Abstract
A molecular model of pokeweed antiviral protein (PAP)-RNA interactions was used to rationally engineer FLP-102(151AA152) and FLP-105(191AA192) as nontoxic PAPs with potent anti-human immunodeficiency virus (anti-HIV) activities. FLP-102 and FLP-105 have been produced in Escherichia coli and tested both in vitro and in vivo. These proteins depurinate HIV type 1 (HIV-1) RNA much better than rRNA and are more potent anti-HIV agents than native PAP or recombinant wild-type PAP. They are substantially less toxic than native PAP in BALB/c mice and exhibit potent in vivo activities against genotypically and phenotypically nucleoside reverse transcriptase inhibitor-resistant HIV-1 in a surrogate human peripheral blood lymphocyte (Hu-PBL) SCID mouse model of human AIDS. Rationally engineered nontoxic recombinant PAPs such as FLP-102 and FLP-105 may provide the basis for effective salvage therapies for patients harboring highly drug-resistant strains of HIV-1. The documented in vitro potencies of FLP-102 and FLP-105, their in vivo antiretroviral activities in the HIV-infected Hu-PBL SCID mouse model, and their favorable toxicity profiles in BALB/c mice warrant the further development of these promising new biotherapeutic agents.
Pokeweed antiviral protein (PAP) is a 29-kDa naturally occurring antiviral agent that can be isolated from the leaves of the pokeweed plant (Phytolacca americana) (6, 36). PAP has a unique ability to depurinate human immunodeficiency virus (HIV) type 1 (HIV-1) RNA (18, 20). PAP exhibits potent antiviral activity against nucleoside analog reverse transcriptase inhibitor (NRTI)-resistant primary clinical HIV-1 isolates (3). Both zidovudine (ZDV)-sensitive and ZDV-resistant clinical HIV-1 isolates were found to be >4 logs more sensitive to PAP than to ZDV (3). We have cloned the gene for PAP and established procedures for the large-scale production and purification of the cloned recombinant PAP (16, 17). We have tested recombinant PAP against a broad panel of viruses in vitro and documented that it is as active as native PAP against both DNA and RNA viruses (16, 17). We were also able to determine the X-ray crystal structure of PAP at a 2.1-Å resolution (8, 9).
More recently, we have used a molecular model of PAP-RNA interactions (19, 21, 22) for the rational design of PAP mutants with potent anti-HIV activities. In the present study, two such recombinant PAPs, FLP-102 (151AA152) and FLP-105 (191AA192), have been engineered, produced, and tested both in vitro and in vivo. These proteins depurinate HIV-1 RNA much better than rRNA and are more potent anti-HIV agents than native PAP or recombinant wild-type PAP. Our preliminary studies indicate that these proteins are substantially less toxic than wild-type PAP and exhibit potent in vivo activities against a genotypically and phenotypically NRTI-resistant clinical HIV-1 isolate in a surrogate human peripheral blood lymphocyte (Hu-PBL) SCID mouse model of human AIDS. We hypothesize that FLP-102 and FLP-105, because of their potent anti-HIV activities and lack of systemic toxicities, may provide the basis for effective salvage therapies for patients harboring highly drug-resistant strains of HIV-1.
MATERIALS AND METHODS
Engineering of recombinant PAPs.
Molecular modeling studies for the rational design of recombinant PAPs were performed as previously described in detail (19, 21, 22). Recombinant wild-type PAP (phosphate-buffered saline [PBS]-PAP) was obtained by subcloning the PAP-I gene (amino acids 22 to 313) into the pBluescript SK− expression vector (17). An uracil-containing template of PAP was obtained by transforming Escherichia coli CJ236 with recombinant plasmid PBS-PAP. The oligonucleotides used for site-directed mutagenesis were synthesized on a 200-nmol scale and purified by high-pressure liquid chromatography (HPLC) by Biosynthesis Inc. (Lewisville, Tex.). The site-directed mutagenesis procedure was performed by using the Mutagene M13 in vitro mutagenesis kit (Bio-Rad, Hercules, Calif.), as described in the manufacturer's manual. DNA sequencing was carried out by the method of Sanger et al. (26) according to the instructions of the manufacturer (U.S. Biochemical Corp., Cleveland, Ohio). Fine chemicals and restriction enzymes were purchased from Roche Molecular Biochemicals (Indianapolis, Ind.).
Expression and purification of recombinant PAPs.
Wild-type and mutant PAPs were expressed in E. coli MV1190 as inclusion bodies and were isolated, solubilized, and refolded as described previously (17). The refolded proteins were analyzed by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE). Protein concentrations were quantitated from the gel by using bovine serum albumin as a standard.
Immunoblot analysis of PAP mutants.
Protein samples were resolved on an SDS-12% polyacrylamide gel and transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) by using a Bio-Rad Trans-Blot apparatus, as described previously (17). The membrane was immunoblotted with rabbit anti-PAP serum (dilution, 1:2,000) and a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG; Sigma Chemical Co., St. Louis, Mo.) as the primary and secondary antibodies, respectively. The blot was developed with 3, 3′-diaminobenzidine (Sigma) as the colorimetric indicator for peroxidase activity.
Aniline cleavage assays of rRNA depurination.
Five micrograms of E. coli 23S and 16S rRNAs (Roche Molecular Biochemicals) was incubated with increasing amounts of wild-type or mutant PAP in 50 μl (final volume) of binding buffer (25 mM Tris-HCl [pH 7.8], 10 mM KCl, 5 mM MgCl2, 2% glycerol) at 37°C for 1 h. The rRNA was extracted with phenol-chloroform (24:24), precipitated with ethanol, and treated with 20 μl of 1 M aniline acetate (pH 4.5) for 30 min on ice. The rRNA was precipitated with ethanol, electrophoresed in a 6% urea-polyacrylamide gel, and stained with ethidium bromide as described previously (19).
HPLC-based RNA depurination assays.
The release of adenine or guanine from E. coli 23S and 16S rRNAs (Roche Molecular Biochemicals) and HIV-1 RNA (ABI Biotechnologies, Columbia, Md.) was measured with an HPLC system (Hewlett-Packard Co., Palo Alto, Calif.) equipped with a diode array detector and a ChemStation software program for data analysis, as described previously (18, 20). Briefly, 2 μg of the RNA substrate was incubated with 2.5 μM wild-type or mutant PAPs for 1 h at 37°C in 50 μl of binding buffer (25 mM Tris-HCl [pH 7.8], 10 mM KCl, 5 mM MgCl2, 2% glycerol). The reaction was stopped by adding 100 μl of HPLC running buffer (50 mM NH4C2H3O2, 5% methanol [pH 5.0]), and 100 μl of the sample was injected automatically into a reverse-phase Lichrospher 100RP-18E analytical column (particle size, 5 mm; 250 by 4 mm; Hewlett-Packard) equilibrated with HPLC running buffer, as described previously (18, 20). Controls included (i) samples containing untreated rRNA and (ii) test samples without rRNA. A calibration curve was generated to establish the linear relationship between the absolute peak area and the quantities of adenine and guanine (Sigma), as described previously (18, 20). Unweighted linear regression analysis of the calibration curve was performed with the CA-Cricket graph III computer program (Computer Association, Inc., Islandia, N.Y.). Intra- and interassay accuracies and precisions were evaluated as described previously (21). Under the chromatographic conditions described above, the retention times for adenine and guanine residues were 11.5 and 5.7 min, respectively, and they eluted from the blank controls without an interference peak. The lowest limit of detection of adenine was 2.5 pmol at a signal-to-noise ratio of ≈3. The average peak areas obtained for 50 and 250 pmol of adenine standard/50 μl were 29 ± 1 and 143 ± 7 milliabsorption units (mAU), respectively. The average peak areas obtained for 50 and 250 pmol of guanine standard/50 μl were 31 ± 2 and 156 ± 8 mAU, respectively. The intra- and interassay coefficients of variation were less than 4%. The overall intra- and interassay accuracies of this method were 98.7% ± 1.7% (n = 5) and 95.7% ± 3.0% (n = 5), respectively.
Cell-free translation assays.
Protein synthesis was assayed in a cell-free system with nuclease-treated rabbit reticulocyte lysates (Promega, Madison, Wis.) and luciferase mRNA, as described previously (17, 19). The 50% inhibitory concentration (IC50s) were calculated by nonlinear regression analysis (Prism-2 software; Graph Pad Software, San Diego, Calif.) by using the average values of three independent experiments. The counts per minute for the control sample to which all the reagents except the test sample were added ranged from 3 × 107 to 4 × 107 cpm/ml, and the level of incorporation for samples treated with all the reagents except PAP was assigned a value of 100%.
