Abstract
DNA gyrase is a bacterial type II topoisomerase which couples the free energy of ATP hydrolysis to the introduction of negative supercoils into DNA. Amino acids in proximity to bound nonhydrolyzable ATP analog (AMP · PNP) or novobiocin in the gyrase B (GyrB) subunit crystal structures were examined for their roles in enzyme function and novobiocin resistance by site-directed mutagenesis. Purified Escherichia coli GyrB mutant proteins were complexed with the gyrase A subunit to form the functional A2B2 gyrase enzyme. Mutant proteins with alanine substitutions at residues E42, N46, E50, D73, R76, G77, and I78 had reduced or no detectable ATPase activity, indicating a role for these residues in ATP hydrolysis. Interestingly, GyrB proteins with P79A and K103A substitutions retained significant levels of ATPase activity yet demonstrated no DNA supercoiling activity, even with 40-fold more enzyme than the wild-type enzyme, suggesting that these amino acid side chains have a role in the coupling of the two activities. All enzymes relaxed supercoiled DNA to the same extent as the wild-type enzyme did, implying that only ATP-dependent reactions were affected. Mutant genes were examined in vivo for their abilities to complement a temperature-sensitive E. coli gyrB mutant, and the activities correlated well with the in vitro activities. We show that the known R136 novobiocin resistance mutations bestow a significant loss of inhibitor potency in the ATPase assay. Four new residues (D73, G77, I78, and T165) that, when changed to the appropriate amino acid, result in both significant levels of novobiocin resistance and maintain in vivo function were identified in E. coli.
Bacterial DNA gyrase is an established target for the development of new antibiotics (19, 28), being an essential type II topoisomerase involved in maintenance of the negative superhelicity of the chromosome during replication and transcription. The enzyme consists of two subunits (subunits A and B) that combine into the heterotetrameric A2B2 complex to form the functional enzyme. The GyrB subunit is comprised of an N-terminal domain (43 kDa) which contains the ATPase active site and a C-terminal domain (47 kDa) which is involved in the interaction with both the GyrA subunit and DNA. A DNA supercoiling model for gyrase was proposed on the basis of the work by Mizuuchi et al. (31) and Roca and Wang (37, 38). Many aspects of the gyrase supercoiling model have been examined experimentally (22-24, 35, 46, 48-50). In brief, gyrase binds to a short segment of DNA and cleaves the double-stranded DNA, which creates a DNA gate. These activities are mediated via GyrA subunits. DNA contiguous with the DNA gate is wrapped around the GyrA subunits and presents a segment of DNA (termed the transport segment or T segment) to the open N-terminal GyrB subunits (termed the ATP-operated clamp). Upon ATP binding, the ATP-operated clamp closes, capturing the T segment. The DNA gate opens, pulling the broken ends of the DNA gate apart and facilitating the passage of the T segment through the gap. To complete the strand-passage cycle, the DNA gate is religated upon closure, the T segment is expelled via opening of the exit gate (C-terminal GyrA subunits), and the ATP-operated clamp reopens upon ATP hydrolysis. These actions by the enzyme change the superhelical state of the DNA substrate.
There are known synthetic (e.g., quinolone) and natural product (e.g., coumarin) inhibitors of bacterial gyrase. These two types of antibiotics act on gyrase through different mechanisms; the quinolones inhibit the strand breakage and rejoining function of the A subunit. The coumarin-containing antibiotics, of which novobiocin and its dimer coumermycin A1 are representative examples, inhibit the ATP-dependent strand-passage function of the B subunit (28). Clinical use of quinolone antibiotics is likely to become restricted due to the emergence of resistant organisms (2, 12, 20). The poor activities of the coumarin-containing antibiotics against gram-negative bacteria, the high frequencies of development of resistance to these antibiotics, and the mammalian cytotoxicities of these antibiotics have limited their use in the clinic (28). However, despite the limitations of the coumarin-containing antibiotics, their potent inhibitory activities against gyrase and their antibacterial activities stress the importance of GyrB as a well-validated antibacterial target that has yet to be fully exploited. In addition, inhibitors of GyrB are likely to be effective against bacteria resistant to GyrA inhibitors, because GyrA subunit mutations are expected to have little effect on GyrB function.
The emergence of bacterial resistance to the presently used antibiotics is prompting a new wave of drug discovery efforts. Bacteria can acquire antibiotic resistance by multiple mechanisms including a reduction in cell permeability, active efflux of the antibiotic, degradation or modification of the antibiotic, antibiotic sequestration by protein binding, metabolic bypass of the reaction inhibited by the antibiotic, overproduction of the antibiotic target, and target-based resistance (8, 33). In the case of target-based resistance, the protein target is altered such that the inhibitor no longer effectively binds to it (43). Target-based resistance results from inhibitors that make essential binding contacts with amino acid residues that are nonessential for enzyme catalysis or function. Under these circumstances, spontaneous resistant mutants with alterations in these nonessential target residues can readily arise, rendering the antibiotic ineffective. Similarly, the emergence of target-based resistance to inhibitors (e.g., novobiocin) which do not entirely overlap the substrate binding site or which extend into nonessential subsites within the enzyme is also more likely (43).
In an effort to design inhibitors of the GyrB subunit ATP binding site and minimize target-based resistance, we were interested in identifying those amino acid residues which are critical for function. We reasoned that target-based resistance to novel GyrB inhibitors which make the strongest contacts with these essential residues may be far less likely and that such inhibitors may have a greater potential for a broad spectrum of action due to the conservation of these residues among different bacterial species. Using site-directed mutagenesis we found seven amino acid residues in the Escherichia coli GyrB ATP active site that are essential for enzyme function in vitro. These seven residues are also essential for in vivo function, as demonstrated by their inability to complement an E. coli gyrB temperature-sensitive mutant. In addition to the known novobiocin resistance mutation at R136 (9, 13, 36, 39), this study identifies four new amino acid positions in E. coli (D73, G77, I78, and T165) that, when altered, allow significant levels of novobiocin resistance while simultaneously maintaining in vivo function.
MATERIALS AND METHODS
Cloning of E. coli GyrA and GyrB subunits.
The gyrA and gyrB genes were amplified from purified E. coli K-12 chromosomal DNA by PCR with Taq polymerase (Qiagen). Oligonucleotide primers complementary to the 5′ and 3′ ends of the open reading frames were designed to introduce the NdeI and BamHI restriction sites, respectively. The PCR products were digested with NdeI and BamHI and ligated to the T7 promoter-based expression vector pET15b (Novagen). The resulting plasmids, pET15b-gyrA and pET15b-gyrB, respectively, produced both Gyr proteins with an amino-terminal His6 tag. The gyr gene in each expression construct was completely sequenced to ensure its integrity, and the sequence of the gyrB gene was identical to the sequence with GenBank accession number AE000447.
Mutagenesis of E. coli GyrB subunit.
Site-directed mutations and a silent subcloning and diagnostic restriction site were introduced into the gyrB gene by PCR by the two-stage overlap extension method. All fragments were completely sequenced to confirm the presence of the site-directed mutations and the absence of unwanted mutations introduced by PCR.
Expression and purification of WT and recombinant proteins.
