Abstract
Three mer transposons from the Murray collection of preantibiotic enterobacteria show >99% sequence identity to current isolates. Tn5073 is most closely related to Tn5036 and Tn1696, and Tn5074 is most closely related to Tn5053. Tn5075 is most closely related to Tn21 but lacks integron In2 and is flanked by insertion elements.
Tn21 encodes mercuric ion resistance (Hgr) and contains the class I integron In2, encoding resistance to sulfonamides (sul) and streptomycin-spectinomycin (aadA) (9, 18). It is carried by the conjugative plasmid NR1 (R100), which was isolated in Japan in the 1950s (21). More recently, Tn21 (18) and other Tn21-like transposons carrying integron-associated antibiotic resistance (7) have been detected in Escherichia coli from agricultural (1, 28) and nonclinical sources (19), as well as from mercury amalgam-exposed, gram-negative gut bacteria (16, 17, 33), clinical bacterial isolates (13, 31, 36), and intercontinental plasmids carried by clinical isolates (10, 22).
It is now thought that Tn21 evolved by the insertion of an In2 ancestor (lacking IS1353) into the urf2M gene of a hypothetical mercury resistance transposon, Tn21Δ (18) (also called TnX [25]), probably catalyzed by transposition proteins encoded in trans (2, 18). A similar event led to the formation of Tn1696 from plasmid R1033, where In4 inserted at the res site of a Tn5036-like mer transposon (25).
Hughes and Datta identified three Hgr bacterial strains, M426, M567, and M634, from a total of 433 strains from the Murray collection of preantibiotic era enterobacteria (11). In this study, we sequenced the mer operons from M426, M567, and M643 in order to investigate the relationships between Hgr sequences from clinical bacteria that had been isolated before antibiotics came into widespread use and present-day Hgr sequences.
The plasmids, bacterial strains, antimicrobial resistance of these strains, and the 16S ribosomal DNA sequence identifications of the three Hgr strains from this study (determined as previously described [32]) are shown in Table 1. All bacteria were grown at 37°C in Luria broth (LB) or on LB agar (27). Hgr plasmids from M426, M567, and M634 were mated with E. coli TG2 (20), and Hgr transconjugants were grown overnight on LB agar plates containing tetracycline (15 μg/ml) and HgCl2 (20 μg/ml).
TABLE 1.
Bacterial strains and plasmids, antibiotic resistance phenotypes, 16S rRNA sequences, and sources of the strains used in this study
| Strain or plasmid | Antibiotic resistancea | Original tube date (yr) | Genotype | 16S rRNA identification | Source or referenceb |
|---|---|---|---|---|---|
| Murray collection | |||||
| Klebsiella sp. strain M426 | Hgr | 1940 | Klebsiella pneumoniae with 99% identity to sequence of accession no. AF144323.1 | Urine NCTC | |
| Proteus morganii M567 | Hgr | 1935 | Morganella morganii with 98% identity to sequence of accession no. AJ301681 | Stool sample, child with dysentery NCTC | |
| Escherichia coli M634 | Hgr | 1931 | Escherichia coli K-12 with 100% identity to sequence of accession no. NC000913 | Cerebrospinal fluid NCTC | |
| Laboratory strains | |||||
| Escherichia coli TG2 | Tcr | K-12 lac-proΔsrl-recA 306::Tn10 supE thi hsdD5 [F′ traD36 proA+B+laclqlacZΔM15] | Laboratory stock (4) | ||
| Escherichia coli KH802 | Rifr | F−gyrA rpoB metB hsdSB (rK− mK+) | P. Strike (34) | ||
| Plasmid RK2 | Apr Tcr Kmr | C. Thomas |
Tcr, tetracycline resistance; Rifr, rifampicin resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance.
NCTC, National Collection of Type Cultures, Colindale, London, United Kingdom.