Ribosome binding assays.
Ribosomes were isolated from rabbit reticulocyte-rich whole blood (Pel-Freez Biologicals, Rogers, Ark.) as described previously (22). Total ribosomes (30 μg) were incubated with 5 μg of wild-type or mutant PAPs to a final volume of 100 μl in binding buffer and incubated at room temperature for 1 h. After incubation, the ribosomes were pelleted by centrifugation at 300,000 × g for 30 min at 4°C. The pellets were washed two times with solution D (10 mM Tris-HCl [pH 7.5], 1 mM KCl, 0.1 M MgCl2) and resuspended in 20 μl of PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1 mM KH2PO4 [pH 7.4]). The protein samples were resolved on an SDS-12% polyacrylamide gel and transferred onto a PVDF membrane (Bio-Rad) with a Bio-Rad Trans-Blot apparatus, as described previously (17, 19). The membrane was immunoblotted with rabbit anti-PAP serum (dilution, 1:2,000) and horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma Chemical) as the primary and secondary antibodies, respectively.
L3 binding assays.
A plasmid containing the cDNA (pJD166.trp) that encodes wild-type Saccharomyces cerevisiae ribosomal protein L3 was a kind gift from Jonathan D. Dinman, University of Medicine and Dentistry of New Jersey, Newark, N.J. Radiolabeled L3 protein was synthesized by a linked transcription-translation system (TNT T3-coupled reticulocyte lysate system; Promega) (22) according to the instructions of the manufacturer (Promega). The translation products were resolved on an SDS-10% polyacrylamide gel, which was dried and autoradiographed. The mouse anti-L3 monoclonal antibody was a kind gift from Jonathan R. Warner, Department of Cell Biology, Albert Einstein College of Medicine, Bronx, N.Y. The in vitro-synthesized L3 protein (8 × 104 cpm) was incubated with 1 μg of wild-type or mutant PAPs in 50 μl (final volume) of binding buffer (10 mM K2HPO4, 5 mM NaCl [pH 8.0]) at 30°C for 30 min. The PAP-L3 complex was coimmunoprecipitated by adding 5 μl of mouse anti-L3 monoclonal antibody (dilution, 1:500) (22). After 60 min of incubation at 30°C, the PAP-L3-antibody complex was precipitated by adding 50 μl of protein A-Sepharose beads that had been pretreated with rabbit anti-mouse IgG (20 μl/ml of beads), and the incubation was continued for another 1 h at 4°C. The beads were washed three times with PBS containing 0.1% Triton X-100, and the proteins were eluted from the Sepharose beads with SDS sample buffer. The proteins were separated through an SDS-12% polyacrylamide gel, transferred to a PVDF membrane, and probed with the polyclonal rabbit anti-PAP antibody (dilution, 1:2,000) and horseradish peroxidase-conjugated goat anti-rabbit IgG (1:1000 dilution) as the primary and secondary antibodies, respectively. The blot was developed with 3,3′-diaminobenzidine (Sigma) as the colorimetric indicator for peroxidase activity. The dried membrane was also exposed to autoradiography to estimate the amounts of L3 protein.
Antiviral susceptibility assays.
Phenotypic susceptibility studies of HIV-1 isolates and strains were performed by measuring the production of the p24 gag protein in peripheral blood mononuclear cells (PBMCs) from seronegative healthy volunteers in the presence of increasing concentrations of the anti-HIV agent (viz., recombinant PAPs) by using the quantitative Coulter HIV-1 p24 antigen enzyme immunoassay (EIA) and the HIV-1 p24 Antigen Kinetic Standard (Beckman Coulter), as described previously (3, 32). Informed consent was obtained from the blood donors according to U.S. Department of Health and Human Services guidelines by using consent forms approved by the Institutional Review Board of the Parker Hughes Institute. In brief, PBMCs were cultured for 72 h in RPMI 1640 medium (Gibco) supplemented with 20% (vol/vol) heat-inactivated fetal bovine serum, 5% human interleukin-2 (Zeptometrix), 2 mM l-glutamine, 25 mM HEPES, 2 g of NaHCO3 per liter, 100 U of penicillin-streptomycin (Gibco) per ml, 50 μg of gentamicin (Gibco) per ml, and 5 μg of phytohemagglutinin-P (Sigma) per ml for 24 to 72 h prior to exposure to HIV at a multiplicity of infection of 0.001 to 0.1 during a 1-h adsorption period at 37 ± 1°C in a humidified 5 to 7% CO2 atmosphere. Subsequently, the cells were cultured in 96-well microtiter plates (100 μl/well; 2 × 106 cells/ml in triplicate wells) in the presence of the recombinant PAPs at a final concentration of 0.001, 0.01, 0.1, 1.0, 10.0, or 100 μg/ml; and 25-μl aliquots of culture supernatants were removed from the wells on day 6 after infection for p24 EIA, as described previously (3, 32). Controls included uninfected and untreated cells (background control) and infected but untreated (virus control) cells. The p24 EIA uses a murine monoclonal antibody to the HIV core protein used to coat microwell strips, to which the antigen present in the test culture supernatants binds. The percent inhibition of viral replication was calculated by comparing the p24 values for the test substance-treated infected cells with the p24 values for the untreated infected cells (i.e., virus controls). The IC50s were determined by using the Statview statistics program (SAS Institute, Inc., Cary, N.C.). In parallel, the effects of various treatments on cell viability were also examined as described previously (10). In brief, noninfected PBMCs were treated with each compound for 5 days under identical experimental conditions. A microculture tetrazolium assay was performed to quantitate cellular proliferation (3, 32). The cytotoxic concentrations which inhibited cellular proliferation by 50% (CC50s) were determined by using the Statview statistics program (SAS Institute, Inc.).
Plaque formation assays.
Plaque formation assays were used to examine the activities of the compounds against viruses other than HIV. The other viruses included in the present study were ganciclovir-sensitive cytomegalovirus (CMV) strain AD169 (ATCC VR-538), an enterovirus (ECHO virus serotype 30 strain Bastianni [ATCC VR-322]), and respiratory syncytial virus (RSV) strain Long (ATCC VR-26). The SF skin fibroblast cell line (ATCC CRL-2097) was used as a target for CMV strain AD169 and ECHO virus 30, and the HEP-2 cell line (ATCC CCL-23) was used as a target for RSV strain Long. These cell lines were cultured at 105 cells/well in 24-well (for all viruses except ECHO virus 30-infected SF cells) or 6-well (for ECHO virus 30-infected SF cells) tissue culture plates with 0.9% methylcellulose or 0.4% SeaPlaque agarose semisolid support. Minimum essential medium with Earle's salts (Gibco), l-glutamine, nonessential amino acids, 2% heat-inactivated fetal bovine serum, 1% penicillin-streptomycin, and 0.05% gentamicin served as the culture medium. The incubation times were 3 days for ECHO virus 30 and RSV Long and 7 days for CMV AD169. Plaque counting was performed with a ×20 dissecting microscope. The fixative agent was crystal violet for all viruses except CMV strain AD169, for which methylene blue was used. Percent inhibition of plaque formation was calculated by comparing the numbers of plaques from the test substance-treated infected cells with the numbers of plaques from untreated infected cells (i.e., virus controls). The IC50s were determined by using the Statview statistics program (SAS Institute, Inc.).
BALB/c mice.
All BALB/c mice used in this study were obtained from the specific-pathogen-free (SPF) breeding facilities of Taconic Laboratories (Germantown, N.Y.) at 5 weeks of age. All husbandry and experimental contacts made with the female BALB/c mice were performed in a controlled environment (12-h light and 12-h dark photoperiod; temperature, 22 ± 1°C; relative humidity, 60% ± 10%) which is fully accredited by the U.S. Department of Agriculture. All mice were housed in MicroIsolator cages (Lab Products, Inc., Maywood, N.Y.) containing autoclaved bedding. The mice were allowed free access to autoclaved pellet food and tap water throughout the experiments. Animal studies were approved by Parker Hughes Institute Animal Care and Use Committee, and all animal care procedures conformed to the Guide for the Care and Use of Laboratory Animals (12a).
Toxicity studies with mice.
