Abstract
The in vitro antibacterial activity of S-3578, a new parenteral cephalosporin, against clinical isolates was evaluated. The MICs of the drug at which 90% of the isolates were inhibited were 4 μg/ml for methicillin-resistant Staphylococcus aureus (MRSA) and 2 μg/ml for methicillin-resistant Staphylococcus epidermidis, which were fourfold higher than and equal to those of vancomycin, respectively. The anti-MRSA activity of S-3578 was considered to be due to its high affinity for penicillin-binding protein 2a (50% inhibitory concentration, 4.5 μg/ml). In time-kill studies with 10 strains each of MRSA and methicillin-susceptible S. aureus, S-3578 caused more than a 4-log10 decrease of viable cells on the average at twice the MIC after 24 h of exposure, indicating that it had potent bactericidal activity. Furthermore, in population analysis of MRSA strains with heterogeneous or homogeneous resistance to imipenem, no colonies emerged from about 109 cells on agar plates containing twice the MIC of S-3578, suggesting the low frequency of emergence of S-3578-resistant strains from MRSA. S-3578 was also highly active against penicillin-resistant Streptococcus pneumoniae (PRSP), with a MIC90 of 1 μg/ml, which was comparable to that of ceftriaxone. S-3578 also had antibacterial activity against a variety of gram-negative bacteria including Pseudomonas aeruginosa, though its activity was not superior to that of cefepime. In conclusion, S-3578 exhibited a broad antibacterial spectrum and, particularly, had excellent activity against gram-positive bacteria including methicillin-resistant staphylococci and PRSP. Thus, S-3578 was considered to be worthy of further evaluation.
Methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant coagulase-negative staphylococci (MRCNS) cause serious problems in nosocomial infection. The major factor in the mechanism of resistance to β-lactams in MRSA and MRCNS is known to be the production of a penicillin-binding protein peculiar to them, designated PBP2′ or PBP2a, which has low affinity for commercially available β-lactams (8). Furthermore, most clinical isolates of MRSA and MRCNS have acquired resistance to not only β-lactams but also various other antibiotics used in clinical settings. The worldwide spread and increased frequency of occurrence of these pathogens have caused difficulties in therapeutic treatment with antibiotics (5, 25).
Some antibiotics such as vancomycin, linezolid, or dalfopristin-quinupristin show clinical efficacy against infections caused by methicillin-resistant staphylococci (1, 6, 14) but have antibacterial activity against only gram-positive bacteria. Much effort has been spent on the development of β-lactams with anti-MRSA activity in laboratories worldwide (10, 12, 24). Some compounds, for example, RWJ-54428 (MC-02479) or BMS-247243, have been reported elsewhere as anti-MRSA agents, but most of them also exhibit narrow-spectrum antibacterial activity (7, 13). On the other hand, broad-spectrum cephalosporins like ceftriaxone or cefepime have been useful against various infectious diseases but are not active against methicillin-resistant staphylococci. BAL9141 (formerly Ro-63-9141), which is under development, has been reported elsewhere to have satisfactory broad-spectrum activity with anti-MRSA activity (9). However, it does not have good water solubility and requires esterification to improve its solubility. With the aim of developing a broad-spectrum cephalosporin with anti-MRSA activity, we selected S-3578, 7β-[2-(2-aminothiadiazol-4-yl)-2(Z)-ethoxyiminoacetamido]-3-(1-N-methylaminopropyl-1H-imidazo[4,5-b]pyridinium-4-yl)-methylcephalosporin, because of its antibacterial activities, aqueous solubility, and crystallinity (26).
In this study, we evaluated the in vitro antibacterial activity of S-3578, particularly focusing on its anti-MRSA activity.
(Part of this work was presented previously [Y. Yamano, H. Miwa, K. Motokawa, T. Yoshida, J. Shimada, and S. Kuwahara, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-371, 2001; T. Fujimura, Y. Yamano, I. Yoshida, T. Yoshida, J. Shimada, and S. Kuwahara, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-372, 2001].)
MATERIALS AND METHODS
Antibiotics.
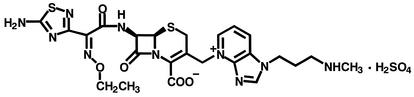
S-3578 was synthesized in the research laboratories of Shionogi & Co., Ltd. (Osaka, Japan). Its chemical structure is shown in Fig. 1. Other compounds were purchased from commercial sources.
FIG. 1.
Chemical structure of S-3578, 7β-[2-(2-aminothiadiazol-4-yl)-2(Z)-ethoxyiminoacetamido]-3-(1-N-methylaminopropyl-1H-imidazo[4,5-b]pyridinium-4-yl)-methylcephalosporin.
Organisms.
Bacterial strains used in this study were clinical isolates collected from hospitals in various parts of Japan in 2000 and 2001 and strains from our laboratory collection. S. aureus SRM710, deficient in PBP2, was used in a binding affinity assay of PBP2a of MRSA (18).
MIC determination.
MICs for aerobic bacteria were determined by a microdilution broth method as recommended by the NCCLS (20) with cation-adjusted Mueller-Hinton broth (CA-MHB) (Becton Dickinson, Sparks, Md.), which was supplemented with 15 μg of NAD/ml, 5% yeast extract (Becton Dickinson), and 5% lysed horse blood to support the growth of Streptococcus pneumoniae and Haemophilus influenzae. CA-MHB with 2% NaCl was used to determine MICs of oxacillin for staphylococci. MICs for Neisseria gonorrhoeae were determined by an agar dilution method with GC medium (Becton Dickinson) containing growth supplement after incubation at 35°C for 20 h in the presence of 6% CO2. MICs for anaerobic bacteria were determined by an agar dilution method with Wilkins-Chalgren medium (Becton Dickinson) after anaerobic incubation at 35°C for 48 h.
Determination of minimum bactericidal concentration (MBC).
MBCs were determined by a macrodilution method with CA-MHB according to the NCCLS recommendations (19).
Time-kill study.
The culture of a test strain at the early or mid-log phase was diluted to about 5 × 105 CFU/ml in 5 ml of fresh CA-MHB. Antibiotics at various concentrations were added to the diluted cultures. Viable cells were counted after shaking at 37°C for 0, 1, 2.5, 4, and 6 h. In the studies of S. aureus, the additional counting was performed at 24 h.
PBP binding affinity.
The affinities of antibiotics for PBPs were determined by a competition assay with [14C]benzylpenicillin (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England) as described previously (17, 23). The binding affinity of each compound was expressed as the concentration required to inhibit 50% of the binding of [14C]benzylpenicillin to each PBP (IC50).
Population analysis and frequency of emergence of resistant colonies.