Plasmid pET15b-gyrA and the wild-type (WT) and mutant pET15b-gyrB plasmids were transformed into recombination-deficient E. coli (recA) expression strain BLR(DE3) (Novagen) to avoid gene rearrangements. Recombinant protein production was carried out as described previously (15). Briefly, 1-liter expression cultures were grown at 30°C for 18 h in 4× YT medium (32 g of Bacto Tryptone [Difco] per liter, 20 g of Bacto Yeast Extract [Difco] per liter, 5 g of NaCl per liter) without induction. Frozen cell pellets were resuspended in 75 ml of buffer A (20 mM Tris HCl [pH 8.0], 10 mM NaCl) containing four tablets of EDTA-free complete protease inhibitor (Roche). Buffer A (45 ml) containing 120 mg of lysozyme was added to each resuspension, followed by the addition of 30 ml of buffer B (50% glycerol, 1.5 M NaCl, 0.5% Triton X-100), and then the mixture was incubated on ice for 30 min. The lysates were sonicated to reduce viscosity and then centrifuged at 10,000 rpm in a Beckman JA-17 rotor for 30 min. The soluble fractions were mixed for 1 h with Talon (Clonetech) resin slurry that had been equilibrated with buffer C (20 mM Tris-HCl [pH 8.0], 300 mM NaCl, 10% glycerol, 0.1% Triton X-100). The Talon resin was recovered by centrifugation. The resin was resuspended in buffer C, and the slurry was poured into a column. The packed column was washed with buffer C containing 0, 10, 50, and 500 mM imidazole. The protein subunits eluted in 500 mM imidazole, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining. The fractions that eluted in 500 mM imidazole were diluted 32-fold in buffer D (50 mM Tris HCl [pH 8.0], 2 mM dithiothreitol (DTT), 1 mM EDTA, 10% glycerol) and then applied to phosphocellulose (Millipore) columns that had been equilibrated in buffer D. The columns were eluted stepwise with buffer D containing 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, and 1 M NaCl. The 90-kDa GyrB proteins were recovered in the steps with 0.3 to 0.5 M NaCl, whereas the 97-kDa GyrA protein was recovered in the steps with 0.2 to 0.4 M NaCl. All protein fractions were concentrated in Ultrafree-15 Biomax-100 membrane centrifugal filter units (Millipore) to 18 to 93 μM protein. The protein concentrations were determined by the Bio-Rad dye binding assay with bovine serum albumin as the standard and by measurement of the absorbance at 280 nM by using the calculated extinction coefficient of the WT GyrB (68,020 M−1 cm−1). Yields of purified protein varied from 4 to 35 mg of protein per liter of culture. To form the functional gyrase A2B2 heterotetrameric enzyme, equal molar amounts of GyrB and GyrA were mixed together at the highest achievable protein concentration (>15 μM) 15 min prior to the start of the assay. The enzyme concentration stated in the assays is calculated on the basis of the molecular mass of the functional gyrase A2B2 heterotetrameric enzyme (374,000 g/mol).
ATPase assay and determination of kinetic parameters.
For all in vitro assays the WT and mutated GyrB subunits were complexed with the WT GyrA subunit in a ratio of 1 to 1 for 15 min on ice at the highest possible protein concentration (>15 μM). This ratio of GyrA and GyrB was chosen because it was optimal in the DNA supercoiling assay (data not shown). Substitutional effects on ATPase activity (V/K values) by each mutation were determined from the slope of the linear portion (i.e., low substrate concentration) of a plot of the rate of ATP hydrolysis (micromolar per second) versus the ATP concentration (micromolar); this value is a good approximation of each enzyme's overall catalytic efficiency (i.e., Vmax/Km) (10). For this study, V/K values (Table 1) are reported in place of Vmax/Km values because many of the proteins did not achieve ATP saturation, and therefore, Vmax and Km values could not be determined directly.
TABLE 1.
Amino acid substitution effects on in vitro gyrase activities and cell viability
| Enzyme | V/K (% of WT)a | ATP Km (μM)b | DNA supercoiling
|
Ts comp.d | Relaxation (+)e | |
|---|---|---|---|---|---|---|
| Result | Enzyme concn (nM)c | |||||
| WT | 100 | 830 | + | 10 | + | + |
| E42A | <2 | − | >400 | − | + | |
| E42D | 103 | 120 | + | 100 | − | + |
| E42Q | <2 | − | >400 | − | + | |
| V43A | 37 | 2,700 | + | 25 | + | + |
| N46A | <2 | − | >400 | − | + | |
| N46D | <2 | − | >400 | − | + | |
| N46E | <2 | − | >400 | − | + | |
| N46Q | <2 | − | >400 | − | + | |
| E50A | 3 | >5,000 | − | >400 | − | + |
| E50D | 3 | >3,600 | − | >400 | − | + |
| E50Q | 18 | 2,900 | − | >400 | − | + |
| D73A | <2 | − | >400 | − | + | |
| D73E | 3 | >3,600 | + | 100 | W | + |
| D73N | <2 | − | >400 | − | + | |
| R76A | 20 | ∼5,000 | − | >400 | − | + |
| R76K | 15 | >3,600 | − | >400 | − | + |
| G77A | 5 | >5,000 | + | 100 | W | + |
| G77S | 10 | >3,600 | + | 100 | W | + |
| I78A | 3 | >5,000 | + | 100 | + | + |
| I78L | 17 | >3,600 | + | 100 | + | + |
| I78V | 56 | 1,600 | + | 25 | + | + |
| P79A | 36 | 2,300 | − | >400 | − | + |
| G81A | 95 | 990 | + | 25 | + | + |
| I94A | 65 | 740 | + | 25 | + | + |
| A100S | 101 | 570 | + | 10 | + | + |
| K103A | 57 | 640 | − | >400 | − | + |
| K103I | 12 | 850 | − | >400 | − | + |
| K110A | 96 | 520 | + | 25 | + | + |
| V120A | 343 | 260 | + | 10 | + | + |
| V120M | 35 | >3,600 | + | 50 | + | + |
| S121A | 79 | 1100 | + | 50 | + | + |
| R136A | 48 | 1300 | + | 50 | + | + |
| R136H | 42 | 1300 | + | 25 | + | + |
| R136L | 27 | >3,000 | + | 25 | + | + |
| R136S | 60 | 940 | + | 25 | + | + |
| T165A | 19 | 4,400 | + | 50 | + | + |
| T165S | 71 | 1,200 | + | 10 | + | + |
| T165V | 59 | 2,600 | + | 25 | + | + |
| V167A | 51 | 1,600 | + | 25 | + | + |
| P171A | 92 | 940 | + | 10 | + | + |
The V/K ratio is expressed as the percentage of that for the WT enzyme (100%) (WT V/K = 0.5 mM−1/s−1).
Km values are apparent.
Enzyme concentrations necessary to completely supercoil 1.25 μg of DNA at 37°C in 20 min.
Ts comp., complementation of the temperature-sensitive strain (+) and noncomplementation (−) by mutations; W, weak growth.
Enyzme at a concentration of 100 to 200 nM was able to completely relax 1.25 μg of negatively supercoiled plasmid DNA at 37°C in 20 min.