The E. coli TG2 Hgr transconjugants from each of the three Murray strains contained an ∼60-kb plasmid that conferred Hgr. Plasmid DNA was isolated by standard methods (27), and PCR was performed with part of the mer operon from each plasmid as described elsewhere (3). PCR products purified by using a QIAquick PCR purification kit (Qiagen, Ltd., Crawley, United Kingdom) were sequenced with the Big Dye terminator cycle sequencing kit (PE Applied Biosystems, Warrington, United Kingdom) and an Applied Biosystems 3700 sequencer, according to the manufacturer's protocols. Further sequence analysis was performed by using primers designed from the sequences so obtained and from merA gene primers (5) (the primers used are described at http://www.biosciences.bham.ac.uk/labs/brown/mer_primers.htm). The transposon terminal inverted repeat DNA and flanking sequences were amplified by inverse PCR (26). Genetic maps of the sequenced mer operons are shown in Fig. 1A. DNA alignments and analysis were performed with the University of Wisconsin Genetics Computer Group version 9.0 suite of programs at the University of Birmingham.
FIG.1.
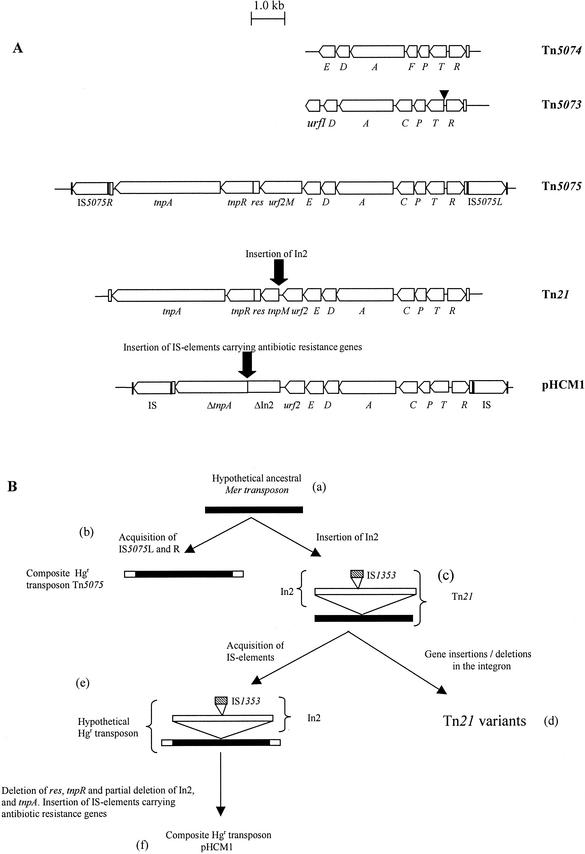
(A) Genetic maps of the sequenced regions of Tn5073, Tn5074, Tn5075, Tn21 (18), and the Tn21-like transposon from pHCM1 (24). Vertical black bars represent the 10- to 12-bp inverted repeat sequences flanking the IS elements. Vertical unfilled bars represent the 38bp inverted repeat sequences at the ends of the class II transposition module. The transposition (tnp) regions consist of the transposase gene (tnpA), the resolvase gene (tnpR), the putative transposon regulator (tnpM), and the resolution site, res. urf2M in Tn5075 is of unknown function. The points at which In2 or antibiotic resistance-encoding IS elements have inserted into Tn21 and pHCM1 are marked with black arrows. The mercury resistance-encoding operons consist of the regulatory genes merR and merD, the mercury transport genes merT, merP, and merC or merF, the putative transporter merE, and the gene encoding mercuric reductase, merA. urfI and urf2 are of unknown function. All mer genes are marked in the figure with a single letter. The position of the multiple DNA sequence repeats found in merT from Tn5073 are indicated (▾). (B) Postulated evolutionary pathway for Tn5075, Tn21, and the Tn21-like transposon in pHCM1. An ancestral mer transposon (a) could acquire either IS5075L and IS5075R to become Tn5075 (b) or an integron related to In2, resulting in the formation of Tn21 (c). Gene insertions and deletions in In2 would lead to Tn21 variants (d), or Tn21 could have acquired IS elements, leading to the formation of a precursor to the Tn21-like transposon in pHCM1 (e). Deletions of transposition genes and partial deletion of In2, followed by insertion of antibiotic resistance gene-carrying IS elements, would result in the formation of the Tn21-like transposon in pHCM1 (f).
