Abstract
The expression of tripartite multidrug efflux pumps such as MexA-MexB-OprM in Pseudomonas aeruginosa contributes to intrinsic resistance to a wide variety of antimicrobials, including β-lactams, chloramphenicol, macrolides, quinolones, and tetracycline. The MexX-MexY linker-pump combination has been shown to be involved in intrinsic resistance to aminoglycosides, but the identity of the cognate outer membrane channel component remains under debate. Fourteen uncharacterized OprM homologs identified in the genome of P. aeruginosa were examined as candidates for this role by assessing the minimum inhibitory concentrations (MICs) of aminoglycosides in P. aeruginosa strain PAK knockout mutants lacking the corresponding genes. Insertional inactivation of OpmG, OpmI, and OpmH resulted in decreases of various degrees in the MICs of streptomycin, kanamycin, and gentamicin. When reintroduced into P. aeruginosa on multicopy plasmids, OpmG was able to complement the susceptibility of an opmG::miniTn5 mutant; however, cloned opmH, the proposed ortholog of Escherichia coli tolC according to our phylogenetic analysis, was able to only partially complement the opmH::miniTn5 mutant. Mini-microarray hybridization analysis demonstrated that opmG disruption does not affect expression of OpmI or OpmH (ruling out such indirect effects on aminoglycoside resistance); however, opmH disruption did have possible effects on expression of OpmG and OpmI. Based on the data, we propose that OpmG is a major outer membrane efflux channel involved in aminoglycoside efflux in P. aeruginosa PAK and that OpmI, its most related paralog, may share an overlapping function.
The combination of tripartite multidrug efflux systems and outer membrane impermeability plays a major role in intrinsic antibiotic resistance in clinically relevant pathogens such as Pseudomonas aeruginosa (15). While healthy individuals are not normally subject to infection by this organism, immunocompromised individuals, including those with burn wounds, immunosuppressed postsurgical patients, and persons suffering from cystic fibrosis, are at risk for contracting Pseudomonas infection (7). While the production of several virulence factors, such as mucous exopolysaccharide, exotoxins, extracellular proteases, extracellular lipases, and endotoxin (6) aid in pathogenesis, recalcitrant P. aeruginosa infections are typically characterized by high-level antibiotic resistance (7).
Although outer membrane impermeability was originally proposed to be responsible for broad-spectrum intrinsic resistance in gram-negative bacteria, it is now well understood that it must be coupled to a secondary mechanism such as active drug efflux (15). The multidrug resistance (MDR) efflux systems of gram-negative bacteria have a three-component structure that allows the expulsion of compounds through two membranes. Three characterized MDR transporters of P. aeruginosa are known to share this structure, which consists of (i) an inner membrane pump-proton antiporter of the resistance-nodulation-cell division (RND) family (e.g., MexB), (ii) a trimeric channel-forming outer membrane pore component that spans both the outer membrane and periplasm (OprM), and (iii) a membrane-anchored periplasmic linker protein (MexA) that is thought to promote interaction between the RND pump and the outer membrane efflux channel (17, 24).
The constitutively active MexA-MexB-OprM system of P. aeruginosa confers intrinsic resistance to a number of classes of antibiotics, including certain β-lactams, chloramphenicol, macrolides, quinolones, and tetracycline (9, 12, 15). Two other RND efflux systems have also been characterized but are not well expressed in wild-type P. aeruginosa. Mutations in the nfxB gene, encoding the repressor for the MexC-MexD-OprJ MDR system, result in overexpression of this system, leading to resistance to specific recent cephalosporins, chloramphenicol, macrolides, quinolones, and tetracycline (12, 15, 16), while mutations leading to the activation of the positive regulator MexT result in the expression of MexE-MexF-OprN and resistance to chloramphenicol, the quinolones, and trimethoprim, as well as the carbapenem subclass of β-lactams through down regulation of OprD (8, 15).
The identification of the AmrA-AmrB-OprA operon in Burkholderia pseudomallei was the first strong evidence that highly charged, hydrophilic compounds, such as the aminoglycoside class of antibiotics, could be substrates for RND efflux pumps (14). Since then, genes have been identified in other organisms that might confer intrinsic aminoglycoside resistance. The AcrD gene product of Escherichia coli was shown to mediate energy-dependent efflux of aminoglycosides (18). Researchers in Japan, Europe, and North America also reported the existence of a fourth MDR pump operon in P. aeruginosa, mexXY (1, 13, 22), which does not encode an outer membrane efflux channel as part of the operon. Although it has been conclusively shown that MexX-MexY plays a role in conferring efflux-mediated aminoglycoside resistance, the identity of the cognate channel for this MDR system remains under debate. OprM can clearly function in combination with MexX-MexY in aminoglycoside efflux (11, 12, 13), but other authors have suggested that another as yet undetermined outer membrane channel is likely the native one for this system (22).