An overnight culture of S. aureus in CA-MHB was diluted with fresh medium to the appropriate bacterial density and spread onto a Mueller-Hinton agar plate containing serial twofold dilutions of antibiotics. The plates were incubated for 3 days at 37°C, and the number of colonies was counted. The frequency of emergence of resistant colonies on an agar plate containing S-3578 was shown as their ratio to the number of colonies grown on an agar plate without antibiotics.
β-Lactamase stability.
β-Lactamases were purified partially from bacterial membrane by ion-exchange chromatography as described previously (16). The stability of antibiotics against hydrolysis by β-lactamases was assessed by determining the hydrolysis rate by a spectrophotometric assay method (21). The kinetics parameters Km and Vmax were calculated from at least two independent experiments with the Michaelis-Menten formulation.
RESULTS
MIC determination.
The antibacterial activity of S-3578 was compared with those of cefepime, ceftriaxone, ceftazidime, and imipenem. For gram-positive bacteria, vancomycin was also used as a comparator, as were oxacillin for staphylococci and penicillin G for S. pneumoniae. The results are shown in Table 1 as the MIC range, MIC50 (the concentration at which the growth of 50% of the isolates was inhibited), and MIC90.
TABLE 1.
In vitro antibacterial activities of S-3578 and reference compounds against clinical isolates
| Organism(s) (no. of isolates) and compound | MIC (μg/ml)
|
||
|---|---|---|---|
| Range | MIC50 | MIC90 | |
| MSSA (84) | |||
| S-3578 | 0.5-2 | 1 | 2 |
| Ceftriaxone | 1-8 | 4 | 4 |
| Cefepime | 1-4 | 4 | 4 |
| Ceftazidime | 4-32 | 16 | 16 |
| Imipenem | ≦0.06-≦0.06 | ≦0.06 | ≦0.06 |
| Vancomycin | 0.5-2 | 1 | 1 |
| Oxacillin | 0.25-2 | 0.5 | 1 |
| MRSA (134) | |||
| S-3578 | 1-8 | 4 | 4 |
| Ceftriaxone | 8->64 | >64 | >64 |
| Cefepime | 8->64 | >64 | >64 |
| Ceftazidime | 32->64 | >64 | >64 |
| Imipenem | ≦0.06->64 | 64 | 64 |
| Vancomycin | 0.5-2 | 1 | 1 |
| Oxacillin | 8->64 | >64 | >64 |
| MSSEa (28) | |||
| S-3578 | ≦0.06-1 | 0.5 | 0.5 |
| Ceftriaxone | 0.25-2 | 1 | 2 |
| Cefepime | 0.13-2 | 0.5 | 1 |
| Ceftazidime | 2-8 | 4 | 8 |
| Imipenem | ≦0.06-≦0.06 | ≦0.06 | ≦0.06 |
| Vancomycin | 0.5-2 | 1 | 2 |
| Oxacillin | ≦0.06-0.13 | 0.13 | 0.13 |
| MRSE (105) | |||
| S-3578 | 0.5-4 | 1 | 2 |
| Ceftriaxone | 4->64 | 32 | >64 |
| Cefepime | 2->64 | 16 | >64 |
| Ceftazidime | 8->64 | 32 | >64 |
| Imipenem | ≦0.06->64 | 8 | 64 |
| Vancomycin | 1-2 | 1 | 2 |
| Oxacillin | 1->64 | 32 | >64 |
| Staphylococcus haemolyticus, methicillin susceptible (7) | |||
| S-3578 | 0.25-0.5 | ||
| Ceftriaxone | 8-32 | ||
| Cefepime | 1-4 | ||
| Ceftazidime | 8-64 | ||
| Imipenem | ≦0.06-≦0.06 | ||
| Vancomycin | 0.5-1 | ||
| Oxacillin | 0.13-0.25 | ||
| Staphylococcus haemolyticus, methicillin resistant (42) | |||
| S-3578 | 0.5-8 | 1 | 4 |
| Ceftriaxone | 16->64 | 64 | >64 |
| Cefepime | 4->64 | 32 | >64 |
| Ceftazidime | 16->64 | >64 | >64 |
| Imipenem | ≦0.06->64 | 0.5 | >64 |
| Vancomycin | 1-4 | 2 | 4 |
| Oxacillin | 0.5->64 | >64 | >64 |
| Staphylococcus capitis (25) | |||
| S-3578 | 0.13-2 | 1 | 1 |
| Ceftriaxone | 0.5->64 | 64 | >64 |
| Cefepime | 0.25->64 | 32 | >64 |
| Ceftazidime | 2->64 | 64 | >64 |
| Imipenem | ≦0.06-64 | 1 | 64 |
| Vancomycin | 0.5-2 | 1 | 2 |
| Oxacillin | ≦0.06->64 | >64 | >64 |
| Staphylococcus saprophyticus (30) | |||
| S-3578 | 0.5-2 | 1 | 1 |
| Ceftriaxone | 8->64 | 16 | 32 |
| Cefepime | 2->64 | 2 | 4 |
| Ceftazidime | 32->64 | 32 | 64 |
| Imipenem | ≦0.06-1 | ≦0.06 | ≦0.06 |
| Vancomycin | 1-2 | 1 | 2 |
| Oxacillin | 0.5->64 | 1 | 2 |
| Staphylococcus warneri (39) | |||
| S-3578 | 0.13-1 | 0.5 | 0.5 |
| Ceftriaxone | 0.5-16 | 2 | 4 |
| Cefepime | 0.25-2 | 1 | 2 |
| Ceftazidime | 4-32 | 8 | 16 |
| Imipenem | ≦0.06-≦0.06 | ≦0.06 | ≦0.06 |
| Vancomycin | 0.5-2 | 1 | 2 |