The three mer operons that we sequenced represent different lineages and are not closely related to each other, but they are closely related to mer transposons isolated since the 1950s. Table 2 shows the percent identities between the sequences of the genes from Tn5073 (Klebsiella pneumoniae M426), Tn5074 (Morganella morganii M567), Tn5075 (E. coli M634) and published DNA sequences.
TABLE 2.
Comparison of DNA sequence identities between Tn5073, Tn5074, Tn5075, and published DNA sequences from other transposons
| Transposon | Genes | % Identity to mer transposon (genes)a | Accession no. | Reference |
|---|---|---|---|---|
| Tn5073 | merRTPCAD | >99.9 Tn5036-like transposon pHCM1 | AL513383 | 24 |
| >99.9 Tn1696 (merAD) | U12338 | 25 | ||
| >99.8 Tn5036 (merRTPCAD) | Y09025 | 35 | ||
| >99.8 pKLH272 (merRTPCAD) | Y08992 | 35 | ||
| Tn5074 | merRTPFADE | >99.3 Tn5053 (merRTPFADE) | L40585 | 14 |
| >99.2 pMER327/419 (merRTPFADE) | X73112 | 8 | ||
| Tn5075 | merRTPCADE | >99.9 Tn21 (merRTPCADE) | AF071413 | 18 |
| >99.9 Tn21-like transposon from pHCM1 (merRTPCADE) | AL513383 | 24 | ||
| res tnpA tnpR | >99.8 Tn21 (res tnpA tnpR) | AF071412 | 18 | |
| urf2M | >97.1 hypothetical urf2m from Tn21Δ | AF071413 | 18 | |
| IS5075L | >99.5 IS5075R | AF457211 | This work | |
| >99.6 Tn21-like transposon from pHCM1 (IS elements) | AL513383 | 24 | ||
| >91.0 pDU1358 5′ to mer operon | M24940 | 6 | ||
| >91.0 R831b 5′ to mer operon | U77087 | 23 | ||
| >91.0 IS4321L, IS4321R, from R751 | U60777 | 30 |
The sequenced merRTPCAD genes (3,788 bp) of the Tn5073 mer operon (Fig. 1A) had the highest identity at the DNA level to those from Tn5036 (35), a Tn5036-like mer transposon from Salmonella enterica serovar Typhi CT18 plasmid pHCM1 (24), and to the sequenced merAD genes of Tn1696, which carries In4 (25) (Table 2). In comparison to the sequence encoded by Tn5036, there were two amino acid differences in the sequence encoded by Tn5073: MerR A17→V and MerA V250→A. The Tn5073 merT gene carries five GTCTGAACCACAAAA duplications at the 5′ end (Fig. 1A). Multiple repeats of this sequence have also been observed in enterobacterial mercury resistance determinants from primates (16) and in Tn5036, Tn5059, and pKLH272 (35).
The sequenced merRTPFADE genes (3,647 bp) from the Tn5074 mer operon (Fig. 1A) had the highest identity at the DNA level to those from Tn5053 (14) and pMER327/419 (8) (Table 2). In comparison to the sequence encoded by Tn5053, there were four amino acid differences in the sequence encoded by Tn5074: MerR A119→S and K121→Q and MerA V232→A and S289→G. The Tn5053 type mer operon was first isolated in environmental bacteria (8, 14) and has also been found in the fecal flora of primates and humans exposed to dental amalgam (16, 20).