The toxicity profiles of the PAPs in BALB/c mice were examined by methods that have previously been reported for other experimental agents (31-33). Female BALB/c mice were injected intravenously with bolus doses of native or recombinant PAPs in HEPES buffer at doses of 2.7, 4.1, 5.5, 6.8, and 8.2 mg/kg of body weight and were monitored daily for lethargy, cleanliness, and morbidity. Control mice were treated with PAP-free HEPES buffer. At the time of death, necropsies were performed and the toxic effects of the PAPs were assessed. For histopathological studies, tissues were fixed in 10% neutral buffered formalin, dehydrated, and embedded in paraffin by routine methods. Glass slides with affixed 6-μm tissue sections were prepared and stained with hematoxylin-eosin. No sedation or anesthesia was used throughout the experiments. The mice were monitored daily for mortality for determination of the day 30 50% lethal doses. The animals were electively killed on day 30 to determine the toxicity of PAP proteins by examining their blood chemistry profiles and blood counts as well as evaluating multiple organs for the presence of toxic lesions. Blood was collected by intracardiac puncture following anesthesia with ketamine-xylazine and was immediately heparinized. The blood chemistry profiles were examined with a Synchron CX5CE chemical analyzer (Beckman Instruments, Inc., Fullerton, Calif.). Blood counts (red blood cell, white blood cell, and platelet counts) were determined with a HESKA Vet ABC-Diff hematology analyzer (HESKA Corporation, Fort Collins, Colo.). Absolute neutrophil counts and absolute lymphocyte counts were calculated from white blood cell counts after the percentages of neutrophils and lymphocytes were determined by manual differential counting.
At the time of necropsy, 21 different tissues from mice (bone, bone marrow, brain, cecum, heart, kidney, large intestine, liver, lung, lymph node, ovary, pancreas, skeletal muscle, skin, small intestine, spleen, stomach, thymus, thyroid gland, urinary bladder, and uterus, as available) were collected within 15 min after killing of the mice, gross pathological findings were documented, and the organs were preserved in 10% neutral phosphate-buffered formalin and processed for histopathological examination. For histopathological studies, formalin-fixed tissues were dehydrated and embedded in paraffin by routine methods. Tissue sections of 4 to 5 μm were affixed to glass slides and stained with hematoxylin-eosin.
SCID mouse model of human AIDS.
All CB-17 SCID mice used in the present study were purchased from Taconic Laboratories at 6 to 8 weeks of age and maintained in the biosafety level 3 containment facility for preclinical research of the Parker Hughes Institute. Specific-pathogen-free conditions were maintained for all husbandry and experimental contacts made with the mice. The mice were housed in MicroIsolator cages containing autoclaved food, water, and bedding. Trimethoprim-sulfamethoxazole (Bactrim) was added to the drinking water of the SCID mice three times a week. Hu-PBL SCID mice were generated by reconstituting SCID mice by intraperitoneal injection of 10 × 106 PBMCs from seronegative volunteer donors, as reported previously (32, 33). Two weeks after inoculation of the cells, the mice were anesthetized with isoflurane and then challenged by intraperitoneal injection of 105 median tissue culture infective doses (TCID50s) of cell-free BR/92/019, a genotypically and phenotypically NRTI-resistant HIV-1 isolate. The SCID mice were infected with BR/92/019 in the biosafety level 3 containment facility, and all manipulations were performed in a biosafety cabinet. PAPs were administered by intraperitoneal injection. ZDV and lamivudine (3TC) were administered simultaneously at 8 and 4 mg, respectively, via gavage twice daily for 5 days/week for 2 weeks. Throughout the experimental period, the mice were monitored daily for overall health and survival. Two weeks after infection, the Hu-PBL SCID mice were electively killed, and their peritoneal lavage cells as well as spleen cells were examined for evidence of infection by an HIV-1 coculture assay (3) and determination of the viral RNA load (log10 number of RNA copies per milligram of spleen tissue or per milliliter of peritoneal lavage fluid) by using an Organon Teknika Nuclisens HIV-1 QT assay kit (bioMerieux, Durham, N.C.). Extraction of RNA was done with silica (50 μl) by using the standard Boom technology and the NucliSens extractor kit (bioMerieux). A standard amplification (nucleic acid sequence-based amplification [NASBA]) and detection assay was performed according to the recommendations of the manufacturer. Detection was based on the use of electrochemiluminescent labels that emit light due to chemical reactions occurring on the surface of an electrode. For histopathological studies, the tissues were fixed in 10% neutral buffered formalin, dehydrated, and embedded in paraffin by routine methods. Tissue sections of 4 to 5 μm were affixed to glass slides, stained with hematoxylin-eosin, and submitted to a veterinary pathologist for examination.
Differences in the proportional response rates with drug treatment were analyzed by a chi-square test of independence (contingency analysis) (33). The data were organized by using a two-way classification table in which data for all drug treatments including the control and the response (number of mice positive or negative) were included. The null hypothesis tested was that there was no difference between the observed and expected distributions in the classification table. The likelihood chi-square value was computed to test the null hypothesis (P < 0.05 was deemed significant). All statistical calculations were performed by using JMP software (SAS Institute, Inc.).
RESULTS
Structure-based design and engineering of nontoxic PAPs with potent anti-HIV activities.
Our molecular modeling studies indicated that rRNA and HIV-1 RNA adopt distinctly different binding modes in their interactions with PAP. In a systematic search for specific mutations that might result in selective enhancement of the anti-HIV activity of PAP, we noticed that the I152 residue on the opposite side of the active site is buried (Fig. 1). Residues K151 and I152 are located on helix α4, which is followed by a loop and helix α5. The side chain of I152 is mostly buried in hydrophobic contact (∼3 to 4 Å) with those of F158 (from the loop), T162 and F166 (helix α5), and Y76 from the middle β strand. In contrast, the side chain of K151 is mostly exposed and may form a hydrogen bond with N148 from the same helix. Residue 152 is approximately 20 Å from catalytic residue R179 and is situated on the opposite side of the PAP active site. It is known that alanine substitution of hydrophobic residues on an α helix would generally not disrupt a helical conformation (1). In many cases, a mutation from a large to a small molecule such as I152A may, in fact, be structurally favored for a helical conformation (37). However, in light of multiple van der Waals contacts between I152 and surrounding residues, the I152A mutation could potentially create a cavity in an otherwise tightly packed hydrophobic region. The bulky isoleucine side chain within the core of the protein confers greater hydrophobic stabilization than is the case for the smaller alanine side chain. The Ile-to-Ala substitution, which can create a large cavity, would be especially destabilizing because it would result in a loss of both hydrophobic and van der Waals interactions. Consequently, the surrounding residues would relax and reduce the volume of the putative cavity. Such mutations may cause local conformational instability and lead to a significant conformational change (12). The side chain of the residue could rotate into a radically different orientation and permit repacking of the core, as exemplified in the S117 mutation observed in T4 lysozyme. Such repacking is associated with adjustments of both the main chain and side chains. The backbone of a protein that contains mutated residues could deviate from the natural conformation. Such conformational changes could reach and affect the dynamic behavior of the catalytic residues that are farther than 25 Å away, as previously shown for the G93A mutation in superoxide dismutase (11, 35). We and other groups have long recognized the dramatic effect that this kind of mutation may have on enzyme activity (not substrate binding) and have been actively pursuing the concept in our protein engineering.
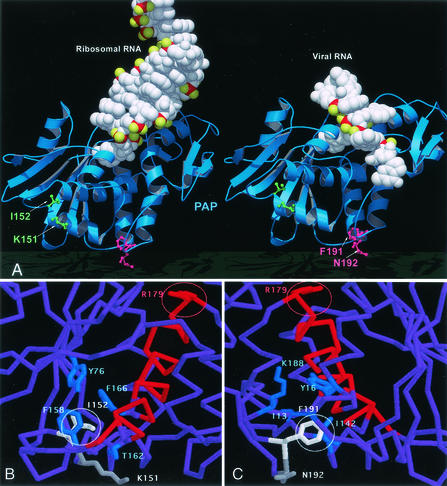
FIG. 1.
Molecular model of PAP-RNA interactions. (A) Ribbon and space-filling representation of PAP (blue) complex models with rRNA (left, in white) and viral RNA (right, in white) molecules. Residues I152 and K151 (green), as shown in the stick model, are mutated in FLP-102. Residues F191 and N192 (red), as shown in the stick model, are mutated in FLP-105. These residues are located on the opposite side of the PAP active site, and the mutation of these residues is not predicted to affect the binding of rRNA or HIV-1 viral RNA substrates. However, residue I152 is buried and the mutation from a large to a small molecule (Ile to Ala) would presumably create a cavity and would permit repacking of the core; it thus can lead to considerable conformational changes in catalytic residues and differentially affect the activity of PAP against the ribosome substrate and the viral RNA. (B) Residue 152 (white) and the surrounding residues (blue) that have van der Waals contacts with I152 are shown, along with helix α5 (red) and R179 (red, in the active site) on the other end of the helix. (C) Residue 191 (white) and its surrounding residues (blue) are shown. The view is almost the opposite from the one in panel B. In contrast, K151 and N192 are mostly exposed.