ATP hydrolysis rates were monitored spectrophotometrically on a Molecular Devices plate reader by monitoring the decrease in NADH absorbance during the reaction in a coupled-enzyme assay, as described previously (11). The standard coupled-enzyme reactions were carried out at 30°C (reaction volume, 100 μl) with the following components: 100 mM Tris-HCl (pH 7.6), 1.5 mM MgCl2, 150 mM KCl, 2.5 mM phosphoenolpyruvate, 0.2 mM NADH, 1 mM DTT, 0.03 mg of pyruvate kinase per ml, 0.01 mg of lactate dehydrogenase per ml, 4% dimethyl sulfoxide (DMSO), 900 μM ATP, and various amounts of gyrase enzyme to generate a rate of 2 μM/min. Novobiocin (Sigma) was dissolved in DMSO. For the spectrophotometric ATPase assay, the level of sensitivity was 2% of the WT activity.
DNA supercoiling and relaxation assays.
The DNA supercoiling reaction mixtures (25 μl) contained the following components: 100 mM Tris-HCl (pH 7.6), 40 mM KCl, 4 mM MgCl2, 1 mM DTT, 250 mM potassium glutamate, 14 nM relaxed pBR322 plasmid, 4% DMSO, 1.5 mM ATP, and E. coli DNA gyrase at concentrations that were ramped from 5 to 400 nM. Inhibitor was added to the buffer, DNA, and enzyme mixture; the mixture was incubated for 10 min at 37°C; and then ATP was added to initiate the reaction for 20 min at 37°C. The reaction was stopped with 5 μl of quench buffer (250 mM EDTA, 50% glycerol, 25 μg of bromophenol blue per ml). The samples were loaded on a 1% agarose gel (12 cm [length] by 14 cm [width]) containing no ethidium bromide and run for 16 h at 20 V in a Buffer Puffer gel box (Owl Scientific) that continuously circulates the 1× Tris-acetate-EDTA (TAE) running buffer. The gels were stained with ethidium bromide for 30 min and then destained in 1× TAE buffer for 4 h. The gels were visualized and quantified with a Bio-Rad gel documentation system.
The DNA relaxation assay mixture had the following components: 100 mM Tris-HCl (pH 7.6), 40 mM KCl, 4 mM MgCl2, 1 mM DTT, 14 nM double-banded CsCl, purified supercoiled plasmid pBR322, and E. coli DNA gyrase enzyme at concentrations that were ramped from 50 to 400 nM. The reaction mixtures (25 μl) were incubated for 30 min at 37°C, and the reactions were initiated with the addition of gyrase enzyme. The reactions were stopped and processed as described above for the DNA supercoiling reaction.
Data analysis.
For the WT and mutant enzymes, the V/K values were derived from the linear portion of a plot of the rate of ATPase hydrolysis (in micromolar per second) versus the ATP substrate concentration (in micromolar) (i.e., the slope of the line). The apparent Km for ATP was determined in cases in which apparent saturation had been achieved by fitting rectangular hyperbolae to a plot of linear rates of ATP hydrolysis (in micromolar per minute) versus increasing Mg · ATP concentrations (in micromolar). The 50% inhibitory concentrations (IC50s) of the competitive inhibitor novobiocin were obtained by measuring the decrease in the linear rates of ATP hydrolysis (in micromolar per minute) with increasing concentrations of inhibitor, and the rates were fit to the data by using a two-parameter hyperbolic fit: y = IC50/[B · (x + IC50)], where B is 1/Vmax, y is the rate of inhibition (in micromolar per minute), and x is the concentration of the inhibitor. In all cases, the ATP concentration did not exceed the apparent Km determined for each mutant enzyme. For all IC50 determinations, the enzyme concentration was less than or equal to twofold the reported IC50.
Complementation of a gyrB(Ts) allele by mutant gyrB genes.
The functions of the mutant gyrB genes were assessed by determination of their abilities to complement the gyrB(Ts) allele in E. coli strain N4177 [strA galK gyrB221 (couR) gyrB203(Ts); Coli Genetic Stock Center] (29). To prevent gene rearrangements due to recombination between chromosomal and plasmid gyrB genes, the recA::Tn10 allele from strain AN193-59 (16) was introduced into N4177 by P1 transduction (30). In order to test pET15b-gyrB constructs for complementation, N4177(DE3)recA, a DE3 lysogen expressing T7 RNA polymerase, was constructed using a Novagen (Madison, Wis.) kit. Unexpectedly, pET15b-gyrB plasmids encoding the His6-tagged WT and mutant gyrB genes did not complement the temperature-sensitive defect at the nonpermissive temperature (42°C) in the presence or absence of the inducer (isopropyl-β-d-thiogalactopyranoside [IPTG]). This suggests that the amino-terminal His6 tag interferes with the enzyme function in vivo, even though the His6-tagged enzymes exhibited no difference from WT protein in an extensive battery of in vitro biochemical assays (see Results and Discussion). To test this hypothesis, the WT gene and all 40 mutant gyrB genes were subcloned into plasmid pMAL-p2X (New England BioLabs) cut with NdeI and BamHI (deleting the malE gene). For each clone the presence of the mutations was confirmed by sequencing. In the resulting pTAC constructs, the GyrB protein is expressed from a tac promoter without an amino-terminal His6 tag; others (21, 42) have successfully used similar constructs to study complementation. N4177(DE3)recA was transformed with WT and mutant plasmids pET15b-gyrB and pTAC-gyrB. Complementation was determined by growth at 30 and 42°C on Luria-Bertani agar plates containing 100 μg of ampicillin per ml. Identical results were obtained with and without 1 mM IPTG for each expression system, suggesting that the lack of complementation with the pET15b-gyrB constructs is due to the His6 tag and is not a result of toxic protein overexpression. An attractive hypothesis is that the His6 tag interferes with the enzyme's ability to associate with other cellular factors like DnaB (41).
RESULTS AND DISCUSSION
Rationale for structure-based mutagenesis study of E. coli GyrB subunit ATP active site.
Target-based resistance has a major role in the emergence of antimicrobial resistance (8, 33). Others (2) have successfully used site-directed mutagenesis to define the essential nature of the enzyme active site, with the goal being to provide a rationale for target-based resistance and, ultimately, to reduce the rate of occurrence of target-based resistance to existing or future antimicrobial agents. In an effort to understand the basis of gyrase resistance and to minimize the antibiotic-based resistance of gyrase, we had two objectives for the mutagenesis study. The first was to determine the essentiality of conserved amino acid residues surrounding the ATP-novobiocin binding site (Fig. 1). The second was to corroborate the GyrB crystal structures containing either the bound ATP analogue (AMP · PNP [adenylyl-imidodiphosphate]) or the known GyrB inhibitor (novobiocin). Initially, alanine substitutions were introduced to assess the importance of the amino acid side chain beyond the β carbon. The following 19 residues were selected for alanine substitution: E42A, V43A, N46A, E50A, D73A, R76A, G77A, I78A, P79A, G81A, I94A, K103A, K110A, V120A, S121A, R136A, T165A, V167A, and P171A (Fig. 1; for clarity, only residues that demonstrated a role in enzyme function are shown). Enzymatic characterization of the alanine mutants identified side chains that were important for function. Alanine substitutions, however, did not reveal the properties of the missing side chain that were important for activity, and therefore, the following 15 conservative amino substitutions were selected: E42D, E42Q, N46D, N46E, N46Q, E50D, E50Q, D73E, D73N, R76K, I78L, I78V, K103I, T165S, and T165V. In addition, we were interested in studying known novobiocin resistance mutations (R136H, R136L, R136S). Finally, the biological significance of these 40 mutations was assessed by complementation of a temperature-sensitive gyrB mutation in E. coli.
