodology, as this has not been done before with in vitro models. With a similar in vitro pharmacokinetic model, it was previously shown that AUC/MIC is the best pharmacokinetic/pharmacodynamic index in terms of a relationship to the antibacterial effect against S. pneumoniae of both gemifloxacin and moxifloxacin when AUBKC is used as the primary antibacterial effect measure (11, 14). Therefore, we related AUC/MIC to AUBKC0-72 by using an Emax model to establish the AUC/MIC ratio required for a 50% effect against either S. pneumoniae or P. aeruginosa. This AUC/MIC ratio for S. pneumoniae was 45 ± 22, while that for P. aeruginosa was not calculable but was greater than the maximum AUC/MIC tested, that is, >427. Although the precise magnitudes of the AUC/MIC ratios needed to predict antibacterial effects are different, very similar data comparing S. pneumoniae and P. aeruginosa have been reported with a nonneutropenic murine thigh infection model (N. L. Jumbo, A. Lovie, W. Liv, M. Deziel, M. H. Miller and G. L. Drusano, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 291, p. 7, 2000). These authors also showed that the AUC/MIC ratios with levofloxacin for static and bactericidal effects were much greater for P. aeruginosa than for S. pneumoniae. With AUBKC as the primary antibacterial effect measure, moxifloxacin AUC/MIC ratios of >150 were required for a maximum effect against S. pneumoniae in this study. Previously, similar AUC/MIC magnitudes were shown to be predictive of the maximum effect of gemifloxacin against S. pneumoniae (14). These AUC/MIC magnitudes are much greater than those reported in animal models with other fluroquinolones (7; W. A. Craig and D. R. Andes, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 289, p. 7, 2000; E. J. Ernst, M. E. Klepser, C. R. Petzold, and G. V. Doern, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 288, p. 6, 2000) or indeed in vitro models with moxifloxacin (10). However, it is known that a number of factors may determine AUC/MIC magnitudes, for example, the presence or absence of white blood cells and the use of bacteriostatic or bactericidal end points (Craig and Andes, 40th ICAAC; Ernst et al., 40th ICAAC). In addition, the inoculum may have an effect on antibacterial effect measures in in vitro models (5), and the impact of growth rate on AUC/MIC values has not been quantified. As our in vitro pharmacokinetic model does not include white blood cells, has an antibacterial effect measure which depends on pathogen clearance, has a large inoculum, and has a population of bacteria not undergoing rapid growth, it is likely that the AUC/MIC ratios required for the maximum effect will be greater than those reported elsewhere. Importantly, however, this model allows 1010 CFU bacteria to be exposed to antibiotic; this bacterial load is larger than that possible in animal models.
Despite the large inoculum and exposure to moxifloxacin for 72 h, it was difficult to show the emergence of resistance to moxifloxacin in S. pneumoniae, even in simulations where little bacterial clearance occurred in the model. Similar data have been reported by Zhanel et al. (18), who failed to detect resistance to moxifloxacin as well as gatifloxacin, levofloxacin, and trovafloxacin in an in vitro model with S. pneumoniae. However, as these authors used S. pneumoniae strains with a susceptibility to moxifloxacin higher that that of some of the strains used here, combined with a smaller inoculum, S. pneumoniae organisms were often cleared from the model, perhaps not allowing resistance to emerge. Furthermore, detection of the emergence of resistance by comparison of pre- and postexposure MICs may not be as sensitive for detecting changes in resistance as PAPs. Animal data obtained in a rabbit tissue cage model with S. pneumoniae and moxifloxacin exposure for 7 days failed to show any emergence of resistance (17). The exception to these observations in both in vitro and animal models appears to be ciprofloxacin; data indicating resistance detected by changes in MICs or PAPs are easily generated by ciprofloxacin exposure (4, 16, 18).