We have previously modeled the complex structure of PAP with ribosomes on the basis of the crystal structures of PAP and the ribosome and the complex structure of PAP with a single-stranded, flexible RNA substrate that could be used as a structural template for HIV RNA (19). The proposed structural model is in good agreement with our mutation studies of residues involved in the active site and the extended substrate-binding site (19). On the basis of our modeling analyses, we have recognized that the binding mode of the ribosome is distinctly different from that of viral RNA, as shown in Fig. 1. Our modeling studies indicated that a substitution of this isoleucine with a smaller residue such as alanine (I152A) would likely create a cavity, causing a repacking of the core and considerable conformational changes of the active site that would be selectively unfavorable for PAP-mediated depurination of rRNA but that possibly would not affect the catalytic depurination of HIV-1 RNA.
A double mutation of both the I152 and the K151 residues was predicted to result in a more substantial loss of ribosome-inhibitory activity than single mutations involving only one of these two residues. Residues F191 and N192 are located in a more flexible environment on the C-terminal end of helix 6 approximately 20 Å away from catalytic residue R179 (Fig. 1). Similar to the binding environment of I152, the side chain of F191 is buried and is in hydrophobic contact with the side chains of I142, I13, and Y16 and the hydrophobic portion of K188, all of which are situated on nearby helices. The side chain of N192 is mostly exposed and shows no significant contact with nearby residues. Substitutions of these residues with alanine or glycine were postulated to cause conformational changes in the active site, leading to a moderately reduced activity against rRNA (but not HIV-1 RNA) substrates, albeit to a much lesser degree than the I152A mutation. However, the K151, I152, F191, and N192 mutations would not cause an impairment in the abilities of the respective PAPs to bind to ribosomes or ribosomal protein L3, which interacts with the partially exposed half of the active-site cleft (Fig. 1).
The recombinant PAP mutants with alanine substitutions of I152, K151, and F191 and glycine substitution of N192 were constructed by site-directed mutagenesis, as described previously (19). Mutant proteins FLP-100 (K151A), FLP-101(I152A), FLP-102 (K151A, I152A), FLP-103 (F191A), FLP-104 (N192G), and FLP-105 (F191A, N192G) were expressed as inclusion bodies in E. coli strain MV1190, purified, solubilized, refolded, and analyzed by SDS-PAGE. Each of the mutant PAPs had an apparent molecular mass of 33 kDa, similar to that of the recombinant wild-type PAP. The refolded recombinant wild-type and mutant proteins were highly immunoreactive with the anti-PAP serum.
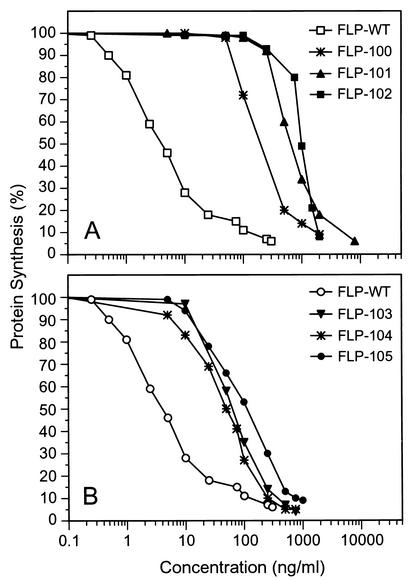
The IC50 of recombinant wild-type PAP in cell-free translation inhibition assays was 3 ng/ml (Fig. 2; Table 1). By comparison, the IC50s for double-substitution mutants FLP-102 and FLP-105 were 994 and 105 ng/ml, respectively. Thus, FLP-102 was 331-fold less toxic than wild-type PAP and FLP-105 was 35-fold less toxic than wild-type PAP (Fig. 3; Table 1). The corresponding single-residue mutants, FLP-100 (K151A), FLP-101 (I152A), FLP-103 (F191A), and FLP-104 (N192G), were also less toxic than the wild-type PAP (Fig. 2; Table 1). As shown in Table 1, most of the 331-fold loss of activity for FLP-102 could be attributed to the I152A mutation, since single-residue-substitution mutant FLP-101(I152A) was 206-fold less toxic than wild-type PAP but single-residue-substitution mutant FLP-100 (K151A) was only 49-fold less toxic than wild-type PAP.
FIG. 2.
Ribosome-inhibitory activity of PAP wild-type and mutants in in vitro rabbit reticulocyte lysate system. Each value is the average of three independent experiments. Protein synthesis was determined by measurement of [35S]methionine incorporation, and the level of incorporation for samples treated with all reagents except PAP was assigned a value of 100%. (A) FLP-102 and single-residue mutants FLP-100 and FLP-101. (B) FLP-105 and single-residue mutants FLP-103 and FLP-104.
TABLE 1.
Sequence identities and biological activities of wild-type and mutant PAPs
| Mutant | Original residue | Substituted residue | IC50 (ng/ml) for translation inhibitiona | IC50 (ng/ml) of FLP-WT |
|---|---|---|---|---|
| FLP-WT | 3.0 (2.5-3.8) | 1 | ||
| FLP-100 | K151 | A151 | 147 (130-167) | 49 |
| FLP-101 | I152 | A152 | 617 (500-710) | 206 |
| FLP-102 | 151KI152 | 151AA152 | 994 (878-1,121) | 331 |
| FLP-103 | F191 | A191 | 57 (52-60) | 19 |
| FLP-104 | N192 | G192 | 54 (44-66) | 18 |
| FLP-105 | 191FN192 | 191AG192 | 105 (94-131) | 35 |
Each value is the mean of at least three experiments. The values in parentheses are 95% confidence limits.
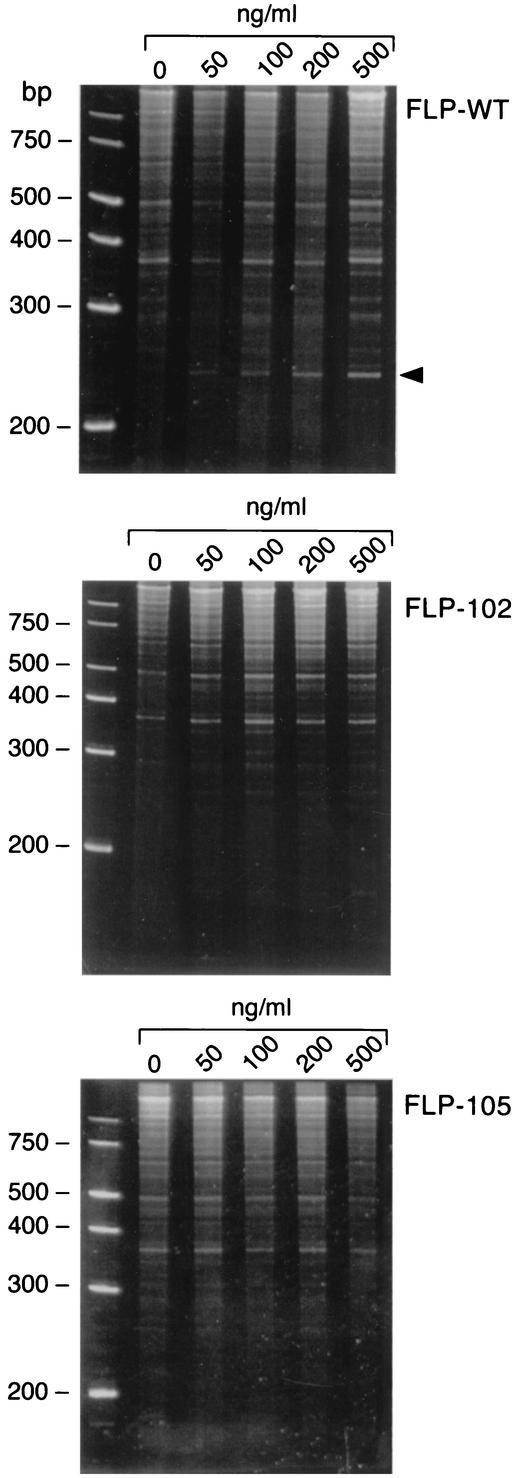
FIG. 3.