| Oxacillin | 0.13-8 | 0.25 | 0.5 |
| Staphylococcus lugdunensis (21) | |||
| S-3578 | 0.5-2 | 1 | 1 |
| Ceftriaxone | 4-64 | 4 | 4 |
| Cefepime | 1-16 | 2 | 2 |
| Ceftazidime | 8-64 | 16 | 16 |
| Imipenem | ≦0.06-0.25 | ≦0.06 | ≦0.06 |
| Vancomycin | 0.5-2 | 1 | 2 |
| Oxacillin | 0.25->64 | 0.5 | 0.5 |
| Staphylococcus caprae (8), Staphylococcus hominis (2), Staphylococcus simulans (1), Staphylococcus schleifrei (1), Staphylococcus cohnii (1), Staphylococcus hyicus (1) | |||
| S-3578 | 0.13-1 | 0.25 | 1 |
| Ceftriaxone | 1->64 | 4 | >64 |
| Cefepime | 0.25->64 | 0.5 | >64 |
| Ceftazidime | 4->64 | 4 | >64 |
| Imipenem | ≦0.06-32 | ≦0.06 | 32 |
| Vancomycin | 0.5-2 | 1 | 1 |
| Oxacillin | 0.13->64 | 0.25 | >64 |
| Streptococcus pyogenes (60) | |||
| S-3578 | ≦0.03-≦0.03 | ≦0.03 | ≦0.03 |
| Ceftriaxone | ≦0.03-≦0.03 | ≦0.03 | ≦0.03 |
| Cefepime | ≦0.03-0.06 | ≦0.03 | ≦0.03 |
| Ceftazidime | 0.13-0.25 | 0.13 | 0.25 |
| Imipenem | ≦0.03-≦0.03 | ≦0.03 | ≦0.03 |
| Vancomycin | 0.5-0.5 | 0.5 | 0.5 |
| Streptococcus agalactiae (68) | |||
| S-3578 | ≦0.03-0.06 | 0.06 | 0.06 |
| Ceftriaxone | ≦0.03-0.13 | 0.06 | 0.06 |
| Cefepime | 0.06-0.13 | 0.06 | 0.13 |
| Ceftazidime | 0.25-1 | 0.5 | 0.5 |
| Imipenem | ≦0.03-≦0.03 | ≦0.03 | ≦0.03 |
| Vancomycin | 0.5-1 | 1 | 1 |
| Streptococcus mitis group (43) | |||
| S-3578 | ≦0.03->32 | 0.13 | 8 |
| Ceftriaxone | ≦0.03-16 | 0.13 | 8 |
| Cefepime | ≦0.03->32 | 0.13 | 4 |
| Ceftazidime | 0.06->32 | 2 | >32 |
| Imipenem | ≦0.03-4 | ≦0.03 | 2 |
| Vancomycin | 0.25-2 | 0.5 | 1 |
| Streptococcus anginosus group (48) | |||
| S-3578 | 0.13-0.5 | 0.25 | 0.5 |
| Ceftriaxone | 0.06-0.5 | 0.25 | 0.25 |
| Cefepime | 0.13-1 | 0.5 | 0.5 |
| Ceftazidime | 1-8 | 4 | 8 |
| Imipenem | ≦0.03-0.06 | ≦0.03 | ≦0.03 |
| Vancomycin | 0.5-1 | 1 | 1 |
| Streptococcus pneumoniae, penicillin susceptible (61) | |||
| S-3578 | ≦0.03-0.5 | 0.06 | 0.25 |
| Ceftriaxone | ≦0.03-0.5 | 0.13 | 0.25 |
| Cefepime | ≦0.03-1 | 0.13 | 0.5 |
| Ceftazidime | 0.06-16 | 2 | 8 |
| Imipenem | ≦0.03-0.06 | ≦0.03 | ≦0.03 |
| Vancomycin | 0.25-0.5 | 0.5 | 0.5 |
| Penicillin G | ≦0.03-0.06 | ≦0.03 | 0.06/PICK> |
| Streptococcus pneumoniae, penicillin intermediate (53) | |||
| S-3578 | ≦0.03-2 | 0.5 | 1 |
| Ceftriaxone | ≦0.03-4 | 0.5 | 1 |
| Cefepime | 0.13-2 | 0.5 | 1 |
| Ceftazidime | 0.25-16 | 8 | 16 |
| Imipenem | ≦0.03-0.13 | 0.06 | 0.13 |
| Vancomycin | 0.25-0.5 | 0.25 | 0.5 |
| Penicillin G | 0.13-1 | 0.5 | 1 |
| Streptococcus pneumoniae, penicillin resistant (35) | |||
| S-3578 | 0.5-2 | 1 | 1 |
| Ceftriaxone | 0.5-2 | 1 | 1 |
| Cefepime | 0.5-2 | 1 | 2 |
| Ceftazidime | 8-32 | 8 | 16 |
| Imipenem | 0.13-0.5 | 0.25 | 0.25 |
| Vancomycin | 0.25-0.5 | 0.5 | 0.5 |
| Penicillin G | 2-4 | 2 | 2 |
| Enterococcus faecalis (61) | |||
| S-3578 | 4->64 | 16 | >64 |
| Ceftriaxone | 64->64 | >64 | >64 |
| Cefepime | 16->64 | 64 | >64 |
| Ceftazidime | >64->64 | >64 | >64 |
| Imipenem | 0.5-8 | 1 | 2 |
| Vancomycin | 0.5-4 | 1 | 2 |
| Peptostreptococcus magnus (4), Peptostreptococcus asaccharolyticus (4), Peptostreptococcus hydrogenalis (3), Peptostreptococcus micros (7) | |||
| S-3578 | 0.06-8 | 0.13 | 4 |
| Ceftriaxone | 0.06-8 | 0.13 | 2 |
| Cefepime | 0.06-16 | 0.25 | 8 |
| Ceftazidime | 0.25-32 | 1 | 8 |
| Imipenem | ≦0.016-0.06 | ≦0.016 | 0.03 |
| Vancomycin | 0.13-1 | 0.5 | 1 |
| Escherichia coli (164) | |||
| S-3578 | 0.06->64 | 0.25 | 0.5 |
| Ceftriaxone | ≦0.016->64 | 0.06 | 0.13 |
| Cefepime | ≦0.016->64 | 0.03 | 0.13 |
| Ceftazidime | 0.06->64 | 0.13 | 0.5 |
| Imipenem | 0.06-2 | 0.13 | 0.25 |
| Klebsiella pneumoniae (87) | |||
| S-3578 | ≦0.06->64 | 0.25 | 1 |
| Ceftriaxone | ≦0.06->64 | ≦0.06 | 0.13 |
| Cefepime | ≦0.06-16 | ≦0.06 | 0.13 |
| Ceftazidime | ≦0.06-4 | 0.13 | 0.5 |
| Imipenem | ≦0.06-0.5 | 0.13 | 0.25 |
| Klebsiella oxytoca (62) | |||
| S-3578 | ≦0.06-8 | 0.13 | 1 |
| Ceftriaxone | ≦0.06-64 | ≦0.06 | 0.13 |
| Cefepime | ≦0.06-2 | ≦0.06 | 0.13 |