The genome of P. aeruginosa was recently completed, and BLAST analysis revealed the presence of 14 previously undiscovered homologs of oprM in this genome sequence (21). It seemed possible to us that one of these putative channels might function with MexX-MexY in the efflux of aminoglycoside antibiotics. Mini-Tn5 insertion mutants in 14 of the genes encoding these OprM homologs were provided by Chiron (formerly PathoGenesis Corporation). By screening the susceptibility of each mutant to aminoglycosides, three candidate outer membrane proteins, OpmG, OpmH, and OpmI, were shown to have possible roles in aminoglycoside efflux.
MATERIALS AND METHODS
Bacterial strains and media.
All strains and plasmids used in this study are listed in Table 1. Strains were grown at 37°C in L broth (1.0% tryptone, 0.5% yeast extract, and either 0.5% [for E. coli] or 0.05% [for P. aeruginosa] NaCl) or on L agar (L broth containing 2% agar). Antibiotics were supplied at the following concentrations: for plasmid maintenance in E. coli, ampicillin at 100 μg/ml; for plasmid maintenance in P. aeruginosa, carbenicillin at 200 μg/ml and tetracycline at 100 μg/ml; for maintenance of insertion mutations in H956-H969, tetracycline at 100 μg/ml; and for maintenance of the insertion mutation in K613, HgCl2 at 15 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli DH5α | supE44 hsdR17 recA1 endA1 gyrA96 thi-I relA1 Δ(lacZYA-argF)U169deoR (φ80ΔlacZdM15) | Hanahan, 1983 |
| P. aeruginosa strains | ||
| H103 | Wild-type PAO1 | Nicas and Hancock, 1980 |
| H911 | Wild-type PAK | Chiron |
| H956 | H911 opmK::miniTn5-Tcr | Chiron |
| H957 | H911 opmA::miniTn5-Tcr | Chiron |
| H958 | H911 opmG::miniTn5-Tcr | Chiron |
| H959 | H911 aprF::miniTn5-Tcr | Chiron |
| H960 | H911 opmI::miniTn5-Tcr | Chiron |
| H961 | H911 opmE::miniTn5-Tcr | Chiron |
| H962 | H911 opmL::miniTn5-Tcr | Chiron |
| H963 | H911 opmD::miniTn5-Tcr | Chiron |
| H964 | H911 opmJ::miniTn5-Tcr | Chiron |
| H965 | H911 opmN::miniTn5-Tcr | Chiron |
| H966 | H911 opmH::miniTn5-Tcr | Chiron |
| H967 | H103 opmF::miniTn5-Tcr | Chiron |
| H968 | H911 opmB::miniTn5-Tcr | Chiron |
| H969 | H911 opmM::miniTn5-Tcr | Chiron |
| H730 (K372) | PAO6609 (met9011 amiE200 rpsL pvd9) Pch− | Poole et al., 1991 |
| H743 (K613) | K372 oprM::ΩHg; OprM-deficient | Poole et al., 1993 |
| Plasmids | ||
| pCR2.1 | TA cloning vector | Invitrogen |
| pJJ104 | pCR2.1 opmG | This study |
| pJJ101 | pCR2.1 opmH | This study |
| pUCP21 | Escherichia-Pseudomonas shuttle vector | Schweizer, 1991 |
| pUCP27 | Escherichia-Pseudomonas shuttle vector | Schweizer, 1991 |
| pJJ106 | pUCP27 opmG | This study |
| pJJ105 | pUCP27 opmH | This study |
| pJJ107 | pUCP21 opmG | This study |
| pJJ109 | pUCP21 opmH | This study |
Recombinant DNA techniques.
Transformation of E. coli, isolation of chromosomal DNA and plasmids, and restriction endonuclease digestions were performed in accordance with standard protocols (19). Electrocompetent P. aeruginosa cells were suspended in magnesium electroporation buffer (1 mM MgCl2, 1 mM HEPES [pH 7.0]) for electroporation. PCR was performed in accordance with the manufacturer's protocols for Taq polymerase, with the addition of 5% dimethyl sulfoxide to the reaction mixture when P. aeruginosa chromosomal DNA was used as the template. Sequences of all PCR primers can be obtained from the authors. PCR primers were also used as sequencing primers. TA cloning was performed in accordance with the manufacturer's protocols accompanying the Topo TA cloning kit (Invitrogen Life Technologies).
Determination of MICs.
Minimum inhibitory concentrations (MICs) were determined by serial twofold dilution in L broth by using the broth microdilution method described by Amsterdam (2). Results were determined after incubation at 37°C for 18 h.
Mini-microarray construction.