In vitro depurination of rRNA by wild-type and mutant recombinant PAPs. E. coli 23S and 16S rRNAs were treated with increasing amounts of PAP, treated with aniline, separated on a 6% urea-polyacrylamide gel, and stained with ethidium bromide. The arrowhead indicates the fragment split by aniline.
Compared with the K151A and I152A mutations in FLP-102, the F191A and N192G mutations in FLP-105 had lesser effects on the activities of PAPs against the ribosomal and viral RNA substrates. Both the N192G and F191A mutations resulted in moderate reductions in ribosomal deactivation activity and slightly improved activity against the HIV RNA substrate. This is consistent with the observation that N192 and F191 are located in an environment more flexible than that in which I152 is located. In light of the moderate changes in PAP activity that occurred as a result of these two mutations, the level of conformational change caused by these two mutations is probably less profound than that caused by I152A. Nevertheless, the F191A mutation probably adopts the same mechanism proposed for the I152A mutation (leading to reduced activity).
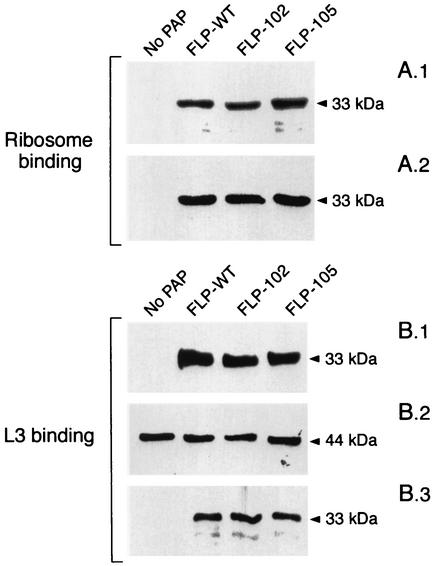
The abilities of the FLP-102 and FLP-105 proteins to depurinate rRNA in rabbit ribosomes were also evaluated by treatment of the ribosomes with the PAPs, subsequent purification of rRNA, and cleavage with aniline. Aniline cleaves the sugar-phosphate backbone of rRNA at PAP depurination sites. Therefore, the release of fragments from aniline-treated rRNA is an indicator of PAP-mediated rRNA depurination. Aniline treatment resulted in the release of a 600-nucleotide (nt) RNA fragment (as indicated by the arrowhead between the 200- and 300-bp markers in Fig. 3) from the rRNA of rabbit ribosomes pretreated with the recombinant wild-type PAP. In contrast, aniline treatment failed to cause the release of detectable amounts of the 600-nt RNA fragment from rRNA isolated from ribosomes treated with mutant PAPs FLP-102 and FLP-105. These findings are consistent with the markedly reduced ribosome-inhibitory activities of these recombinant PAPs in in vitro translation assays (Fig. 2). In accord with the predictions of our modeling studies, the reduced ribosome-inhibitory activities of FLP-102 and FLP-105 were not due to reduced binding to the ribosome, ribosomal protein L3, which serves as a docking site for PAP, or the SR loop (Fig. 4). By comparison, active center cleft mutants FLP-4 (69AA70) and FLP-7 (90AAA92), which were used as controls, exhibited markedly reduced levels of binding to the ribosomes as well as the L3 protein (22), consistent with the notion that the active center cleft of PAP plays an important role in the PAP-ribosome and PAP-L3 interactions.
FIG. 4.
(A) Association of PAP mutants with ribosomes isolated from rabbit reticulocyte-enriched blood. (A.1) Total ribosomal protein (5 μg) was incubated with 1 μg of PAP and the ribosome-PAP complexes were isolated by ultracentrifugation. The ribosome-PAP complexes were separated through an SDS-12% polyacrylamide gel, electroblotted onto a PVDF membrane, and immunoblotted with a polyclonal antibody to PAP. (A.2) Prior to separation of the PAP-ribosome complex, a fraction (5 μl) of the reaction mixture was removed, separated through an SDS-12% polyacrylamide gel, transferred to a PVDF membrane, and immunoblotted with a polyclonal antibody to PAP. The results show that equal amounts of PAPs were added to the reaction mixture. (B) Association of wild-type and mutant PAPs with in vitro-synthesized ribosomal protein L3. (B.1) Coimmunoprecipitated PAP revealed by immunoblotting with anti-PAP antibody. 35S-labeled L3 was incubated with wild-type and mutant PAPs and coimmunoprecipitated with protein A-Sepharose beads precoated with monoclonal antibody to L3. The PAP-L3 complexes were separated through SDS-12% polyacrylamide gels, transferred to a PVDF membrane, and immunoblotted with a polyclonal anti-PAP antibody. (B.2) The blot was exposed to X-ray film, which shows equal amounts of labeled L3 protein in each reaction mixture. (B.3) Prior to coimmunoprecipitation, a fraction (5 μl) was removed from the reaction mixture, separated through an SDS-12% polyacrylamide gel, transferred to a PVDF membrane, and immunoblotted with a polyclonal antibody to PAP. The results show that equal amounts of PAP were added to each reaction mixture.
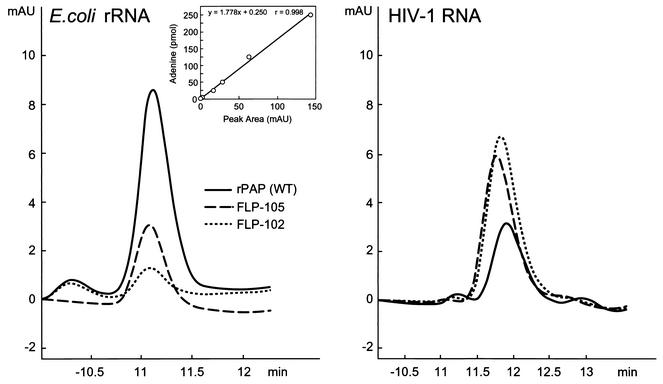
We next compared the abilities of mutant PAPs FLP-102 and FLP-105 to depurinate rRNA versus their abilities to depurinate HIV-1 RNA to the depurination abilities of recombinant wild-type PAP using HPLC-based quantitative adenine release assays. While recombinant wild-type PAP depurinated rRNA more efficiently than it did HIV-1 RNA, both FLP-102 and FLP-105 were more efficient in depurinating HIV-1 RNA, and their activities against HIV-1 RNA were superior to that of wild-type PAP (Table 2). FLP-102 exhibited the most promising selective anti-HIV activity: whereas the release of adenine from E. coli rRNA caused by treatment with wild-type PAP was 76 ± 4 pmol/μg of RNA/μmol of protein, FLP-102 protein was 10-fold less active and resulted in the release of only 7 ± 2 pmol of adenine/μg of RNA/μmol of protein (P < 0.05) (Table 2; Fig. 5). However, FLP-102 was more potent in deadenylating HIV-1 RNA than wild-type PAP was: whereas wild-type PAP released 36 ± 3 pmol of adenine/μg of RNA/μmol of protein, FLP-102 released 100 ± 7 pmol of adenine/μg of RNA/μmol of protein (P < 0.05) (Table 2; Fig. 5). Similar results were obtained in guanine release assays (Table 2).
TABLE 2.
RNA depurinating activities and in vivo anti-HIV activities of wild-type and mutant PAPs
| Mutant and RNA source | Purine release (pmol/μg of RNA/μmol of PAP)
|
IC50 (μg/ml) for inhibition of HIV-1 HTLV IIIB replication | Cytotoxicity (CC50 [μg/ml]) | |
|---|---|---|---|---|
| Adenine | Guanine | |||
| FLP-WT | ||||
| E. coli | 76 ± 4 | 71 ± 3 | 2.9 ± 1.3 | 3.1 ± 1.9 |
| HIV-1 | 36 ± 3 | 48 ± 3 | ||
| FLP-102 | ||||
| E. coli | 7 ± 2 | 7 ± 2 | 0.2 ± 0.0 | >10 |
| HIV-1 | 100 ± 7 | 207 ± 11 | ||
| FLP-105 | ||||
| E. coli | 17 ± 3 | 10 ± 4 | 0.7 ± 0.2 | >10 |
| HIV-1 | 86 ± 7 | 180 ± 10 | ||
FIG. 5.
Depurination of E. coli and HIV-1 RNA by wild-type and mutant PAPs. Two micrograms of E. coli rRNA and HIV-1 RNA was incubated with 2.5 μM wild-type and mutant PAPs for 1 h at 37°C in 50 μl of binding buffer. The reaction was stopped by adding 100 μl of HPLC running buffer, and 100 μl of the sample was injected into the column as described in Materials and Methods. Control samples of RNA were treated with PBS instead of PAP. (Inset) Standard curve of adenine and guanine standards.