The emergence of resistance to moxifloxacin was much more marked with P. aeruginosa than with S. pneumoniae. Previous data obtained with a dose fractionation design involving 1,200 mg of ciprofloxacin every 24 h, 600 mg every 12 h, or 400 mg every 8 h and recovery of resistant bacteria on agar containing zero, 0.5, 1, or 4 mg of ciprofloxacin/liter indicated that the single large dose was most effective at preventing resistance, thus highlighting the role of Cmax/MIC (15a). These data were not altogether confirmed by Madaras-Kelly et al. (15), who related the emergence of resistance as measured by changes in the MICs for Pseudomonas before and after exposure to either AUC/MIC or Cmax/MIC by using a sigmoid Emax model. Our data indicate that AUC/MIC is related to the emergence of resistance, as shown by the similarity of the PAPs for the simulations with 400 mg every 12 h and 800 mg every 24 h and the results of multiple regression and time-to-event analyses. However, due to the limited nature of the dose fractionation performed, Cmax/MIC and AUC/MIC are very closely related; thus, some caution needs to be exercised with these data. These data clearly show the effect of drug exposure, bacterial species, initial pathogen susceptibility, and time of exposure on the emergence of resistance. Three patterns can be described. First, the pathogen is cleared from the model, and no increase in resistance is detected. This pattern is seen in the simulations with the strains of S. pneumoniae that were more susceptible to moxifloxacin. The second pattern is little pathogen eradication in the model but no emergence of resistance. This pattern is observed with the least susceptible strain of S. pneumoniae and with P. aeruginosa strains with the highest AUC/MIC ratios. The third pattern is no clearance from the model and emergence of resistance, as detected by changing PAPs, which tend to show increased resistance as the duration of exposure increases. This pattern is seen with the two P. aeruginosa strains with the lowest AUC/MIC ratios.
In conclusion, these data show that the magnitude of the fluoroquinolone AUC/MIC needed to predict equivalent antibacterial effects is smaller for S. pneumoniae than for P. aeruginosa and that the emergence of resistance to fluroquinolones in in vitro models depends on species (P. aeruginosa producing more resistance than S. pneumoniae), duration of exposure, and drug exposure, as indicated by Cmax or AUC, and on initial pathogen susceptibility, as denoted by MIC.
Acknowledgments
We thank Bayer AG for financial support of this study.
REFERENCES
- 1.Blaser, J., B. B. Stone, M. C. Groner, and S. H. Zinner. 1987. Comparative study with enoxacin and netilmicin in a pharmacodynamic model to determine importance of ratio of antibiotic peak concentrations to MIC for bacterial activity and emergence of resistance. Antimicrob. Agents Chemother. 31:1054-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.British Society for Antimicrobial Chemotherapy Working Party. 1991. A guide to sensitivity testing. British Society for Antimicrobial Chemotherapy, Birmingham, United Kingdom. [PubMed]
- 3.Broughall, J. M. 1978. Aminoglycosides, p. 194-206. In D. S. Reeves, I. Phillips, J. D. Williams, and R. Wise (ed.), Laboratory methods in antimicrobial chemotherapy. Churchill Livingstone, Edinburgh, United Kingdom.