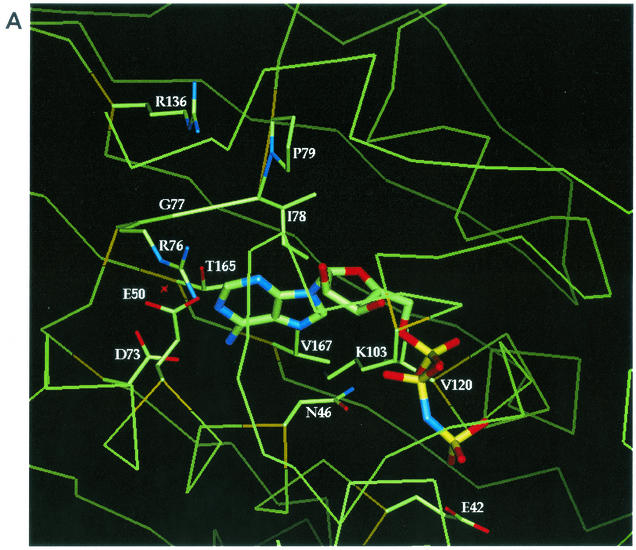
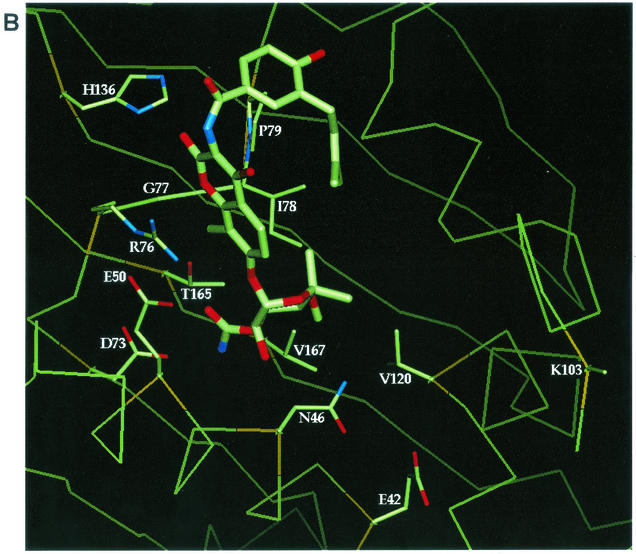
FIG. 1.
Locations of selected amino acids in the E. coli GyrB ATP binding site relative to AMP · PNP and novobiocin. (A) AMP · PNP complexed in the E. coli N-terminal 43-kDa fragment based on the structural data for the sequence in the Protein Data Bank with identification number 1EI1. (B) Novobiocin complexed in the E. coli N-terminal 24-kDa fragment (novobiocin-resistant mutant R136H) based on the structural data for the sequence in the Protein Data Bank with identification number 1AJ6. The locations of the side chains for residues E42, N46, E50, D73, R76, G77, I78, P79, K103, V120, R136 or H136, T165, and V167 are shown and labeled in both panels A and B. The α-carbon of the mutant residues is shown in gold. The protein backbone is shown as a thin green line.
His6-tagged versus nontagged gyrase protein.
It is known that the ATPase kinetics of gyrase and other type II topoisomerases are non-Michaelis-Menten kinetics, in part due to the nonlinear dependence of the enzyme activity on the enzyme concentration (3). However, Michaelis-Menten kinetics can be and have been used by others (7, 42) successfully to study the kinetics of gyrase under conditions in which there is an observed linear dependence of the rate of ATPase hydrolysis on the enzyme concentration. Both Cullis et al. (7) and Smith and Maxwell (42) have used Michaelis-Menten kinetics to obtain the apparent Vmax and Km values for WT and mutant gyrase enzymes. We have followed a similar strategy in the evaluation of gyrase mutant proteins. To demonstrate that the His6 tag does not affect gyrase, we determined the kinetic parameters of ATPase for the WT His6-tagged gyrase (A2B2) (Km = 830 ± 30 μM; kcat = 1.2 ± 0.1 s−1) under conditions appropriate for Michaelis-Menten kinetics. We found the values for these kinetic parameters for ATP hydrolysis to be in agreement with the values reported in the literature for the non-His6-tagged WT gyrase enzyme (Km = 210 to 500 μM; kcat = 0.6 to 1.4 s−1), as summarized by Maxwell and Gellert (27). Similarly, using as a control non-His6-tagged E. coli gyrase (A2B2) from a commercial source (purchased from John Innes Enterprises Ltd.), we obtained similar kinetic parameters (Km = 600 ± 20 μM; kcat = 3.8 ± 0.1 s−1) with comparable amounts of enzyme in the ATPase, DNA supercoiling, and DNA relaxation assays (data not shown), demonstrating that the His6 tag does not substantially affect the kinetic properties of the enzyme. All data argue that the His6-tagged enzyme is equivalent to the non-His6-tagged enzyme in all in vitro biochemical assays.
Structure-function relationships of the GyrB subunit ATP active site.
In Table 1, we present experimental evidence that several amino acid residues in the GyrB ATP active site are essential for enzyme catalysis in vitro and in vivo. We discovered that two of these residues (P79A and K103A) play an essential role in the coupling of ATP hydrolysis to the DNA supercoiling activities of the enzyme. In the following paragraphs, we review and compare the results of our mutagenesis experiments with those reported by other groups and relate our findings to the reported GyrB crystal structures.
Replacement of E42 by alanine reduced activity to undetectable levels. A similar loss of activity was seen with E42Q replacement, which substitutes a hydroxyl group for an amide group. The replacement of E42 by a functionally equivalent side chain that is one carbon shorter (E42D) restores the ATPase V/K value to the values for the WT, demonstrating that the carboxylic acid is important for catalysis. Interestingly, the Km for ATP for E42D decreased, suggesting that a shorter side chain improves binding and compensates for a lower rate (20%) of ATP hydrolysis in the WT enzyme. Structural data place the E42 residue in proximity to the γ-phosphate of the bound ATP molecule (Fig. 1A). Our results agree well with those of others (21), who have proposed that E42 functions as a general base in the activation of water for a nucleophilic attack on the γ-phosphate of ATP. The most conservative substitution, E42D, however, did not allow in vivo viability, demonstrating that the glutamine side chain is essential. Considering that the E42D mutant GyrB had in vitro activities which were equal to or better than those of the GyrB proteins of other mutants which were functional in vivo, it is not clear why the E42D mutant GyrB is not functional. This result suggests that this side chain may have an additional functional role in the protein.
All mutations at N46 to either alanine or conservative substitutions (D, E, or Q) abolished ATPase and supercoiling activities. The absence of ATPase activity by an N46D mutation has also been reported by Kampranis et al. (25) Crystallographic data illustrate that the N46 residue coordinates the Mg2+ in the ATP binding site (26, 47). The N46 residue is totally conserved among all type II topoisomerases. Our data demonstrate that the N46 side chain is essential for enzyme function and cannot be replaced by another conserved residue.