In total, 11,298 bp of Tn5075 were sequenced. The 3,962-bp mer operon from Tn5075 carrying merRTPCADE has the highest identity at the DNA level to the equivalent regions of Tn21 (18) and the Tn21-like mer transposon from S. enterica serovar Typhi CT18 plasmid pHCM1 (24) (Fig. 1A) (Table 2). The Tn5075 res site and the transposition genes (3,529 bp) tnpR and tnpA again had the highest identity to those of Tn21. In comparison to the sequence encoded by Tn21, there were three amino acid differences in the sequence encoded by Tn5075: MerA, Q558→H; TnpR, T165→A; and TnpA R455→A. Most importantly, Tn5075 did not carry the integron In2, which in Tn21 is located between urf2 and the putative transposition gene tnpM (12) (Fig. 1A). Instead, in Tn5075, there is a single 945-bp open reading frame (urf2M) of unknown function rather than the predicted 987-bp hypothetical urf2M from Tn21Δ proposed by Liebert et al. (18). Tn5075 urf2M is 97.1% identical at the DNA level to the hypothetical urf2M that Liebert et al. (18) postulated to exist in the ancestor of Tn21. The reason for this lower percent identity is a 23-bp deletion in the Tn5075 urf2M sequence compared to the tnpM sequence in Tn21, resulting in a frameshift and a 314-amino-acid protein, rather than the 328-amino-acid protein predicted previously (18).
The insertion (IS) elements IS5075L and IS5075R (which are 1,351 bp and >99.5% identical to each other) flank Tn5075, forming a composite transposon (Fig. 1A). IS5075L and IS5075R belong to the IS110 family and are >99.6% identical to the IS elements flanking the Tn21-like mer transposon from plasmid pHCM1 (Fig. 1A). IS5075L and IS5075R are between 91 and 93% identical to IS element sequences flanking other mercury resistance-encoding genes from gram-negative bacteria (Table 2).
Transposition of the Hgr phenotype was determined by a mate-out assay (15) with E. coli TG2 carrying plasmid RK2, conjugated with Hgr plasmids from M426, M567, or M634, into E. coli KH802. Transconjugants were selected on LB agar plates containing HgCl2 (20 μg/ml) and carbenicillin (200 μg/ml), and the donor strain was counterselected with rifampin (50 μg/ml). Purified plasmid RK2 DNA from the transconjugants was analyzed for transposon insertion by PstI digestion and gel electrophoresis (27). Tn5073 transposed into plasmid RK2 at a frequency of 6.3 × 10−5 per donor cell. Tn5074 transposed into RK2 at a frequency of 2.9 × 10−4 per donor cell. We found no transposition of Tn5075 in our assays, which could detect frequencies of >10−7.
In conclusion, the internal genetic structure of Tn5075 is consistent with the recently proposed structure of Tn21Δ (Fig. 1B) (2, 18), and Tn5073 is closely related to Tn5036 and Tn1696 (25). The sequence data from Tn5075 support the hypothesis that Tn21 evolved from an Hgr transposon similar to Tn5075, rather than from Tn2613 (29). Tn1696 and Tn21 represent independent lineages of mer transposons that have acquired integrons (25). The dates of isolation of Tn5073 (1940) and Tn5075 (1931) and the close relationship of these transposons to other lineages (Tn5073 merAD genes are >99.9% identical to those of Tn1696; Tn5075 is >99.6% identical to Tn21 except where In2 is not present) are consistent with the idea that integrons transposed into preexisting clinical Hgr transposons. The Tn5074 mer operon, isolated from a clinical source, has the greatest DNA identity to the Tn5053 and pMER327/419 mer operons, which have been isolated from both environmental and nonclinical (16, 20) sources.