- 4.Coyle, E. A., G. W. Kaatz, and M. J. Ryback. 2001. Activities of newer fluoroquinolones against ciprofloxacin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:1654-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Firsov, A. A., S. N. Vostrov, O. V. Kononenko, S. H. Zinner, and Y. A. Portnoy. 1999. Prediction of the effects of inoculum size on the antimicrobial action of trovafloxacin and ciprofloxacin against Staphylococcus aureus and Escherichia coli in an in vitro dynamic model. Antimicrob. Agents Chemother. 43:498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heushberger, E., and M. J. Rybak. 2000. Activities of trovafloxacin, gatifloxacin, clinafloxacin, sparfloxacin, levofloxacin, and ciprofloxacin against penicillin-resistant Streptococcus pneumoniae in an in vitro model of infection. Antimicrob. Agents Chemother. 44:598-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacy, M. K., W. Lu, X. Xu, P. R. Tessier, D. P. Nicolau, R. Quintilliani, and C. H. Nightingale. 1999. Pharmacodynamic comparisons of levofloxacin, ciprofloxacin, and ampicillin against Streptococcus pneumoniae in an in vitro model of infection. Antimicrob. Agents Chemother. 43:672-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister, P. D., and C. C. Sanders. 1999. Pharmacodynamics of levofloxacin and ciprofloxacin against Streptococcus pneumoniae. J. Antimicrob. Chemother. 43:79-86. [DOI] [PubMed] [Google Scholar]
- 9.Lister, P. D., and C. C. Sanders. 1999. Pharmacodynamics of trovafloxacin, ofloxacin, and ciprofloxacin against Streptococcus pneumoniae in an in vitro pharmacokinetic model. Antimicrob. Agents Chemother. 43:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lister, P. D., and C. C. Sanders. 2001. Pharmacodynamics of moxifloxacin, levofloxacin and sparfloxacin against Streptococcus pneumoniae. J. Antimicrob. Chemother. 47:811-818. [DOI] [PubMed] [Google Scholar]
- 11.MacGowan, A., C. Rogers, H. A. Holt, M. Wootton, and K. Bowker. 2000. Assessment of different antibacterial effect measures used in in vitro models of infection and subsequent use in pharmacodynamic correlations for moxifloxacin. J. Antimicrob. Chemother. 46:73-78. [DOI] [PubMed] [Google Scholar]
- 12.MacGowan, A. P., K. E. Bowker, M. Wootton, and H. A. Holt. 1999. Activity of moxifloxacin, administered once a day, against Streptococcus pneumoniae in an in vitro pharmacodynamic model of infection. Antimicrob. Agents Chemother. 43:1560-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacGowan, A. P. 1999. Moxifloxacin (Bay 12-8039): a new methoxyl quinolone antibacterial. Expert Opin. Investig. Drugs 8:181-199. [DOI] [PubMed] [Google Scholar]
- 14.MacGowan, A. P., C. A. Rogers, H. A. Holt, M. Wootton, and K. E. Bowker. 2001. Pharmacodynamics of gemifloxacin against Streptococcus pneumoniae in an in vitro pharmacokinetic model of infection. Antimicrob. Agents Chemother. 45:2916-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madaras-Kelly, K. J., B. E. Ostergaard, L. B. Horde, and J. C. Rotschafer. 1996. Twenty-four-hour area under the concentration-time curve/MIC ratio as a generic predictor of fluoroquinolone antimicrobial effect by using three strains of Pseudomonas aeruginosa in an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 40:627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Marchbanks, C. R., J. R. McKiel, D. H. Gilbert, N. J. Robillard, B. Painter, S. H. Zinner, and M. N. Dudley. 1993. Dose ranging and fractionation of intravenous ciprofloxacin against Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro model of infection. Antimicrob. Agents Chemother. 37:1756-1763. [DOI] [PMC free article] [PubMed]
- 16.Thorburn, C. E., and D. I. Edwards. 2001. The effect of pharmacokinetics on the bactericidal activity of ciprofloxacin and sparfloxacin against Streptococcus pneumoniae and emergence of resistance. J. Antimicrob. Chemother. 48:15-22. [DOI] [PubMed] [Google Scholar]
- 17.Xuan, D., M. Zhong, H. Mattoes, K. Q. Bui, J. McNabb, D. P. Nicolau, R. Quintilliani, and C. H. Nightingale. 2001. Streptococcus pneumoniae response to repeated moxifloxacin or levofloxacin exposure in a rabbit tissue cage model. Antimicrob. Agents Chemother. 45:794-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhanel, G. G., M. Walters, N. Laing, and D. J. Hoban. 2001. In vitro pharmacodynamic modelling simulating free serum concentrations of fluoroquinolones against multidrug-resistant Streptococcus pneumoniae. J. Antimicrob. Chemother. 47:435-440. [DOI] [PubMed] [Google Scholar]