| Ceftazidime | ≦0.06-1 | 0.13 | 0.5 |
| Imipenem | ≦0.06-2 | 0.13 | 0.25 |
| Enterobacter cloacae (84) | |||
| S-3578 | ≦0.06->64 | 0.5 | 16 |
| Ceftriaxone | ≦0.06->64 | 0.5 | >64 |
| Cefepime | ≦0.06-32 | ≦0.06 | 1 |
| Ceftazidime | ≦0.06->64 | 0.5 | 64 |
| Imipenem | ≦0.06-4 | 1 | 2 |
| Enterobacter aerogenes (51) | |||
| S-3578 | ≦0.06-8 | 0.25 | 2 |
| Ceftriaxone | ≦0.06->64 | 0.25 | 32 |
| Cefepime | ≦0.06-4 | ≦0.06 | 0.5 |
| Ceftazidime | 0.13->64 | 0.5 | 64 |
| Imipenem | 0.13-2 | 1 | 2 |
| Citrobacter freundii (42) | |||
| S-3578 | 0.13-16 | 0.25 | 8 |
| Ceftriaxone | ≦0.06->64 | 0.25 | >64 |
| Cefepime | ≦0.06-2 | ≦0.06 | 1 |
| Ceftazidime | 0.13->64 | 0.5 | >64 |
| Imipenem | ≦0.06-1 | 0.25 | 0.5 |
| Serratia marcescens (93) | |||
| S-3578 | 0.13->64 | 0.5 | 8 |
| Ceftriaxone | ≦0.06->64 | 0.25 | 32 |
| Cefepime | ≦0.06->64 | 0.13 | 2 |
| Ceftazidime | 0.13->64 | 0.25 | 4 |
| Imipenem | 0.13-16 | 0.5 | 2 |
| Proteus mirabilis (62) | |||
| S-3578 | 0.5->64 | 2 | 4 |
| Ceftriaxone | ≦0.06->64 | ≦0.06 | ≦0.06 |
| Cefepime | ≦0.06->64 | ≦0.06 | ≦0.06 |
| Ceftazidime | ≦0.06-0.25 | ≦0.06 | 0.13 |
| Imipenem | 0.5-8 | 2 | 4 |
| Proteus vulgaris (46) | |||
| S-3578 | 1->64 | 8 | 64 |
| Ceftriaxone | ≦0.06->64 | >64 | >64 |
| Cefepime | ≦0.06-8 | 0.13 | 0.5 |
| Ceftazidime | ≦0.06-2 | ≦0.06 | 0.25 |
| Imipenem | 0.5-8 | 2 | 4 |
| Morganella morganii (59) | |||
| S-3578 | 0.25-32 | 0.5 | 16 |
| Ceftriaxone | ≦0.06-8 | ≦0.06 | 4 |
| Cefepime | ≦0.06-0.25 | ≦0.06 | ≦0.06 |
| Ceftazidime | ≦0.06-32 | 0.5 | 16 |
| Imipenem | 0.5-4 | 2 | 4 |
| Providencia rettgeri (27) | |||
| S-3578 | ≦0.06->64 | 0.25 | 8 |
| Ceftriaxone | ≦0.06->64 | ≦0.06 | >64 |
| Cefepime | ≦0.06->64 | ≦0.06 | 2 |
| Ceftazidime | ≦0.06->64 | 0.13 | >64 |
| Imipenem | 0.5-2 | 1 | 2 |
| Haemophilus influenzae, ampicillin susceptible (56) | |||
| S-3578 | ≦0.06-0.25 | 0.13 | 0.25 |
| Ceftriaxone | ≦0.06-0.13 | ≦0.06 | ≦0.06 |
| Cefepime | ≦0.06-0.25 | ≦0.06 | 0.13 |
| Ceftazidime | ≦0.06-0.5 | 0.13 | 0.25 |
| Imipenem | 0.13-4 | 1 | 2 |
| Ampicillin | 0.13-1 | 0.25 | 0.5 |
| Haemophilus influenzae, β-lactamase producing (7) | |||
| S-3578 | ≦0.06-0.5 | ||
| Ceftriaxone | ≦0.06-≦0.06 | ||
| Cefepime | ≦0.06-0.25 | ||
| Ceftazidime | ≦0.06-0.5 | ||
| Imipenem | 0.13-2 | ||
| Ampicillin | 4->64 | ||
| Haemophilus influenzae, BLNARb (37) | |||
| S-3578 | 0.25-8 | 2 | 8 |
| Ceftriaxone | ≦0.06-0.5 | ≦0.06 | 0.25 |
| Cefepime | ≦0.06-2 | 0.5 | 2 |
| Ceftazidime | ≦0.06-1 | 0.25 | 0.5 |
| Imipenem | 0.25-64 | 4 | 16 |
| Ampicillin | 2-32 | 2 | 16 |
| Moraxella catarrhalis (53) | |||
| S-3578 | ≦0.06-4 | 0.5 | 1 |
| Ceftriaxone | ≦0.06-32 | 0.5 | 2 |
| Cefepime | ≦0.06-16 | 1 | 2 |
| Ceftazidime | ≦0.06-8 | 0.13 | 0.25 |
| Imipenem | ≦0.06-0.13 | ≦0.06 | 0.13 |
| Pseudomonas aeruginosa (108) | |||
| S-3578 | 0.5->64 | 8 | 64 |
| Ceftriaxone | 2->64 | 64 | >64 |
| Cefepime | 0.5->64 | 4 | 16 |
| Ceftazidime | 1->64 | 2 | 32 |
| Imipenem | ≦0.06->64 | 1 | 16 |
| Burkholderia cepacia (21) | |||
| S-3578 | 32->64 | >64 | >64 |
| Ceftriaxone | 16->64 | 64 | >64 |
| Cefepime | 16->64 | 32 | >64 |
| Ceftazidime | 2-32 | 4 | 16 |
| Imipenem | 4-32 | 16 | 16 |
| Acinetobacter baumannii (40) | |||
| S-3578 | 0.13->64 | 1 | 16 |
| Ceftriaxone | 8->64 | 16 | 64 |
| Cefepime | 0.5->64 | 4 | 32 |
| Ceftazidime | 2->64 | 8 | 32 |
| Imipenem | 0.13-64 | 0.25 | 0.25 |
| Neisseria gonorrhoeae (21) | |||
| S-3578 | 0.06->64 | 4 | 32 |
| Ceftriaxone | 0.008-0.5 | 0.06 | 0.25 |
| Cefepime | 0.016-16 | 0.5 | 8 |
| Ceftazidime | 0.06-4 | 1 | 2 |
| Imipenem | 0.06-2 | 0.5 | 2 |
| Penicillin G | 0.25->64 | 4 | 16 |
| Bacteroides fragilis (41) | |||
| S-3578 | 4->128 | 64 | >128 |
| Ceftriaxone | 4->128 | 16 | 128 |
| Cefepime | 4->128 | 64 | >128 |
| Ceftazidime | 4->128 | 32 | >128 |
| Imipenem | 0.06-4 | 0.13 | 1 |
MSSE, methicillin-susceptible S. epidermidis.