Six-hundred-base-pair amplicons corresponding to internal fragments of each of the OprM-family homologs were PCR amplified from PAO1 genomic DNA by using gene-specific primers. Amplicons were purified by using the Qiagen PCR purification kit and then resuspended in spotting solution (0.4 M NaOH, 10 mM EDTA [pH 8.2] in RNase-free distilled water [dH2O]) at a concentration of 20 ng/μl. The amplicons were then denatured at 100°C for 10 min, immediately placed on ice, transferred to a 96-well microtiter plate, and spotted onto positively charged nylon membranes in 0.5-μl spots by using a 96-well groove-pin replicator. After air drying, the membranes were soaked in alkaline denaturing solution (1.5 M NaCl, 0.5 M NaOH in RNase-free dH2O) for 10 min and then transferred to neutralizing solution (1 M NaCl, 0.5 M Tris HCl [pH 7.0] in RNase-free dH2O) for 5 min. Membranes were air dried, baked for 30 min at 80°C, wrapped in transparent plastic wrap, and exposed to UV light for 30 s to cross-link the DNA to the membrane. The membranes were stored between filter papers at 4°C.
RNA isolation and reverse transcription-PCR.
Cultures of P. aeruginosa strains H911, H958, and H966 were grown overnight with appropriate selection and then subcultured into fresh medium and allowed to grow to an optical density at 650 nm of 0.5. All manipulations of RNA were carried out in designated RNase-free areas, and all solutions were treated overnight with 1% diethylpyrocarbonate (DEPC) and then autoclaved to inhibit RNases. RNA was isolated by using the RNeasy mini RNA isolation kit (Qiagen) in accordance with the manufacturer's protocols. Contaminating genomic DNA was removed from the RNA sample by using the DNA-free kit (Ambion) in accordance with the manufacturer's protocols. Purified RNA was quantitated spectrophotometrically and stored at −20°C. Reverse transcription reactions were performed in accordance with the protocol for the use of Superscript II RNase H− reverse transcriptase (Invitrogen Life Technologies). The 5′ primer pool for the first-strand reverse transcriptase reaction consisted of a 10-μM mixture of each of 19 gene-specific reverse primers. cDNA was stored at −20°C. [α-32P]dCTP was incorporated into DNA during 15 cycles of PCR (95°C for 30 s, 63°C for 45 s, and 72°C for 1 min) carried out as follows. The PCR mixture (50 μl total volume) contained 5 μl of 10× amplification buffer, 1 mM MgSO4, 50 μM each dATP, dGTP, and dTTP, 50 μCi [α-32]dCTP (Amersham Pharmacia Biotech), 200 μM each of the forward (5′) and reverse (3′) primer pools (each containing a mixture of the 19 primers used in the original amplification), 5% dimethyl sulfoxide, 10 ng of cDNA template, and 1.25 U of Platinum Pfx DNA polymerase (Invitrogen Life Technologies). Unincorporated [α-32P]dCTP was purified from labeled PCR product by using MicroSpin G-50 columns (Amersham Pharmacia Biotech). Incorporation of the radioactive nucleotide was confirmed by adding 1 μl of PCR product to 5.5 ml of scintillation fluid and measuring radioactivity on a scintillation counter.
Hybridization of radioactive cDNA.
Prior to hybridization with labeled DNA probe, membranes were placed in glass hybridization tubes and incubated at 42°C for 3 h with 5 ml of prehybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt's solution, 50% [wt/vol] formamide, 1% [wt/vol] sodium lauryl sulfate [SDS]) and 100 μg of denatured salmon sperm DNA/ml to block nonspecific binding to the membranes. α-32P-Labeled DNA probe was denatured at 100°C for 5 min and then chilled on ice. Radioactive probe (14 μl) was added to the hybridization tube. Following an overnight incubation at 42°C, membranes were washed twice in 5 ml of each of 2× SSC-0.1% SDS and 0.2× SSC-0.1% SDS while rotating at room temperature and then twice in 0.2× SSC-0.1% SDS while rotating at 42°C. Membranes were subsequently blotted dry by using filter paper and wrapped in transparent plastic wrap.
Autoradiographic imaging.
Membranes were placed onto an MD Storage phosphor screen (Molecular Dynamics) for 72 h. Autoradiographic imaging was performed on the Molecular Dynamics PhosphorImager SI. Quantification of hybridization spots was performed by using the ImageQuant version 1.1 software (Molecular Dynamics). Local background was subtracted from the value of each hybridization spot. The calculated spot density values were normalized by dividing by the density value for the uvrD internal control. Comparisons between conditions (wild-type and mutant strains) were performed by taking the ratio of spot density values of the wild type over the mutant in three independent experiments.
RESULTS
Phylogenetic analysis of OprM and OprM homologs.