We next examined in six independent experiments, each of which was performed in triplicate, the abilities of FLP-102 and FLP-105 to inhibit the replication of HIV-1 strain HTLV IIIB in human PBMCs (Table 2). Both proteins inhibited HIV-1 replication in a concentration-dependent fashion, with IC50s of 0.2 ± 0.0 μg/ml for FLP-102 and 0.7 ± 0.2 μg/ml for FLP-105. By comparison, the wild-type protein FLP-WT was less active than either protein and inhibited HIV-1 replication with an IC50 of 2.9 ± 1.3 μg/ml (Table 2) (P < 0.001). The CC50s were 3.1 ± 1.9 μg/ml for FLP-WT and >10 μg/ml for both FLP-102 and FLP-105.
We also examined the antiviral activities of wild-type and mutant PAPs against viruses other than HIV. FLP-WT inhibited the plaque formation caused by CMV strain AD169, the enterovirus ECHO virus 30, and RSV strain Long, with IC50s of 0.8 ± 0.2, 7.2 ± 1.5, and 0.7 ± 0.4 μg/ml, respectively. By comparison, the IC50s of both FLP-102 and FLP-105 against these viruses were >10 μg/ml. Thus, the conformational changes associated with the mutations of these recombinant PAPs result in selective enhancement of their anti-HIV activities and impair their ribosome-inhibitory activities as well as their broad-spectrum antiviral activities.
In vivo toxicity profiles and anti-HIV activities of FLP-102 and FLP-105.
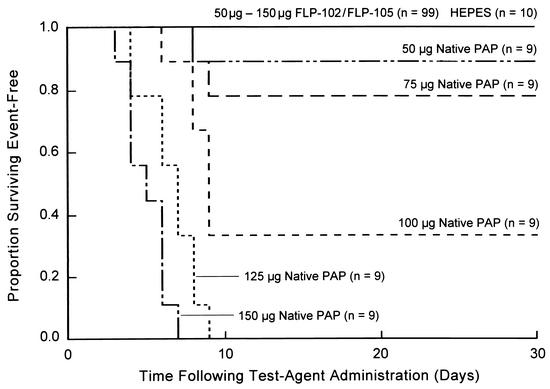
BALB/c mice were treated with 50 μg (n = 10), 75 μg (n = 10), 100 μg (n = 10), 125 μg (n = 10), or 150 μg (n = 10) of FLP-102 or FLP-105. FLP-102 and FLP-105 were nontoxic to BALB/c mice even at a dose level of 150 μg/mouse (∼8.2 mg/kg; average weight, 18.2 g), whereas wild-type native PAP exhibited toxicity at a dose level of 50 μg/mouse and was invariably fatal at a dose level of ≥125 μg/mouse (Fig. 6). Control mice (n = 10) were treated with intraperitoneal injections of the PAP-free vehicle solution. All 49 FLP-102-treated mice and all 50 FLP-105-treated mice remained healthy throughout the 30-day observation period, with no evidence of morbidity. Blood tests done on day 30 did not suggest any significant systemic toxicity. In particular, even at the highest cumulative dose level of 150 μg/mouse, neither FLP-102 nor FLP-105 caused (i) anemia, neutropenia, or lymphopenia suggestive of hematologic toxicity; (ii) elevations of blood urea nitrogen and creatinine levels or electrolyte disturbances suggestive of renal toxicity; (iii) elevations of aspartate transaminase, alanine transaminase, alkaline phosphatase, lactate dehydrogenase, or bilirubin levels suggestive of hepatotoxicity; or (iv) elevation of amylase levels suggestive of pancreas toxicity. No toxic lesions were found in any of the 21 organs from the 25 FLP-102-treated mice (5 mice/dose group) or 25 FLP-105-treated mice (5 mice/dose group) killed on day 30. Similarly, no toxic lesions were found in the organs of 10 vehicle-treated control mice. In contrast, myocardial necrosis, necrotizing hepatitis, necrotic myositis, and acute tubular necrosis were found when organs from native PAP-treated (100 to 150 μg/mouse) control mice were examined. Similar experiments were performed to look at the toxicities of PAPs at an earlier time following administration. No laboratory findings suggestive of any systemic toxicity were found on day 3 in any of the mice treated with dose levels of 50 μg (n = 3), 75 μg (n = 3), 100 μg (n = 3), 125 μg (n = 3), or 150 μg (n = 3) of FLP-102 or FLP-105. Taken together, these experiments demonstrated that FLP-102 and FLP-105 are nontoxic to BALB/c mice at dose levels as high as 8.2 mg/kg.
FIG. 6.
Toxicities of FLP-102 and FLP-105 for mice. BALB/c mice were treated with single intraperitoneal bolus injections of mutant recombinant PAP FLP-102 or FLP-105 or native PAP at the indicated dose levels. Survival curves for mice treated with the specified PAPs at specific dose levels are depicted. None of the 99 mice treated with FLP-102 or FLP-105 at dose levels ranging from 50 to 150 μg/mouse became sick or died within the 30-day observation period.
We next sought to determine if FLP-102 and FLP-105 exhibit any in vivo anti-HIV activities in the Hu-PBL SCID mouse model of human AIDS. Control mice were treated either with vehicle alone (negative control treatment) or with ZDV plus 3TC (positive control treatment). FLP-102 and FLP-105 were used at daily dose levels of 20 μg/mouse (2 mg/kg; five mice) or 40 μg/mouse (4 mg/kg; five mice) for 5 days/week for 2 weeks (cumulative dose levels, 200 and 400 μg/mouse, respectively) and were administered by intraperitoneal bolus injections. ZDV and 3TC were used at dose levels of 8 and 4 mg, respectively, and were administered via gavage twice daily for 5 days/week for 2 weeks. All mice were reconstituted with 10 × 106 PBMCs and infected with 1 × 105 TCID50s of the genotypically and phenotypically NRTI-resistant HIV-1 BR/92/019 isolate 2 weeks after reconstitution. Treatments were started immediately after inoculation of the HIV-1 isolate. Spleen specimens from 9 of 10 (90%) vehicle-treated control mice were HIV-1 positive by NASBA and had an HIV RNA burden of 3.9 ± 0.3 logs, whereas spleen specimens from only 3 of 10 (30%) ZDV-3TC-treated mice were HIV-1 positive by NASBA and had an HIV RNA burden of 3.2 ± 0.3 logs (Table 3). The wild-type recombinant PAP FLP-WT was not active in this model system, and spleen specimens from 7 of 10 (70%) FLP-WT-treated mice (average HIV RNA burden, 4.1 ± 0.5 logs) were HIV-1 positive by NASBA. By comparison, spleens from only 2 of 10 (20%) FLP-102-treated mice (average HIV RNA burden, 2.9 logs; chi-square = 11.0; P = 0.0009) and 3 of 10 (30%) FLP-105-treated mice (average HIV RNA burden, 3.2 ± 0.3 logs) were HIV-1 NASBA positive (chi square = 8.2; P = 0.004). Thus, both FLP-102 and FLP-105 were at least as effective as the ZDV-3TC combination in preventing in vivo HIV-1 replication in the spleens of Hu-PBL SCID mice. Peritoneal lavage specimens from 7 of 8 vehicle-treated control mice, 6 of 10 ZDV-3TC-treated mice, and 10 of 10 FLP-WT treated mice were HIV-1 positive by NASBA. In contrast, none of the peritoneal lavage specimens from nine FLP-102 treated mice (chi square = 17.0; P = 0.0001) and the peritoneal lavage specimens from only three of the nine FLP-105-treated mice showed evidence of HIV-1 infection by NASBA (chi-square = 5.6; P = 0.02) (Table 3). The in vivo anti-HIV activities of the mutant PAPs were further confirmed by culture of spleen specimens for HIV-1. Whereas 9 of 10 spleen specimens and 9 of 10 spleen and peritoneal lavage (mixed) specimens from vehicle-treated control mice were HIV-1 culture positive, only 2 of 10 spleen specimens and 3 of 10 spleen and peritoneal lavage specimens from FLP-102-treated mice were culture positive for HIV-1. By comparison, 3 of 10 spleen specimens and 5 of 10 spleen and peritoneal lavage specimens from FLP-105-treated mice were culture positive for HIV-1. Thus, both PAP mutants were at least as effective as the ZDV-3TC combination, and FLP-102 was the most active PAP in preventing HIV-1 replication in Hu-PBL SCID mice.