The DNA sequence data suggest that Tn5075, the Tn21-like mer transposon in pHCM1, and Tn21 had a common ancestor and may have evolved as shown in Fig. 1B; i.e., an ancestral mer transposon acquired IS5075L and IS5075R, leading to the formation of Tn5075. Alternatively, an integron related to In2 inserted into the ancestral mer transposon, leading to the formation of Tn21. Gene insertions and deletions within the integron in Tn21 could lead to the formation of Tn21 variants (7). Acquisition of IS5075-like elements by Tn21 could have led to the formation of a hypothetical mer transposon. Deletion of res and tnpR and partial deletion of In2 and tnpA from this transposon followed by insertion of antibiotic resistance-carrying IS elements, could have led to the formation of the Tn21-like transposon in pHCM1 (24). Although the Tn21-like transposon in pHCM1 is flanked by IS elements which are 99.6% identical to IS5075L and IS5075R from Tn5075, it is more closely related to Tn21 than to Tn5075 because it contains a vestige of the In2 sequence, which Tn5075 does not and is 100% identical to Tn21 across the mer genes and tnpA.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the mer sequences determined in this study are as follows: Tn5073 (strain M426), AF461013; Tn5074 (strain M567), AF461012; and Tn5075 (strain M634), AF457211. The 16S rRNA gene sequence accession number for strain M567 is AF461011.
Acknowledgments
We thank Julian Davies, Didier Mazel, and Barry Holmes for their help. We also thank Chris Thomas and Peter Strike for the gifts of plasmids and strains and Gennady Kholodii and anonymous referees for suggested improvements to the paper.
This work was supported by a postgraduate research scholarship to A.M.E. from the Egyptian Ministry of Higher Education, and the work was partially supported by a BBSRC grant 6/G07943 to N.L.B. and a BBSRC/Wellcome Trust Joint Infrastructure Fund Grant (6/JIF13209). Support in bioinformatics was from MRC grant G.4600017.
REFERENCES
- 1.Bass, L., C. A. Liebert, M. D. Lee, A. O. Summers, D. G. White, S. G. Thayer, and J. G. Maurer. 1999. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce, K. D., A. M. Osborn, A. J. Pearson, P. Strike, and D. A. Ritchie. 1995. Genetic diversity within mer genes directly amplified from communities of noncultivated soil and sediment bacteria. Mol. Ecol. 4:605-612. [DOI] [PubMed] [Google Scholar]
- 4.Gibson, T. 1984. Studies on the Epstein-Barr virus genome. Ph.D. thesis. University of Cambridge, Cambridge, United Kingdom.
- 5.Glendinning, K. G. 2000. Studies on mercuric reductase and thermophilic mercury resistance. Ph.D. thesis. The University of Birmingham, Birmingham, United Kingdom.
- 6.Griffin, H. G., T. J. Foster, S. Silver, and T. K. Misra. 1987. Cloning and DNA sequence of mercuric and organomercurial-R determinants of plasmid pDU1358. Proc. Natl. Acad. Sci. USA 84:3112-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grinsted, J., F. de la Cruz, and R. Schmitt. 1990. The Tn21 subgroup of bacterial transposable elements. Plasmid 26:163-189. [DOI] [PubMed] [Google Scholar]
- 8.Hobman, J., G. Kholodii, V. Nikiforov, D. A. Ritchie, P. Strike, and O. Yurieva. 1994. The sequence of the mer operon of pMER327/419 and transposon ends of pMER327/419, 330, and 05. Gene 277:73-78. [DOI] [PubMed] [Google Scholar]
- 9.Hobman, J. L., and N. L. Brown. 1997. Bacterial mercury-resistance genes, p. 527-568. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems. Marcel Dekker, Inc., New York, N.Y. [PubMed]
- 10.Hopkins, J. D., T. F. O'Brien, and M. Syvanen. 1988. Functional and structural map of pLST1000: a multiresistance plasmid widely distributed in Enterobacteriaceae. Plasmid 20:163-166. [DOI] [PubMed] [Google Scholar]
- 11.Hughes, V. M., and N. Datta. 1983. Conjugative plasmids in bacteria of the ′pre-antibiotic' era. Nature 302:725-726. [DOI] [PubMed] [Google Scholar]
- 12.Hyde, D. R., and C. P. D. Tu. 1985. tnpM: a novel regulatory gene that enhances Tn21 transposition and suppresses cointegrate resolution. Cell 42:629-638. [DOI] [PubMed] [Google Scholar]