BLNAR, β-lactamase-negative ampicillin-resistant strain.
S-3578 showed consistent antibacterial activity against staphylococci including methicillin-resistant strains and inhibited the growth of all clinical isolates of staphylococci at 8 μg/ml. The MIC90s of S-3578 for MRSA and methicillin-resistant Staphylococcus epidermidis (MRSE) were 4 and 2 μg/ml, respectively, whereas those of other β-lactam compounds were 64 μg/ml or more. S-3578 was fourfold less active against MRSA than was vancomycin, but its activity against MRSE was equal to that of vancomycin. For methicillin-susceptible S. aureus (MSSA) and methicillin-susceptible S. epidermidis, the MIC90s of S-3578 were 2 and 0.5 μg/ml, respectively. It was characteristic that the MICs of S-3578 for methicillin-resistant staphylococcus strains were only two- to fourfold higher than those for methicillin-susceptible staphylococcus strains.
S-3578 had activities against penicillin-susceptible, -intermediate, and -resistant S. pneumoniae, with MIC90s of 0.25, 1, and 1 μg/ml, respectively, which were comparable to those of ceftriaxone and cefepime. However, S-3578 was not as active against Enterococcus faecalis as were other cephalosporins, though it showed the lowest MIC50 among the cephalosporins tested.
S-3578 was also active against a variety of gram-negative bacteria, although its activity was not superior to those of cefepime and imipenem, except that imipenem was less active than was S-3578 against H. influenzae. The MIC90s of S-3578 for Escherichia coli, Klebsiella pneumoniae, and Klebsiella oxytoca were 1 μg/ml or less, which was comparable to those of ceftazidime but two- to eightfold higher than those of cefepime, ceftriaxone, and imipenem. Some S-3578-resistant strains of these species were also resistant to other cephalosporins. The MIC90s of S-3578 for Enterobacter aerogenes, Citrobacter freundii, Serratia marcescens, Proteus mirabilis, and Providencia rettgeri ranged from 2 to 8 μg/ml, and most strains of Proteus vulgaris were not susceptible to S-3578. S-3578 was active against H. influenzae except for β-lactamase-negative ampicillin-resistant strains with a MIC90 of 0.25 μg/ml, regardless of the β-lactamase production. Its activity was comparable to those of cefepime and ceftazidime but lower than that of ceftriaxone. The activity of S-3578 against Moraxella catarrhalis (MIC90, 1 μg/ml) was higher than those of ceftriaxone and cefepime but fourfold lower than that of ceftazidime. S-3578 showed activity against Pseudomonas aeruginosa with a MIC50 and a MIC90 of 8 and 64 μg/ml, respectively, which were two- to fourfold higher than those of ceftazidime and cefepime. The activity of S-3578 against Acinetobacter baumannii was the highest among the cephalosporins tested, though its MIC50 and MIC90 were 1 and 16 μg/ml, respectively. S-3578, however, had the same potential against N. gonorrhoeae as did penicillin G, though other compounds were more active.
The antibacterial activities against anaerobic pathogens were determined. S-3578 inhibited growth of all strains of Peptostreptococcus spp. at 8 μg/ml, which meant that it was as active as ceftriaxone was. Bacteroides fragilis was not susceptible to S-3578.
Bactericidal activity.
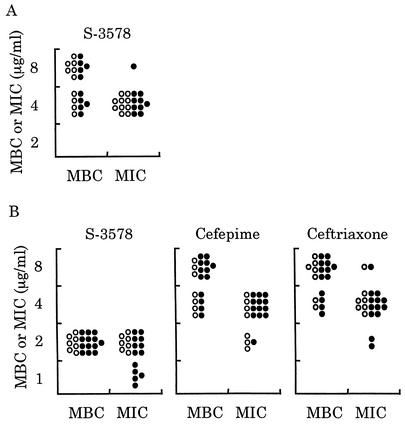
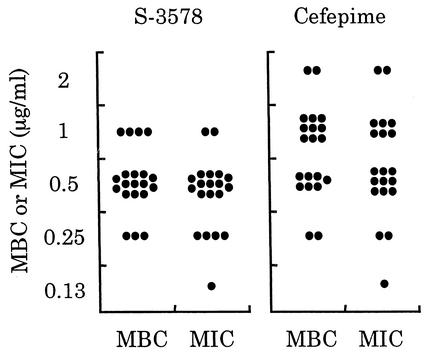
We first determined MBCs for 20 clinical isolates each of MRSA and MSSA including β-lactamase producers. The MBC/MIC ratios for S-3578 against all strains were 1 or 2 (Fig. 2). In particular, the number of MSSA strains showing the ratio of 1 for S-3578 was larger than the number of those showing the same ratio for cefepime and ceftriaxone, suggesting that S-3578 was more bactericidal than the reference agents were. The production of β-lactamase did not have any influence on both MBCs and MICs of S-3578 as well as of the reference compounds.
FIG. 2.
Distribution of MBCs and MICs for 20 clinical isolates each of MRSA (A) or MSSA (B). The activity of S-3578 was compared with those of cefepime and ceftriaxone against MSSA. Open and closed circles indicate β-lactamase-producing and nonproducing strains, respectively.