TABLE 3.
Anti-HIV activities of FLP-102 and FLP-105 against BR/92/019 in Hu-PBL SCID mice
| Treatmenta | No. of mice HIV positive by NASBA
|
Log10 viral load in NASBA- positive tissues
|
HIV culture positive
|
||||
|---|---|---|---|---|---|---|---|
| Spleenb | PLc | Spleen | PL | Spleen | PL | Spleen + PL | |
| Control (vehicle) | 9/10 | 7/8 | 3.9 ± 0.3 | 4.3 ± 0.2 | 9/10 | NDd | 9/10 |
| ZDV-3TC | 3/10 | 6/10 | 3.2 ± 0.3 | 4.0 ± 0.4 | ND | ND | ND |
| FLP-WT | 7/10 | 10/10 | 4.1 ± 0.5 | 4.8 ± 0.2 | ND | ND | ND |
| FLP-102 | 2/10 | 0/9 | ∼2.9 (1.6, 4.3) | NAe | 2/10 | ND | 3/10 |
| FLP-105 | 3/10 | 3/9 | 3.2 ± 0.3 | 4.2 ± 0.6 | ND | ND | 5/10 |
FLP-102 and FLP-105 were used at a daily dose level of 20 μg/mouse for 5 days/week for 2 weeks and were administered by intraperitoneal bolus injections. ZDV and 3TC were used at dose levels of 8 and 4 mg, respectively, and were administered via gavage twice daily for 5 days/week for 2 weeks. All mice were reconstituted with 10 × 106 PBMCs and infected with 1 × 105 TCID50s of the HIV-1 BR/92/019 isolate 2 weeks after reconstitution. The treatments were initiated immediately after inoculation of the HIV-1 isolate.
Chi-square values for spleen HIV positivity by NASBA were as follows: ZDV-3TC versus the control, 8.2 (P = 0.004); FLP-102 versus the control, 11.0 (P = 0.0009); FLP-105 versus the control, 8.2 (P = 0.004); FLP-102 versus FLP-WT, 5.3 (P = 0.021); FLP-105 versus FLP-WT, 3.3 (P = 0.07).
Chi-square values for peritoneal lavage specimen (PL) HIV positivity by NASBA were as follows: ZDV-3TC versus the control, 1.8 (P = 0.182); FLP-102 versus the control, 17.0 (P = 0.0001); FLP-105 versus the control 5.6 (P = 0.02); FLP-102 versus FLP-WT, 26.3 (P = 0.0001); FLP-105 versus FLP-WT, 12.2 (P = 0.0001).
ND, not determined.
NA, not applicable.
DISCUSSION
Combination antiretroviral therapy has become the standard of care for patients with HIV infection in the United States (4, 5,23, 24, 27, 28, 29). Antiretroviral treatment regimens that use combinations of drugs from at least two of the three classes of antiretroviral therapy, namely, NRTIs, nonnucleoside analog reverse transcriptase inhibitors, and protease inhibitors, exhibit potent and sustained antiviral effects and confer consistent long-term viral suppression in patients with HIV infection (4, 5, 23, 24, 27, 28, 29). However, the individual components of these combination regimens can select for drug-resistant viruses, and the emergence of antiviral drug resistance limits their clinical benefit (2, 7, 10, 13-15, 25, 30, 34). This resistance is a consequence of the high mutation rate and fast replication of HIV and the selective effects of these drugs, which favor the emergence of mutations that can establish clinical drug resistance. Recent results indicate that the failure of highly active antiretroviral therapy, which typically includes at least two NRTIs and a protease inhibitor or a nonnucleoside analog reverse transcriptase inhibitor, results from the multiplicity of mutations that confer genotypic resistance to almost all available antiretroviral drugs (2, 7, 10, 13-15, 25, 30, 34). In these patients, genotypic resistance tests confirm the lack of alternative salvage therapy strategies based on the presently available antiretroviral drugs (7, 10, 13-15, 23, 25, 30, 34). Patients who fail highly active antiretroviral therapy constitute a reservoir of multidrug-resistant HIV that may limit treatment options in the future. The frequency of genotypic and phenotypic drug-resistant HIV is increasing among therapy-naive HIV-infected seroconverters (2, 7, 10, 13-15, 25, 30, 34). Thus, the transmission of drug-resistant HIV is a serious problem that merits further attention by public health officials as well as virologists and clinicians. Therefore, there is an urgent need for new anti-HIV agents capable of inhibiting the replication of drug-resistant HIV. Rationally engineered nontoxic recombinant PAPs such as FLP-102 and FLP-105 may provide the basis for effective salvage therapies for patients harboring highly drug-resistant strains of HIV-1. The documented in vitro potencies, in vivo antiretroviral activities in HIV-infected Hu-PBL SCID mice, and favorable toxicity profiles of those antiviral proteins warrant their further development for possible clinical use. The preliminary information regarding the relative safety of the mutant PAPs compared to that of wild-type PAP remains to be confirmed in detailed pharmacodynamic studies which will require the establishment of validated methods for measurement of the levels of the recombinant PAPs in serum.
Acknowledgments
This work was supported by the Defense Advanced Research Projects Agency under grant N65236-99-1-5422 (awarded to F.M.U.).
REFERENCES
- 1.Bell, J. A., W. J. Becktel, U. Sauer, W. A. Baase, and B. W. Matthews. 1992. Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59. Biochemistry 31:3590-3596. [DOI] [PubMed] [Google Scholar]
- 2.Briones, C., V. Soriano, and J. Gonzalez-Lahoz. 2000. Prevalence of drug-resistant HIV-1 genotypes in heavily pre-treated patients on current virological failure. AIDS 14:1659-1660. [DOI] [PubMed] [Google Scholar]
- 3.Erice, A., H. H. Balfour, Jr., D. E. Myers, V. L. Leske, K. J. Sannerud, V. Kuebelbeck, J. D. Irvin, and F. M. Uckun. 1993. Anti-human immunodeficiency virus type 1 activity of an anti-CD4 immunoconjugate containing pokeweed antiviral protein. Antimicrob. Agents Chemother. 37:835-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freedberg, K. A., E. Losina, M. C. Weinstein, A. D. Paltiel, C. J. Cohen, G. R. Seage, D. E. Craven, H. Zhang, A. D. Kimmel, and S. J. Goldie. 2001. The cost effectiveness of combination antiretroviral therapy for HIV disease. N. Engl. J. Med. 344:824-831. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb, M. S. 2001. AIDS—past and future. N. Engl. J. Med. 344:1788-1791. [DOI] [PubMed] [Google Scholar]
- 6.Irvin, J. D., and F. M. Uckun. 1992. Pokeweed antiviral protein: ribosome inactivation and therapeutic applications. Pharmacol. Ther. 55:279-302. [DOI] [PubMed] [Google Scholar]
- 7.Izopet, J., A. Bicart-See, C. Pasquier, K. Sandres, E. Bonnet, B. Marchou, J. Puel, and P. Massip. 1999. Mutations conferring resistance to zidovudine diminish the antiviral effect of stavudine plus didanosine. J. Med. Virol. 59:507-511. [PubMed] [Google Scholar]
- 8.Kurinov, I., F. Rajamohan, T. Venkatachalam, and F. M. Uckun. 1999. X-ray crystallographic analysis of the structural basis for the interaction of pokeweed antiviral protein with guanine residues of ribosomal RNA. Protein Sci. 8:2399-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurinov, I. V., D. E. Myers, J. D. Irvin, and F. M. Uckun. 1999. X-ray crystallographic analysis of the structural basis for the interactions of pokeweed antiviral protein with its active site inhibitor and ribosomal RNA substrate analogs. Protein Sci. 8:1765-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuritzkes, D. R., A. Sevin, B. Young, M. Bakhtiari, H. Wu, M. St. Clair, E. Connick, A. Landay, J. Spritzler, H. Kessler, M. M. Lederman, et al. 2000. Effect of zidovudine resistance mutations on virologic response to treatment with zidovudine-lamivudine-ritonavir: genotypic analysis of human immunodeficiency virus type 1 isolates from AIDS Clinical Trials Group Protocol 315. J. Infect. Dis. 181:491-497. [DOI] [PubMed] [Google Scholar]
- 11.Liu, R., R. K. Narla, I. Kurinov, B. Li, and F. M. Uckun. 1999. Increased hydroxyl radical production and apoptosis in PC12 neuron cells expressing the gain-of-function mutant G93A SOD1 gene. Radiation Res. 151:133-141. [PubMed] [Google Scholar]
- 12.Matthews, B. W. 1995. Studies on protein stability with T4 lysozyme. Adv. Protein Chem. 46:249-278. [DOI] [PubMed] [Google Scholar]
- 12a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, D.C.