Crystallographic data show a salt-bridge interaction between residues E50 and R76 that forms a portion of the adenine ring-novobiocin binding pocket. Replacement of E50 by alanine reduced the ATPase V/K to 3% of that for the WT enzyme, whereas replacement of E50 by E50D and E50Q reduced the ATPase V/K values to 3 and 18% of that for the WT enzyme, respectively. The data for E50D and E50Q argue that both the side chain length and the carboxylic acid group are important contributors to enzyme activity. Similarly, replacement of R76 with either alanine or lysine results in an enzyme with low levels of ATPase activity and no DNA supercoiling activity. Hence, there is a strict requirement for arginine at this position. The biochemical and biological data corroborate the crystallographic data by demonstrating that the salt-bridge interaction between E50 and R76 is important for ATPase activity and novobiocin binding and is essential for DNA supercoiling activity and in vivo function.
D73 forms a direct hydrogen bond with the adenosine amino group of AMP · PNP (Fig. 1A). In our study, the replacement of D73 by alanine or asparagine abolished both ATPase and DNA supercoiling activities. The lack of ATPase activity has also been reported for the D73N mutation (25). Interestingly, we found that the D73E substitution restored both ATPase and DNA supercoiling activities in vitro and conferred a weak function in vivo (Table 1). However, the Km for ATP for D73E is significantly higher, suggesting that the larger glutamine side chain impedes the correct orientation of the binding at the adenine ring of ATP. This observation is strengthened by the fact that the same D73E mutation also has a negative effect on the binding of novobiocin (Table 2). Both the structural and the biochemical data on the role of the D73 residue are in agreement.
TABLE 2.
Comparison of novobiocin IC50 for ATPases of WT and mutant proteinsa
| Enzyme | Novobiocin IC50 (μM) | IC50 ratiob |
|---|---|---|
| WT | 0.046 | |
| V43A | 0.12 | 3 |
| E50A | 7.7 | 170 |
| D73E | 2.5 | 54 |
| R76A | >10 | >770 |
| G77A | 4.4 | 96 |
| G77S | 65 | 1,400 |
| I78A | 1.4 | 30 |
| I78L | 0.64 | 14 |
| I78V | 0.072 | 2 |
| P79A | 9.7 | 210 |
| I94A | 0.16 | 3 |
| A100S | 0.080 | 2 |
| K103A | 0.12 | 3 |
| V120A | 0.048 | 1 |
| V120M | 0.052 | 1 |
| R136A | 5.0 | 110 |
| R136H | 1.3 | 28 |
| R136L | 6.6 | 140 |
| R136S | 3.8 | 83 |
| T165A | 0.85 | 18 |
| T165S | 0.058 | 1 |
| T165V | 0.61 | 13 |
| V167A | 0.063 | 1 |
IC50s were determined as described in Materials and Methods.
IC50 for mutant/IC50 for wild type.
Structural data place the G77 residue ∼4.3 Å from the C-2 carbon and ∼5.1 Å from the N-3 nitrogen of the adenine ring (Fig. 1A). The simple addition of a methyl group in the enzyme with the G77A mutation has a dramatic effect on ATP binding (Km > 5,000 μM) and results in 5% of the ATP hydrolysis activity of the WT enzyme (Table 1). Surprisingly, this mutant gene weakly complements gyrB(Ts), which may be explained by the residual activity of this enzyme in the supercoiling of DNA in vitro, albeit at a higher enzyme concentration.
From the crystallographic data, the I78 side chain lies ∼4.5 Å above the plane of the adenine ring (Fig. 1A). Replacement of I78 by alanine results in an enzyme which has lost significant affinity for the ATP substrate and hydrolysis activity but yet retains sufficient DNA supercoiling activity to remain functional in vivo. Restoration of a portion of the hydrophobic side chain to either a leucine or a valine substitution improves the ATPase activity, demonstrating that the hydrophobic contacts are important in ATP binding (Table 1). The proteins with the I78A, D73E, and G77A mutations all showed disproportionate reductions in ATPase activities relative to their abilities to supercoil relaxed plasmid DNA (Table 1), suggesting that these substitutions affected ATPase activity to a far greater extent than they do DNA supercoiling activity.
The catalytic introduction of negative supercoils into relaxed plasmid DNA requires the gyrase enzyme to have the ability to hydrolyze ATP. Two interesting substitutions were P79A and K103A. Enzymes with these mutations retained significant levels of ATPase activity yet demonstrated no DNA supercoiling ability. These enzymes were also assayed for DNA supercoiling activity at 30°C in the event that the mutations rendered the enzyme temperature sensitive; however, no supercoiled DNA was detected (data not shown). These results suggest that the P79 and K103 side chains are required for the coupling of ATP hydrolysis to DNA supercoiling activity. Both mutations were capable of relaxing supercoiled plasmid DNA in the absence of ATP to a similar extent as the WT enzyme was (data not shown), suggesting that the N-terminal GyrB subunit (i.e., the ATP-operated clamp) was not locked in the closed position. The data suggest, however, that the ATP-operated clamp in these mutants may not be capable of closing or functioning properly, giving rise to the lack of DNA supercoiling ability. In this study mutants with mutations of other residues with significantly less ATPase activities can supercoil DNA, arguing that both P79A and K103A generate sufficient ATPase activity for supercoiling but that this energy is not used properly in the enzymes with these mutations. To further support the uncoupling hypothesis we have examined the mutants in an ATP-stimulated quinolone-mediated DNA cleavage assay using both linear and supercoiled pBR322 DNA as substrates (6, 22). The mutants with the P79A and K103A mutations did not demonstrate ATP-stimulated quinolone-mediated DNA cleavage, whereas the gyrase activity of the WT was stimulated (data not shown). All three proteins had comparable quinolone-mediated DNA cleavage activities, suggesting that the mutants were simply defective in ATP-stimulated quinolone-mediated cleavage. These results provide additional evidence that the P79A and K103A mutations have a role in the coupling of ATP hydrolysis to DNA supercoiling activity. Both P79 and K103 are completely conserved in all GyrB subunits sequenced to date. Structural studies show that K103 forms a salt bridge with the β-phosphate of AMP · PNP (Fig. 1A). K103 is also located in a flexible loop region of the GyrB subunit which spans amino acid residues 97 to 119. These residues have been crystallized in several different conformations, demonstrating the flexibility of the loop (S. Bellon, unpublished data). Replacement of K103 by isoleucine resulted in an enzyme with one-fifth of the ATPase activity of the enzyme with the alanine substitution. The reduction or loss of ATPase activity by the mutants with the larger K103 substitutions is in agreement with the results of a previous study that examined glutamic acid, isoleucine, and threonine at position 103 (34). Considering the interactions that K103 makes with the bound ATP molecule and the enzymatic activity of the mutant with the alanine substitution, it appears that the side chain has a role in enzyme conformational changes and/or the coupling of ATP hydrolysis to DNA supercoiling activity, perhaps by acting as a sensor for detection of when ATP hydrolysis has occurred. The P79A mutation data suggest that this residue has a role similar to that of K103, although structural data place the side chain farther away (∼5.5 Å from the N-3 adenine ring).
Removal of the hydrophobic side chain in the mutant with the V120A mutation increased the affinity for ATP by decreasing the Km for ATP to one-third the value for the WT without perturbing the DNA supercoiling activity (Table 1). The calculated V/K value (1.7 mM−1 s−1) for V120A at ATP concentrations below 140 μM is also in agreement with the Km for ATP being one-third of that for the WT enzyme (V/K value, 0.5 mM−1 s−1). Structural data place the valine side chain ∼3.3 Å from the α-phosphate of ATP, and removal of the hydrophobic valine could explain the small increase in the affinity for ATP. Similarly, when a larger hydrophobic side chain, V120M, was used, the Km for ATP increased to >3,600 μM, yet the ATP hydrolysis, DNA supercoiling, and DNA relaxation activities were only mildly affected, demonstrating that V120 contributes predominately to ATP binding.