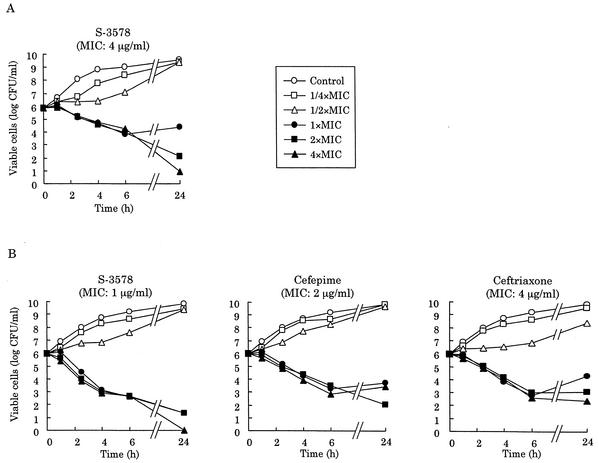
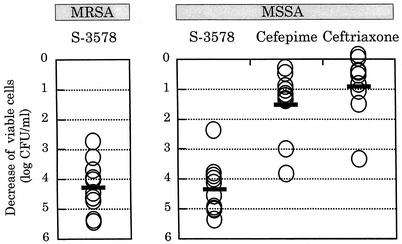
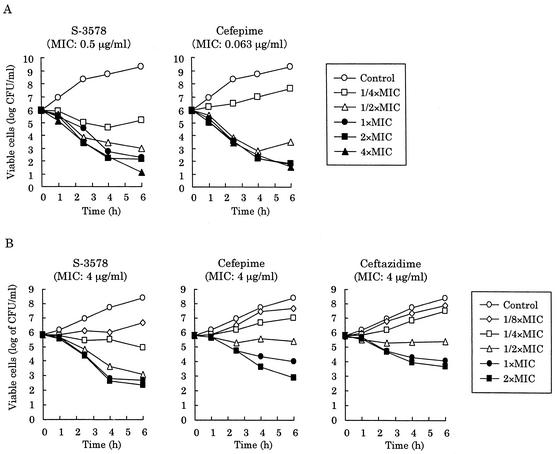
Next, we constructed the killing kinetics curves for MRSA and MSSA strains. S-3578 caused a time-dependent decrease of viable cells of the MSSA strain Smith at one or more times the MIC until 6 h, as did cefepime and ceftriaxone (Fig. 3B). Furthermore, the number of viable cells after 24-h exposure to S-3578 was significantly lower than the number after exposure to the reference compounds. S-3578 also showed similar bactericidal activity against the MRSA strain SR3637 at two or more times the MIC (Fig. 3A). Moreover, the decreases of viable cells with 10 clinical isolates each of MRSA or MSSA were compared after 24-h exposure to twice the MIC of each antibiotic. S-3578 caused more than a 4-log10-CFU/ml reduction on the average for both MRSA and MSSA, while cefepime and ceftriaxone demonstrated only less than a 2-log10-CFU/ml reduction for MSSA (Fig. 4). These results indicated that the bactericidal activity of S-3578 was more potent than those of cefepime and ceftriaxone against S. aureus, irrespective of methicillin resistance.
FIG. 3.
Killing kinetics curves for S-3578 and reference compounds against S. aureus SR3637 (MRSA) (A) and Smith (MSSA) (B).
FIG. 4.
Decrease of viable cells in 10 clinical isolates each of MRSA or MSSA after 24-h exposure to twice the MIC of S-3578. Cefepime and ceftriaxone were used as comparators for MSSA. An open circle indicates decrease of viable cell counts of each strain compared to the initial inoculum, and a bar indicates the average of those of 10 strains.
The MBCs of S-3578 for 20 clinical isolates of E. coli for which MICs were 0.13 to 1 μg/ml ranged from 0.25 to 1 μg/ml, while the MBCs and MICs of cefepime ranged from 0.25 to 2 μg/ml and from 0.13 to 2 μg/ml, respectively (Fig. 5). All strains tested were β-lactamase producers, indicating that the activity of S-3578 was not affected by β-lactamase. Furthermore, the killing kinetics curves for E. coli NIHJ JC-2 and P. aeruginosa SR24706 indicated that S-3578 also had time-dependent bactericidal activity against these species which was comparable to those of the reference compounds (Fig. 6).
FIG. 5.
Distribution of MBCs and MICs for 20 clinical isolates of E. coli. The activity of S-3578 was compared with that of cefepime. All strains produced β-lactamases.
FIG. 6.
Killing kinetics curves for S-3578 and reference compounds against E. coli NIHJ JC-2 (A) and P. aeruginosa SR24706 (B).
Binding affinity for PBPs.
S-3578 was found to have high affinity for PBP2a of MRSA, with an IC50 of 4.5 μg/ml (Table 2). This characteristic seemed to reflect the anti-MRSA activity of S-3578. In contrast, cefepime, which was not active against MRSA, had a very low affinity for PBP2a. S-3578 also showed high affinities for PBP1 and -2 of S. aureus with IC50s of less than 0.5 μg/ml, as did ceftriaxone and cefepime.
TABLE 2.
Binding affinity for PBPs of S. aureus
| Compound | IC50 (μg/ml)a
|
MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|
| PBP1 | PBP2a | PBP2 | PBP3 | PBP4 | Smith | SRM710 | |
| S-3578 | 0.28 | 4.5 | 0.34 | 4.2 | 16 | 1 | 2 |
| Cefepime | 0.37 | 410 | 0.12 | >16 | >16 | 2 | 64 |
| Ceftriaxone | 0.31 | NTb | 0.11 | 2.0 | >16 | 4 | NT |
IC50 was determined by a competition assay with the membrane fraction of S. aureus SRM710 (MRSA) for PBP2a and that of S. aureus Smith (MSSA) for other PBPs.
NT, not tested.
Cefepime had high affinities for PBP2 and -3 of E. coli with IC50s of less than 1 μg/ml, and ceftriaxone had affinities for PBP1A, -1B, -2, and -3 (Table 3). On the other hand, S-3578 had high affinity only for PBP3. Although the IC50s of S-3578 for PBP1A, -3, and -4 of P. aeruginosa ranged from 0.31 to 0.43 μg/ml (Table 4), its binding affinity was not as high as those of cefepime and ceftazidime, the IC50s of which for PBP3 were 10-fold less than that of S-3578.
TABLE 3.
Binding affinity for PBPs of E. coli
| Compound | IC50 (μg/ml)
|
MIC (μg/ml)a | ||||||
|---|---|---|---|---|---|---|---|---|
| PBP1A | PBP1B | PBP2 | PBP3 | PBP4 | PBP5 | PBP6 | ||
| S-3578 | 5.8 | 4.3 | >16 | 0.18 | >16 | >16 | >16 | 0.25 |
| Cefepime | >4 | 3.4 | 0.29 | 0.021 | >4 | >4 | >4 | 0.03 |
| Ceftriaxone | 0.092 | 0.24 | 0.30 | 0.013 | >4 | >4 | >4 | 0.06 |
MIC for E. coli NIHJ JC-2, the membrane fraction of which was employed for determination of IC50s.
TABLE 4.
Binding affinity for PBPs of P. aeruginosa
| Compound | IC50 (μg/ml)
|
MIC (μg/ml)a | ||||||
|---|---|---|---|---|---|---|---|---|
| PBP1A | PBP1B | PBP2 | PBP3 | PBP4 | PBP5 | PBP6 | ||
| S-3578 | 0.34 | 5.0 | >16 | 0.31 | 0.43 | >16 | >16 | 1 |
| Cefepime | 0.076 | 7.6 | >16 | 0.024 | 0.79 | >16 | >16 | 0.5 |
| Ceftazidime | 0.11 | 5.1 | 5.3 | 0.022 | 1.6 | >16 | >16 | 1 |
MIC for P. aeruginosa ATCC 25619, the membrane fraction of which was employed for determination of IC50s.
Frequency of emergence of resistant colonies.