- 13.O'Brien, W. A. 2000. Resistance against reverse transcriptase inhibitors. Clin. Infect. Dis. 30(Suppl. 2):S185-S192. [DOI] [PubMed] [Google Scholar]
- 14.Picard, V., E. Angelini, A. Maillard, E. Race, F. Clavel, G. Chene, F. Ferchal, and J. M. Molina. 2001. Comparison of genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 isolates from patients treated with stavudine and didanosine or zidovudine and lamivudine. J. Infect. Dis. 184:781-784. [DOI] [PubMed] [Google Scholar]
- 15.Pillay, D., S. Taylor, and D. D. Richman. 2000. Incidence and impact of resistance against approved antiretroviral drugs. Rev. Med. Virol. 10:231-253. [DOI] [PubMed] [Google Scholar]
- 16.Rajamohan, F., S. Doumbia, C. Engstrom, S. Pendergrass, D. Maher, and F. M. Uckun. 2000. Expression of biologically active recombinant pokeweed antiviral protein in methylotrophic yeast Pichia pastoris. Protein Expression Purif. 18:193-201. [DOI] [PubMed] [Google Scholar]
- 17.Rajamohan, F., C. R. Engstrom, T. J. Denton, L. A. Engen, I. Kourinov, and F. M. Uckun. 1999. High level expression and purification of biologically active recombinant pokeweed antiviral protein. Protein Expression Purif. 16:359-368. [DOI] [PubMed] [Google Scholar]
- 18.Rajamohan, F., I. Kurinov, T. Venkatachalam, and F. M. Uckun. 1999. Deguanylation of human immunodeficiency virus (HIV-1) RNA by recombinant pokeweed antiviral protein. Biochem. Biophys. Res. Commun. 263:419-424. [DOI] [PubMed] [Google Scholar]
- 19.Rajamohan, F., M. Pugmire, I. Kurinov, and F. M. Uckun. 2000. Modeling and alanine-scanning mutagenesis studies of recombinant pokeweed antiviral protein. J. Biol. Chem. 275:3382-3390. [DOI] [PubMed] [Google Scholar]
- 20.Rajamohan, F., T. K. Venkatachalam, J. D. Irvin, and F. M. Uckun. 1999. Pokeweed antiviral protein isoforms PAP-I, PAP-II, and PAP-III depurinate RNA of human immunodeficiency virus (HIV-1). Biochem. Biophys. Res. Commun. 260:453-458. [DOI] [PubMed] [Google Scholar]
- 21.Rajamohan, F., C. Mao, and F. M. Uckun. 2001. Binding interactions between the active center cleft of recombinant pokeweed antiviral protein and the α-sarcin/ricin stem loop of ribosomal RNA. J. Biol. Chem. 276:24075-24081. [DOI] [PubMed] [Google Scholar]
- 22.Rajamohan, F., Z. Ozer, C. Mao, and F. M. Uckun. 2001. Active center cleft residues of pokeweed antiviral protein mediate its high affinity binding to the ribosomal protein L3. Biochemistry 40:9104-9114. [DOI] [PubMed] [Google Scholar]
- 23.Rey, D., M. P. Schmitt, M. Partisani, G. Hess-Kempf, V. Krantz, E. de Mautort, C. Bernard-Henry, M. Priester, C. Cheneau, and J. M. Lang. 2001. Efavirenz as a substitute for protease inhibitors in HIV-1-infected patients with undetectable plasma viral load on HAART: a median follow-up of 64 weeks. J. Acquir. Immune Defic. Syndr. 27:459-462. [DOI] [PubMed] [Google Scholar]
- 24.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 25.Ross, L., A. Scarsella, S. Raffanti, K. Henry, S. Becker, R. Fisher, Q. Liao, A. Hirani, N. Graham, M. St. Clair, and J. Hernandez. 2001. Thymidine analog and multinucleoside resistance mutations are associated with decreased phenotypic susceptibility to stavudine in HIV type 1 isolated from zidovudine-naive patients experiencing viremia on stavudine-containing regimens. AIDS Res. Hum. Retrovir. 17:1107-1115. [DOI] [PubMed] [Google Scholar]
- 26.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sepkowitz, K. A. 2001. AIDS—the first 20 years. N. Engl. J. Med. 344:1764-1772. [DOI] [PubMed] [Google Scholar]
- 28.Shafer, R. W., and D. A. Vuitton. 1999. Highly active antiretroviral therapy (HAART) for the treatment of infection with human immunodeficiency virus type 1. Biomed. Pharmacother. 53:73-86. [DOI] [PubMed] [Google Scholar]
- 29.Starr, S. E., C. V. Fletcher, S. A. Spector, F. H. Yong, T. Fenton, R. C. Brundage, D. Manion, N. Ruiz, M. Gersten, M. Becker, J. McNamara, L. M. Mofenson, L. Purdue, S. Siminski, B. Graham, D. M. Kornhauser, W. Fiske, C. Vincent, H. W. Lischner, W. M. Dankner, P. M. Flynn, et al. 1999. Combination therapy with efavirenz, nelfinavir, and nucleoside reverse-transcriptase inhibitors in children infected with human immunodeficiency virus type 1. N. Engl. J. Med. 341:1874-1881. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki, K., G. R. Kaufmann, M. Mukaide, P. Cunningham, C. Harris, L. Leas, M. Kondo, M. Imai, S. L. Pett, R. Finlayson, J. Zaunders, A. Kelleher, and D. A. Cooper. 2001. Novel deletion of HIV type 1 reverse transcriptase residue 69 conferring selective high-level resistance to nevirapine. AIDS Res. Hum. Retrovir. 17:1293-1296. [DOI] [PubMed] [Google Scholar]
- 31.Uckun, F. M., R. Narla, T. Zeren, Y. Yanishevski, D. E. Myers, B. Waurzyniak, O. Ek, E. Schneider, Y. Messinger, L. M. Chelstrom, R. Gunther, and W. Evans. 1998. In vivo toxicity, pharmacokinetics, and anti-cancer activity of genistein conjugated to human epidermal growth factor. Clin. Cancer Res. 4:1125-1134. [PubMed] [Google Scholar]
- 32.Uckun, F. M., L. M. Chelstrom, L. Tuel-Ahlgren, I. Dibirdik, J. D. Irvin, M. C. Langlie, and D. E. Myers. 1998. TXU (anti-CD7)-pokeweed antiviral protein as a potent inhibitor of human immunodeficiency virus. Antimicrob. Agents Chemother. 42:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uckun, F. M., S. Qazi, S. Pendergrass, E. Lisowski, B. Waurzyniak, C. L. Chen, and T. Venkatachalam. 2002. In vivo toxicity, pharmacokinetics, and anti-HIV activity of stavudine-5′[P-bromophenyl methoxyalaninyl phosphate] (stampidine) in mice. Antimicrob. Agents Chemother. 46:3428-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venturi, G., L. Romano, M. Catucci, M. L. Riccio, A. De Milito, A. Gonnelli, M. Rubino, P. E. Valensin, and M. Zazzi. 1999. Genotypic resistance to zidovudine as a predictor of failure of subsequent therapy with human immunodeficiency virus type-1 nucleoside reverse-transcriptase inhibitors. Eur. J. Clin. Microbiol. Infect. Dis. 18:274-282. [DOI] [PubMed] [Google Scholar]
- 35.Yim, H. S., J. H. Kang, P. B. Chock, E. R. Stadtman, and M. B. Yim. 1997. A familial amyotrophic lateral sclerosis-associated A4V Cu, Zn-superoxide dismutase mutant has a lower Km for hydrogen peroxide. Correlation between clinical severity and the Km value. J. Biol. Chem. 272:8861-8863. [DOI] [PubMed] [Google Scholar]
- 36.Zarling, J. M., P. A. Moran, O. Haffar, J. Sias, D. D. Richman, C. A. Spina, D. E. Myers, V. Kuebelbeck, J. A. Ledbetter, and F. M. Uckun. 1990. Inhibition of HIV replication by pokeweed antiviral protein targeted to CD4+ cells by monoclonal antibodies. Nature 347:92-95. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, X. J., W. A. Baase, and B. W. Matthews. 1992. Multiple alanine replacements within alpha-helix 126-134 of T4 lysozyme have independent, additive effects on both structure and stability. Protein Sci. 1:761-776. [DOI] [PMC free article] [PubMed] [Google Scholar]