- 13.Jones, M. E., E. Peters, A.-M. Weersink, A. Fluit, and J. Verhoef. 1997. Widespread occurrence of integrons causing multiple antibiotic resistance in bacteria. Lancet 349:1742-1743. [DOI] [PubMed] [Google Scholar]
- 14.Kholodii, G. Y., S. Z. Mindlin, I. A. Bass, O. V. Yurieva, S. V. Minakhina, and V. G. Nikiforov. 1995. Four genes, two ends, and a res region are involved in the transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol. Microbiol. 17:1189-1200. [DOI] [PubMed] [Google Scholar]
- 15.Kholodii, G., O. Yurieva, S. Mindlin, Z. Gorlenko, V. Rybochkin, and V. Nikiforov. 2000. Tn5044, a novel Tn3 family transposon coding for temperature-sensitive mercury resistance. Res. Microbiol. 151:291-312. [DOI] [PubMed] [Google Scholar]
- 16.Liebert, C. A., J. Wireman, T. Smith, and A. O. Summers. 1997. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 63:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liebert, C. A., J. Wireman, T. Smith, and A. O. Summers. 1997. The impact of mercury released from dental “silver” fillings on antibiotic resistances in the primate oral and intestinal bacterial flora, p. 441-460. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems. Marcel Dekker, Inc., New York, N.Y. [PubMed]
- 18.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mindlin, S., G. Kholodii, Z. Gorlenko, S. Minakhina, L. Minakhin, E. Kalyaeva, A. Kopteva, M. A. Petrova, O. V. Yurieva, and V. Nikiforov. 2001. Mercury resistance transposons of gram-negative bacteria, and their classification. Res. Microbiol. 152:811-822. [DOI] [PubMed] [Google Scholar]
- 21.Nakaya, R., A. Nakamura, and Y. Murata. 1960. Resistance transfer agents in Shigella. Biochem. Biophys. Res. Commun. 3:654-659. [DOI] [PubMed] [Google Scholar]
- 22.O'Brien, T. F., M. del Pilar Pla, K. H. Mayer, H. Kishi, E. Gilleece, M. Syvanen, and J. D. Hopkins. 1985. Intercontinental spread of a new antibiotic resistance gene on an epidemic plasmid. Science 230:87-88. [DOI] [PubMed] [Google Scholar]
- 23.Ogawa, H. I., C. L. Tolle, and A. O. Summers. 1984. Physical and genetic map of the organomercury resistance (Omr) and inorganic mercury resistance (Hgr) loci of the IncM plasmid R831b. Gene 32:311-320. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. G. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. Ó'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 25.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riley, J., R. Butler, R. Finniear, D. Jenner, S. Powell, R. Anand, J. C. Smith, and A. F. Markham. 1990. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 18:2887-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sunde, M., and H. Sørum. 2001. Self-transmissible multidrug resistance plasmids in Escherichia coli of the normal intestinal flora of healthy swine. Microb. Drug Resist. 7:191-196. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, M., T. Yamamoto, and T. Sawai. 1983. Evolution of complex resistance transposons from an ancestral mercury transposon. J. Bacteriol. 153:1432-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 31.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wintzingerode, F. V., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 33.Wireman, J., C. A. Liebert, T. Smith, and A. O. Summers. 1997. Association of mercury resistance with antibiotic resistance in the gram-negative fecal bacteria of primates. Appl. Environ. Microbiol. 63:4494-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood, W. B. 1966. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J. Mol. Biol. 16:118-133. [DOI] [PubMed] [Google Scholar]
- 35.Yurieva, O., G. Kholodii, L. Minakhin, Z. Gorlenko, E. Kalyaeva, S. Mindlin, and V. Nikiforov. 1997. Intercontinental spread of promiscuous mercury resistance transposons in environmental bacteria. Mol. Microbiol. 24:321-329. [DOI] [PubMed] [Google Scholar]
- 36.Zühlsdorf, M. T., and B. Weidemann. 1992. Tn21-specific structures in gram-negative bacteria from clinical isolates. Antimicrob. Agents Chemother. 36:1915-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]