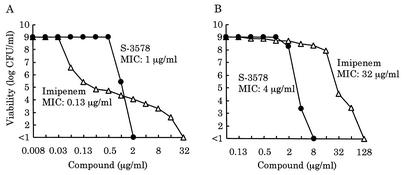
In order to estimate the frequency of resistance development, we first carried out population analysis on two clinical isolates of MRSA, the low-level-methicillin-resistant strain SR19754 and the high-level-methicillin-resistant strain SR19760. As shown in Fig. 7, strain SR19754 had a subpopulation resistant to imipenem at a frequency of about 10−6 in spite of the low MIC of imipenem for this strain, that is, it exhibited a heteroresistance profile. In contrast, the viabilities of both strains against S-3578 sharply decreased with increase of the antibiotic concentration. In particular, it was notable that no colonies emerged on agar plates containing twice the MIC of S-3578. Furthermore, about 109 bacterial cells each of 20 clinical isolates of MRSA were spread on agar plates containing S-3578 and incubated for 3 days at 35°C. For all isolates, no colonies appeared on the plates at twice the MIC, and emergence from an isolate for which the MIC was 8 μg/ml did not occur even at the MIC (data not shown). From these results, the frequency at which resistant colonies emerged was estimated to be below 10−9.
FIG. 7.
Population analysis profiles of two clinical isolates of MRSA. Strains SR19754 (A) and SR19760 (B) are low-level- and high-level-methicillin-resistant strains, respectively. The MICs of S-3578 and imipenem for each strain are indicated in the figure. Closed circles and open triangles indicate viable cell counts on plates containing S-3578 or imipenem, respectively.
β-Lactamase stability.
The hydrolysis of S-3578 and cefepime by a β-lactamase purified from S. aureus SR5644 was not detected at 100 μg of the compounds per ml (data not shown), indicating that S-3578 was stable against hydrolysis by the β-lactamase from S. aureus. S-3578, as well as cefepime, was stable against TEM-1 and a class A β-lactamase from K. pneumoniae GN69 (22) because their hydrolysis rates were less than 0.1% of that of ampicillin at the substrate concentration of 100 μg/ml (data not shown). Although the Km values of S-3578 against various AmpC enzymes from C. freundii, Enterobacter cloacae, and P. aeruginosa were lower than those of cefepime, its relative Vmax/Km values were comparable to those of cefepime. These results indicated that S-3578 had higher affinity for AmpC enzymes but was as stable as cefepime was against hydrolysis (Table 5). On the other hand, S-3578 was not as stable as cefepime was against a Toho-1-like extended-spectrum β-lactamase and a class B IMP-1-like enzyme (data not shown).
TABLE 5.
Stability of S-3578 against AmpC β-lactamases
| Enzyme source | Compound | Km (μM) | Relative Vmaxa | Relative Vmax/Kma |
|---|---|---|---|---|
| Citrobacter freundii SR19 | S-3578 | 26 | 0.008 | 0.20 |
| Cefepime | 340 | 0.030 | 0.056 | |
| Cephaloridine | 640 | 100 | 100 | |
| Enterobacter cloacae SR4321 | S-3578 | 19 | 0.006 | 0.16 |
| Cefepime | 500 | 0.14 | 0.14 | |
| Cephaloridine | 500 | 100 | 100 | |
| Pseudomonas aeruginosa SR24-12 | S-3578 | 60 | 0.030 | 0.35 |
| Cefepime | 560 | 0.11 | 0.14 | |
| Cephaloridine | 700 | 100 | 100 |
The value is expressed relative to an arbitrary value of 100 for cephaloridine.
DISCUSSION
One of the most striking features of S-3578 was its activity against methicillin-resistant staphylococci. S-3578 inhibited growth of all clinical isolates of Staphylococcus spp. including methicillin-resistant strains at 8 μg/ml, although other β-lactam compounds including imipenem were not active against most strains of methicillin-resistant staphylococci. We further characterized the anti-MRSA activity of S-3578 as follows. First, this activity of S-3578, like those of other anti-MRSA β-lactams (3, 9, 15), was considered to be due to the high affinity for PBP2a. This was consistent with reports of a correlation between the affinity for PBP2a and the MIC for MRSA strains with a high level of resistance to β-lactams (2, 11). Second, S-3578 showed potent bactericidal activity against MSSA and MRSA strains. The superiority of S-3578 over cefepime and ceftriaxone in bactericidal activity against MSSA was represented by a significant decrease of viable cells after 24-h exposure to S-3578. The results in Fig. 3 and 4 indicate that S-3578 had potent bactericidal activity against not only MSSA but also MRSA. Third, the frequency of emergence of resistant colonies from MRSA strains was extremely low. Moreover, S-3578 showed remarkable reduction of the viability of the heteroresistant strain in contrast with imipenem, which easily selected its resistant subpopulation (Fig. 7). These results suggested that S-3578-resistant strains would rarely emerge during antibiotic treatment. The two latter features would contribute to in vivo therapeutic efficacy against MRSA infection.
In addition to the activity against methicillin-resistant staphylococci, S-3578 was highly active against streptococci including penicillin-resistant S. pneumoniae, though most E. faecalis strains were not susceptible to this compound. On the other hand, S-3578 was not as active against gram-negative bacteria as cefepime was, but it showed antibacterial activity comparable to that of ceftazidime against most members of the family Enterobacteriaceae, H. influenzae, and M. catarrhalis. The antipseudomonal activity of S-3578, with a MIC50 of 8 μg/ml, was two- to fourfold less than those of ceftazidime and cefepime.
In conclusion, S-3578 is a novel parenteral broad-spectrum cephalosporin antibiotic with high activity against methicillin-resistant staphylococci. It should be noted that S-3578 is soluble in water at more than 100 mg/ml (26). This water solubility is a great advantage because most anti-MRSA cephalosporins have been reported elsewhere to be insoluble in water (4, 9). The pharmacokinetic profile of S-3578 in mice is similar to that of cefepime, and S-3578 shows good therapeutic efficacy in murine models of infection by MRSA, penicillin-resistant S. pneumoniae, or P. aeruginosa (M. Tsuji, M. Takema, H. Miwa, J. Shimada, and S. Kuwahara, submitted for publication). Thus, S-3578 is considered to be a promising compound for further evaluation as an anti-MRSA broad-spectrum cephalosporin antibiotic.
Acknowledgments
We are grateful to M. Doi, Y. Jinushi, Y. Kimura, H. Motoyama, K. Motokawa, T. Munekage, and K. Uotani for their superb technical assistance.