The structural data for E. coli show that the guanidinium group of R136 forms a hydrogen bond with the main-chain carbonyl group of Y5, which comes from the N-terminal portion of the other GyrB subunit (47). The hydroxyl group of Y5 forms a hydrogen bond with the 2′-hydroxyl ribose of the bound nucleotide (47). The N-terminal arm of the other dimer provides ∼14 amino acids that are essential for GyrB subunit dimerization and activation of the ATPase catalytic center (4). The loss of the arginine side chain in the mutant with the R136A mutation could presumably lessen the interaction with the Y5 residue, which might explain the less efficient ATP hydrolysis activity of the mutant.
A key residue in ATP binding is T165, which is totally conserved in all GyrB subunits and type II topoisomerases. Replacement of T165 with alanine results in a gyrase enzyme with a fivefold increase in its Km for ATP (Table 1), whereas replacement of the side chain with serine nearly restores the Km for ATP to that for the WT enzyme, suggesting that the hydroxyl group plays a functional role in ATP binding. Biochemical data support the structural data which place the T165 hydroxyl group ∼3.2 Å from the N-7 nitrogen in the adenine ring and ∼3.5 Å from the primary amine on the bound AMP · PNP molecule (26). Perhaps even more importantly, the T165 side chain hydroxyl participates in a tight hydrogen bond network involving a crystallographically conserved water molecule with the N-7 nitrogen in the adenine ring (26).
The temperature-sensitive strain used in this study, strain N4177(DE3)recA, has two amino acid changes (R136C and P171S) (1). Since the R136C change is a coumarin resistance mutation (5), it implies that the P171S mutation is responsible for thermosensitivity. In this study, we have changed P171 to alanine and found levels of enzyme activity that were nearly those of the WT enzyme. We also saw no indication of thermosensitivity, since the P171A mutation complemented the temperature-sensitive strain at 42°C, implying that only a change from proline to serine at this position confers the temperature-sensitive phenotype. In other bacteria (e.g., Staphylococcus aureus), the equivalent GyrB P171 residue is also alanine.
Finally, as an important control to confirm that all of the mutant GyrB proteins were properly folded and capable of forming tetrameric A2B2 enzymes, we assayed each mutant gyrase A2B2 enzyme for its ability to relax negatively supercoiled DNA independently of ATP binding and hydrolysis. Gyrase-mediated relaxation of supercoiled DNA is thought to occur by releasing the energy bound in the negatively supercoiled helical DNA structure (50). To achieve DNA relaxation, the enzyme is required to bind to the DNA substrate, create a double-stranded DNA break, facilitate the passage of uncut DNA through the enzyme and the double-stranded DNA break, and subsequently, reseal the DNA break to alter the linking number of the plasmid. To investigate the integrities and stabilities of the mutant gyrase enzymes, the DNA relaxation activities of all 40 gyrase mutants were determined by ramping the enzyme concentration. The concentrations at which all of the mutant proteins relaxed negatively supercoiled plasmid DNA were within twofold of the concentration for the WT enzyme (Table 1), confirming that the mutant GyrB proteins were properly folded and able to form stable functional gyrase A2B2 complexes. As a control, individual GyrA subunit and all WT and mutated GyrB subunits by themselves did not promote DNA relaxation when the enzyme concentration tested was 400 nM (data not shown).
Novobiocin resistance mutations in the GyrB subunit.
A variety of techniques including gel filtration, isothermal titration calorimetry, and IC50 determinations have been used to study gyrase-inhibitor binding interactions (5, 7, 14, 17, 45). Contreras and Maxwell (5) have successfully used IC50 determinations to quantify the differences in novobiocin binding affinities between WT and mutant gyrase enzymes. We also chose to quantify the contribution that individual residues make to novobiocin binding by determining the IC50s for the ATPases of the mutant GyrB proteins (Table 2). The enzyme concentrations for this experiment ranged from 100 and 200 nM for the mutants with all mutations with the exception of those with the E50A, D73E, G77A, and I78A mutations, for which 1 μM enzyme was used in order to obtain a sufficient rate of ATP hydrolysis for the determination. As novobiocin is a competitive inhibitor, the IC50s for each gyrase mutant were determined by use of ATP concentrations no greater than the reported Km for ATP. Only mutant GyrB proteins which retained sufficient ATP hydrolysis activity could be quantified for their role in novobiocin binding.
Since gyrase ATP hydrolysis activity is essential for cell viability, those amino acid substitutions that result in enzymes without measurable activity are not viable and are not expected to arise from exposure to GyrB inhibitors. This statement is supported by our data showing that none of the enzymes that lacked ATPase or DNA supercoiling activity could complement the temperature-sensitive gyrB mutation (Table 1). Any single amino acid substitution which both retains a threshold level (i.e., a level necessary for bacterial survival) of gyrase activity and allows reduced inhibitor binding is likely to arise as a resistance mutation upon exposure to a GyrB inhibitor. Our study has identified five amino acid residues in the ATPase active site (D73, G77, I78, R136, T165) whose substitution allows the gyrase enzyme to function in vivo and that also result in significantly increased IC50s of novobiocin for ATPase. How these five residues contribute to resistance with respect to structural data and whether the mutations at these positions have previously been identified as novobiocin resistance mutations are discussed in the following paragraphs.
The most commonly identified novobiocin resistance mutations are at R136 and have been seen in many bacteria (18, 40, 44), including E. coli (5, 13, 36, 39). The replacement of R136 by cysteine, histidine, leucine, and serine has been found in novobiocin-resistant E. coli isolates (9, 13, 36, 39). Replacement of R136 by alanine reduced the level of ATPase activity to ∼50% of that for the WT enzyme and resulted in a less than twofold increase in the Km for ATP (Table 1). We showed that previously isolated spontaneous mutations at R136 to histidine, leucine, and serine for novobiocin resistance in E. coli result in significant increases in the novobiocin IC50s for ATPase (Table 1), and all of these mutations were functional in vivo (Table 2). In the WT structure, R136 directly forms a hydrogen bond with novobiocin; the coumarin carbonyl oxygen is ∼3.0 and ∼2.6 Å from the NH-1 and NH-2 groups of guanidinium, respectively. In contrast, the gyrase mutant with the R136H mutation lacks this hydrogen bond (17), and the volume vacated by the guanidinium group contains an ordered water molecule. The water molecule forms hydrogen bonds with the coumarin carbonyl oxygen (∼2.8 Å) and with the carbonyl oxygen of G77 on the protein. In addition to losing a direct hydrogen bond between the coumarin moiety and R136H, it is thought that immobilization of the water molecule in the mutant enzyme upon complexation of the ligand leads to significant entropic costs. This contributes to a further reduction in the affinity of binding to novobiocin over that for the WT enzyme (17). In comparison to the mutation with the histidine substitution, the IC50s for the other R136 substitutions of ATPase are even greater (three- to fivefold), suggesting that further truncation of the side chain or that replacement by a more hydrophobic side chain is detrimental. Contreras and Maxwell (5) have previously reported the IC50s of novobiocin for WT ATPase (90 nM) versus those for the enzymes with the R136H (700 nM) and R136S (4,300 nM) mutations, which also showed approximately sixfold increases for mutants with shorter side chains. The present results are in good agreement with those results, and these observations may be explained by the fact that R136H gains stabilizing interactions with the neighboring R76 and T80 residues (17) which are not available to either the shorter or the hydrophobic side chain.
D73 forms a direct hydrogen bond with the carbamoyl group of novobiocin (26) (Fig. 1B). The D73E substitution retains the functional carboxylic acid group and extends the side chain by one carbon. Such a subtle change has a profound effect on enzyme activity, yet the substitution permits weak growth in vivo (Table 1). Similarly, the IC50 of novobiocin for ATPase increased ∼50-fold over that for the WT enzyme, demonstrating that the interaction with D73 is essential. To our knowledge, the D73E mutation has not been isolated as a novobiocin resistance mutation, which may suggest that the weak in vivo growth is insufficient and prevents the selection of the mutation.
Mutations outside of R136 have been identified in novobiocin-resistant isolates of S. aureus; however, the genotype-phenotype association in S. aureus remains to be confirmed (44). These S. aureus mutations (in parentheses) correspond to the following E. coli residues: G77 (G85S), I94 (I102S), A100 (A108S), V120 (S128L), and T165 (T173N). In Streptococcus pneumoniae, an S127L substitution (equivalent to E. coli V120) has also been implicated in novobiocin resistance (32). On the basis of our data on the novobiocin IC50 for ATPase, the substitutions at I94, A100, and V120 had insignificant effects on novobiocin binding to E. coli gyrase (Table 2), suggesting that the residues may be resistance mutations only in the S. aureus and S. pneumoniae gyrases. In contrast, the G77A or G77S substitution and the T165A or T165V substitution significantly affected novobiocin binding to E. coli gyrase (Table 2).
The G77S or G77A substitution has a profound effect on ATPase activity and DNA supercoiling that results in only weak growth in vivo (Table 1). The IC50 of novobiocin for the ATPase of the mutant with the G77S mutation jumps dramatically to ∼1,400-fold over that for the WT enzyme, demonstrating that any side chains larger than hydrogen are not well tolerated (Table 2). Examination of the protein backbone at position G77 shows that only glycine can be tolerated without a resulting distortion of the backbone. Such a distortion is consistent with a reduced affinity for novobiocin. The corresponding G77S substitution has been identified as a resistance mutation in S. aureus (44) but has not been isolated in E. coli. Perhaps in E. coli the weak in vivo growth phenotype is insufficient to select for resistant mutants, whereas in S. aureus these effects are somewhat ameliorated.
From crystallographic data the I78 side chain is in proximity to the oxygen of the methoxyl group on the novobiose sugar (∼3.2 Å) (Fig. 1B) and the isopentenyl group (∼4.4 Å) as it wraps back in the direction of the sugar (26). From conservative substitutions we deduce that the bifurcation of the hydrophobic side chain at the β-carbon is critical for both enzyme activity (Table 1) and novobiocin binding (Table 2). These results complement structural data that suggest that the I78 side chain mediates a hydrophobic interaction between the two halves of the bound novobiocin molecule. Neither I78A nor I78L has been identified as a novobiocin resistance mutation in any bacteria to date.
Mutations at T165 that remove the side chain hydroxyl group significantly affect novobiocin binding by increasing the novobiocin IC50 for ATPase between 13- and 18-fold (Table 2). The novobiocin-GyrB crystal structure shows that T165 forms a water-mediated hydrogen bond with the carbamoyl moiety of the novobiose sugar (17, 26). Our results demonstrate that T165 is an important residue in gyrase activity but is not essential for gyrase function, thereby making it a good candidate for contributing to novobiocin resistance, but so far this has been implicated only in S. aureus (44).
Conclusions.
Our results satisfied the two objectives of the study, which were to determine the essentiality of conserved amino acid residues that surround the ATP-novobiocin binding site and to corroborate our GyrB crystal structures containing either the bound ATP analogue (AMP · PNP) or novobiocin. We have identified and provided evidence that two GyrB residues (P79A and K103A) have a role in coupling the energy of ATP hydroysis to DNA supercoiling. Detailed biochemical study of these mutants to determine the specific mechanism of this observation is under way. This structure-function approach has identified GyrB residues (D73, G77, I78, R136, T165) that have the potential to become novobiocin target-based resistance mutations; however, our data do not predict the clinical relevance of this potential. Finally, our data on the IC50 of novobiocin for ATPase suggest that subtle differences between GyrB amino acid side chains from different species may lead to organism-specific target-based resistance; gyrB mutations I94A, A100S, and V120L are found in novobiocin-resistant S. aureus strains (44), and V120L is found in novobiocin-resistant S. pneumoniae strains (32) but not in novobiocin-resistant E. coli strains.
Acknowledgments
We thank S. Pazhanisamy, M. Namchuk, C. Gates, K. Tanner, N. Mani, D. Stamos, J. Thomson, M. Murcko, J. Partaledis, and M. Partridge for discussions and comments on the manuscript. Our special thanks go to Juliana Shan-Min Sun for technical assistance.
Jonathan D. Parsons and Christian H. Gross contributed equally to this work.
REFERENCES
- 1.Adachi, T., M. Mizuuchi, E. A. Robinson, E. Appella, M. H. O'Dea, M. Gellert, and K. Muzuuchi. 1987. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 15:771-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akasaka, T., M. Tanaka, A. Yamaguchi, and K. Sato. 2001. Type II topoisomerase mutation in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob. Agents Chemother. 45:2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali, J. A., A. P. Jackson, A. J. Howells, and A. Maxwell. 1993. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry 32:2717-2724. [DOI] [PubMed] [Google Scholar]
- 4.Brino, L., A. Urzhumtsev, M. Mousli, C. Bronner, A. Mitschler, P. Oudet, and D. Moras. 2000. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 275:9468-9475. [DOI] [PubMed] [Google Scholar]
- 5.Contreras, A., and A. Maxwell. 1992. GyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol. Microbiol. 6:1617-1624. [DOI] [PubMed] [Google Scholar]
- 6.Critchlow, S. E., M. H. O'Dea, A. J. Howells, M. Couturier, M. Gellert, and A. Maxwell. 1997. The interaction of the F-plasmid killer protein, CcdB, with DNA gyrase: induction of DNA cleavage and blocking transcription. J. Mol. Biol. 273:826-839. [DOI] [PubMed] [Google Scholar]
- 7.Cullis, P. M., A. Maxwell, and D. P. Weiner. 1997. Exploiting nucleotide thiophosphates to probe mechanistic aspects of Escherichia coli DNA gyrase. Biochemistry 36:6059-6068. [DOI] [PubMed] [Google Scholar]
- 8.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 9.del Castillo, I., J. L. Vizan, M. C. Rodriguez-Sainz, and F. Moreno. 1991. An unusual mechanism for resistance to the antibiotic coumermycin A1. Proc. Natl. Acad. Sci. USA 88:8860-8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fersht, A. 1985. Enzyme structure and mechanism, 2nd ed., p. 99. W. H. Freeman & Co., New York, N.Y.