REFERENCES
- 1.Antony, S. J., E. Diaz-Vasquez, and C. Stratton. 2001. Clinical experience with linezolid in the treatment of resistant gram-positive infections. J. Natl. Med. Assoc. 93:386-391. [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers, H. F., and M. Sachdeva. 1990. Binding of β-lactam antibiotics to penicillin-binding proteins in methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 161:1170-1176. [DOI] [PubMed] [Google Scholar]
- 3.Chambers, H. F. 1995. In vitro and in vivo antistaphylococcal activities of L-695,256, a carbapenem with high affinity for the penicillin-binding protein PBP 2a. Antimicrob. Agents Chemother. 39:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho, A., T. W. Glinka, M. Ludwikow, A. T. Fan, M. Wang, and S. J. Hecker. 2001. New anti-MRSA cephalosporins with a basic aminopyridine at the C-7 position. Bioorg. Med. Chem. Lett. 11:137-140. [DOI] [PubMed] [Google Scholar]
- 5.Diekema, D. J., M. A. Pfaller, J. Turnidge, J. Verhoef, J. Bell, A. C. Fluit, G. V. Doern, R. N. Jones, and the SENTRY Participants Group. 2000. Genetic relatedness of multidrug-resistant, methicillin (oxacillin)-resistant Staphylococcus aureus bloodstream isolates from SENTRY antimicrobial resistance surveillance centers worldwide, 1998. Microb. Drug Resist. 6:213-221. [DOI] [PubMed] [Google Scholar]
- 6.Drew, R. H., J. R. Perfect, L. Srinath, E. Kurkimilis, M. Dowzicky, and G. H. Talbot. 2000. Treatment of methicillin-resistant Staphylococcus aureus infections with quinupristin-dalfopristin in patients intolerant of or failing prior therapy. J. Antimicrob. Chem. 46:775-784. [DOI] [PubMed] [Google Scholar]
- 7.Fung-Tomc, J. C., J. Clark, B. Minassian, M. Pucci, Y. Tsai, E. Gradelski, L. Lamb, I. Media, E. Huczko, B. Kolek, S. Chaniewski, C. Ferraro, T. Washo, and D. P. Bonner. 2002. In vitro and in vivo activities of a novel cephalosporin, BMS-247243, against methicillin-resistant and susceptible staphylococci. Antimicrob. Agents Chemother. 46:971-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebeisen, P., I. Heinze-Krauss, P. Angehrn, P. Hohl, M. G. P. Page, and R. L. Then. 2001. In vitro and in vivo properties of Ro63-9141, a novel broad-spectrum cephalosporin with activity against methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 45:825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinze-Krauss, I., P. Angehrn, P. Guerry, P. Hebeisen, C. Hubschwerlen, I. Kompis, M. G. P. Page, H. G. F. Richter, V. Runtz, H. Stalder, U. Weiss, and C. Wei. 1996. Synthesis and structure-activity relationship of (lactamylvinyl)cephalosporins exhibiting activity against staphylococci, pneumococci, and enterococci. J. Med. Chem. 39:1864-1871. [DOI] [PubMed] [Google Scholar]
- 11.Higashi, Y., A. Wakabayashi, Y. Matsumoto, Y. Watanabe, and A. Ohno. 1999. Role of inhibition of penicillin binding proteins and cell wall cross-linking by beta-lactam antibiotics in low- and high-level methicillin resistance of Staphylococcus aureus. Chemotherapy 45:37-47. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa, T., K. Kamiyama, Y. Nakayama, Y. Iizawa, K. Okonogi, and A. Miyake. 2001. Studies on anti-MRSA parenteral cephalosporins. III. Synthesis and antibacterial activity of 7β-[2-(5-amino-1,2,4-thiadiazol-3-yl)-2(Z)-alkoxyiminoacetamido]-3-[(E)-2-(1-alkylimidazo[1,2-b]pyridazinium-6-yl)thiovinyl]-3-cephem-4-carboxylates and related compounds. J. Antibiot. 54:257-277. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, A. P., M. Warner, M. Carter, and D. M. Livermore. 2002. In vitro activity of cephalosporin RWJ-54428 (MC-02479) against multidrug-resistant gram-positive cocci. Antimicrob. Agents Chemother. 46:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowy, F. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto, M., H. Tamaoka, H. Ishikawa, and M. Kikuchi. 1998. In vitro and in vivo antibacterial activities of OPC-20011, a novel parenteral broad-spectrum 2-oxaisocephem antibiotic. Antimicrob. Agents Chemother. 42:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami, K., and T. Yoshida. 1985. Covalent binding of moxalactam to cephalosporinase of Citrobacter freundii. Antimicrob. Agents Chemother. 27:727-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami, K., K. Nomura, M. Doi, and T. Yoshida. 1987. Production of low-affinity penicillin-binding protein by low-and high-resistance groups of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 31:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami, K., K. Nomura, M. Doi, and T. Yoshida. 1987. Increased susceptibility to cephamycin-type antibiotics of methicillin-resistant Staphylococcus aureus defective in penicillin-binding protein 2. Antimicrob. Agents Chemother. 31:1423-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 1999. Methods for determining bactericidal activity of antimicrobial agents; approved guideline. M26-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard, 5th ed. M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.O'Callaghan, C. H., P. W. Muggleton, and G. W. Ross. 1969. Effects of β-lactamase from gram-negative organisms on cephalosporins and penicillins, p. 57-63. Antimicrob. Agents Chemother. 1968. [PubMed]
- 22.Sawai, T., S. Yamagishi, and S. Mitsuhashi. 1973. Penicillinases of Klebsiella pneumoniae and their phylogenetic relationship to penicillinases mediated by R factors. J. Bacteriol. 115:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spratt, B. G. 1977. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur. J. Biochem. 72:341-352. [DOI] [PubMed] [Google Scholar]
- 24.Springer, D. M., B. Luh, and J. J. Bronson. 2001. Anti-MRSA cephems. Part 1: C-3 substituted thiopyridinium derivatives. Bioorg. Med. Chem. Lett. 11:797-801. [DOI] [PubMed] [Google Scholar]
- 25.Witte, W. 1999. Antibiotic resistance in Gram-positive bacteria: epidemiological aspects. J. Antimicrob. Chemother. 44:A1-A9. [DOI] [PubMed] [Google Scholar]
- 26.Yoshizawa, H., H. Itani, K. Ishikura, T. Irie, K. Yokoo, T. Kubota, K. Minami, T. Iwaki, H. Miwa and Y. Nishitani. 2002. S-3578, a new broad spectrum parenteral cephalosporin exhibiting potent activity against both methicillin-resistant Staphylococcus aureus (MRSA) and Pseudomonas aeruginosa: synthesis and structure-activity relationships. J. Antibiot., 55:975-992. [DOI] [PubMed]