- 11.Fox, T., J. T. Coll, X. Xie, P. J. Ford, U. Germann, M. D. Porter, S. Pazhanisamy, M. A. Fleming, V. Galullo, M. S. S. Su, and K. P. Wilson. 1998. A single amino acid substitution makes ERK2 susceptible to pyridinyl imidazole inhibitors of p38 MAP kinase. Protein Sci. 7:2249-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman, S. M., T. Lu, and K. Drlica. 2001. Mutation in the DNA gyrase A gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob. Agents Chemother. 45:2378-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellert, M., M. H. O'Dea, T. Itoh, and J. Tomizawa. 1976. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. USA 73:4474-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gormley, N. A., G. Orphanides, A. Meyer, P. M. Cullis, and A. Maxwell. 1996. The interaction of coumarin antibiotics with fragments of the DNA gyrase B protein. Biochemistry 35:5083-5092. [DOI] [PubMed] [Google Scholar]
- 15.Grossman, T. H., E. S. Kawasaki, S. R. Punreddy, and M. S. Osburne. 1998. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene 209:95-103. [DOI] [PubMed] [Google Scholar]
- 16.Grossman, T. H., M. Tuckman, S. Ellestad, and M. S. Osburne. 1993. Isolation and characterization of Bacillus subtilis genes involved in siderophore biosynthesis: relationship between B. subtilis sfp0 and Escherichia coli entD genes. J. Bacteriol. 175:6203-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holdgate, G. A., A. Tunnicliffe, W. H. J. Ward, S. A. Weston, G. Rosenbrock, P. T. Barth, I. W. F. Taylor, R. A. Pauptit, and D. Timms. 1997. The entropic penalty of ordered water accounts for weaker binding of the antibiotic novobiocin to a resistant mutant of DNA gyrase: a thermodynamic and crystallographic study. Biochemistry 36:9663-9673. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, M. L., and M. L. Dyall-Smith. 1991. Mutations in DNA gyrase result in novobiocin resistance in halophilic archaebacteria. J. Bacteriol. 173:642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooper, D. C. 1998. Bacterial topoisomerases, anti-topoisomerase resistance. Clin. Infect. Dis. 27(Suppl. 1):S54-S63. [DOI] [PubMed] [Google Scholar]
- 20.Hooper, D. C. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist. Update 2:38-55. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, A. P., and A. Maxwell. 1993. Identifying the catalytic residue of the ATPase reaction of DNA gyrase. Proc. Natl. Acad. Sci. USA 90:11232-11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kampranis, S. C., A. D. Bates, and A. Maxwell. 1999. A model for the mechanism of strand passage by DNA gyrase. Proc. Natl. Acad. Sci. USA 96:8414-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kampranis, S. C., and A. Maxwell. 1996. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. USA 93:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kampranis, S. C., and A. Maxwell. 1998. Hydrolysis of ATP at only one subunit: gyrB subunit is sufficient to promote supercoiling by DNA gyrase. J. Biol. Chem. 273:26305-26309. [DOI] [PubMed] [Google Scholar]
- 25.Kampranis, S. C., N. A. Gormley, R. Tranter, G. Orphanides, and A. Maxwell. 1999. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemistry 38:1967-1976. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, R. J., O. M. P. Singh, C. V. Smith, T. Skarzynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and cyclothilidine revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 27.Maxwell, A., and M. Gellert. 1984. The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 259:14472-14480. [PubMed] [Google Scholar]
- 28.Maxwell, A. 1997. DNA gyrase as a drug target. Trends Microbiol. 5:102-109. [DOI] [PubMed] [Google Scholar]
- 29.Menzel, R., and M. Gellert. 1983. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell 34:105-113. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Mizuuchi, K., L. M. Fisher, M. H. O'Dea, and M. Gellert. 1980. DNA gyrase action involves the introduction of transient double-stranded breaks into DNA. Proc. Natl. Acad. Sci. USA 77:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz, R., M. Bustamante, and A. G. De La Campa. 1995. Ser-127 to Leu substitution in the DNA gyrase B subunit of Streptococcus pneumoniae is implicated in novobiocin resistance. J. Bacteriol. 177:4166-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 276:382-388. [DOI] [PubMed] [Google Scholar]
- 34.O'Dea, M. H., J. K. Tamura, and M. Gellert. 1996. Mutations in the B subunit of Escherichi coli DNA gyrase that affect ATP-dependent reactions. J. Biol. Chem. 271:9723-9729. [DOI] [PubMed] [Google Scholar]
- 35.Orphanides, G., and A. Maxwell. 1994. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 22:1567-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orr, E., N. F. Fairweather, I. B. Holland, and R. H. Pritchard. 1979. Isolation and characterization of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol. Gen. Genet. 177:103-112. [DOI] [PubMed] [Google Scholar]
- 37.Roca, J., and J. C. Wang. 1994. DNA transport by type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell 77:609-616. [DOI] [PubMed] [Google Scholar]
- 38.Roca, J., and J. C. Wang. 1992. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71:833-840. [DOI] [PubMed] [Google Scholar]
- 39.Ryan, M. J. 1976. Coumermycin A1: a perferential inhibitor of replicative DNA synthesis in Escherichia coli. I. In vivo characterization. Biochemistry 15:3769-3777. [DOI] [PubMed] [Google Scholar]
- 40.Samuels, D. S., R. T. Marconi, W. M. Huang, and C. F. Garon. 1994. gyrB mutations in coumermycin A1-resistant Borrelia burgdorferi. J. Bacteriol. 176:3072-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smelkova, N., and K. J. Marians. 2001. Timely release of both replication forks from oriC requires modulation of origin topology. J. Biol. Chem. 276:39186-39191. [DOI] [PubMed] [Google Scholar]
- 42.Smith, C. E., and A. Maxwell. 1998. Identification of a residue involved in transition-state stabilization in the ATPase reaction of DNA gyrase. Biochemistry 37:9658-9667. [DOI] [PubMed] [Google Scholar]
- 43.Spratt, B. G. 1994. Resistance to antibiotics mediated by target alterations. Science 264:388-393. [DOI] [PubMed] [Google Scholar]
- 44.Stieger, M., P. Angehrn, B. Wohlgensinger, and H. Gmunder. 1996. GyrB mutations in Staphylococcus aureus strains resistant to cyclothialidines, coumermycin, and novobiocin. Antimicrob. Agents Chemother. 40:1060-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugino, A., P. Higgins, P. O. Brown, C. L. Peebles, and N. R. Cozzarelli. 1978. Energy coupling in DNA gyrase and mechanism of action of novobiocin. Proc. Natl. Acad. Sci. USA 75:4838-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tingey, A. P., and A. Maxwell. 1996. Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 24:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wigley, D. B., G. J. Davies, E. J. Dodson, A. Maxwell, and G. Dudson. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624-629. [DOI] [PubMed] [Google Scholar]
- 48.Williams, N., A. J. Howells, and A. Maxwell. 2001. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 306:969-984. [DOI] [PubMed] [Google Scholar]
- 49.Williams, N., and A. Maxwell. 1999. Locking the DNA gate of DNA gyrase: investigating the effects on DNA cleavage and ATP hydrolysis. Biochemistry 38:14157-14164. [DOI] [PubMed] [Google Scholar]
- 50.Williams, N., and A. Maxwell. 1999. Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry 38:13502-13511. [DOI] [PubMed] [Google Scholar]