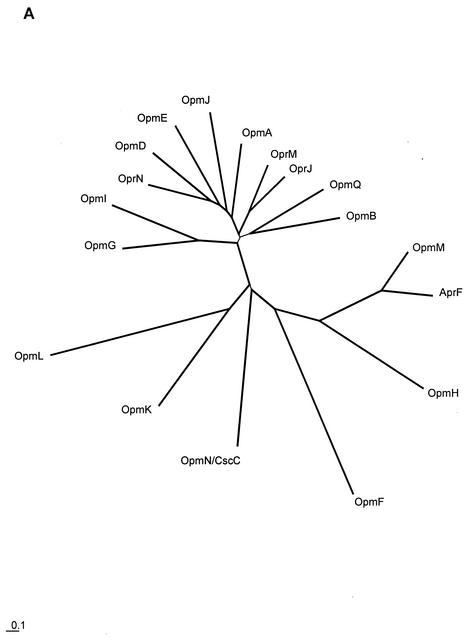
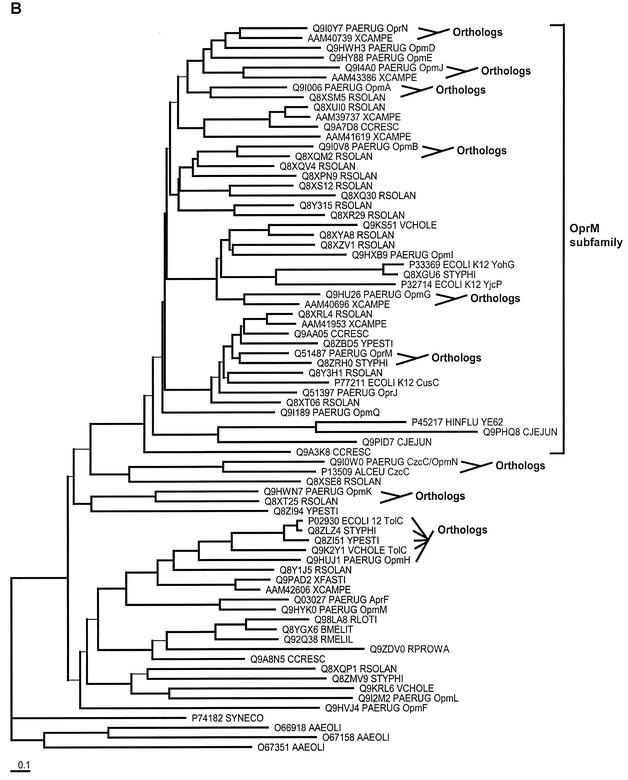
OprM shares significant primary sequence similarity with OprJ, OprN, and the alkaline protease type I secretion protein, AprF. In addition to this, the first P. aeruginosa genome sequence revealed the presence of 14 more homologs of OprM (all were given names starting with Opm, for outer membrane protein M family). The evolutionary relationships between all of these proteins were investigated by phylogenetic analysis (Fig. 1). Efflux proteins OprM, OprJ, and OprN appeared to be members of a distinct group (Fig. 1A), which we termed the OprM subfamily, involving 11 of the 18 total family members. A second more divergent group included the type I secretion protein AprF. A more extensive analysis, which included homologs from the genomes of other representative bacteria, was also performed to assess which of these protein family members are orthologous with genes from other organisms. This identification of orthology can be useful, since orthologs (i.e., genes that diverged only due to speciation) are more likely to share common functions than paralogs (i.e., genes that diverged due to gene duplication). Our analyses indicated that most of the OprM protein family members have orthologs in other organisms studied to date, with some sharing orthology with functionally studied proteins. OpmH was found to be most closely related to E. coli TolC (17) (and TolC orthologs in other organisms), and the level of sequence divergence was consistent with OpmH being the ortholog of TolC. P. aeruginosa OpmN (CzcC) was found to be most similar to, and the probable ortholog of, Alcaligenes eutrophus CzcC, which is involved in cation resistance.
FIG. 1.
Phylogenetic analysis of the 18-member family of outer membrane proteins of P. aeruginosa that appear to be homologous to OprM. OprM, OprN, OprJ, and AprF are the previously experimentally characterized proteins, while the rest of the family members (those with names beginning with Opm) were previously uncharacterized. Phylogenetic analysis was performed on these family members alone (A) and on these family members plus homologs from other selected organisms (B). The latter permitted us to investigate the orthology of these family members with some of the proteins functionally studied in other organisms. To ease visual interpretation with notable functionally studied proteins, only a representative set of organisms was included in this analysis (SWISS-PROT numbers are provided in panel B for all protein sequences). To permit unfinished genome projects to first report their results, genes from organisms with incomplete genomes were avoided. Genes from complete well-described genomes were included, and Alcaligenes eutrophus CzcC was included because of its clear orthology in one case. Trees were created by using the neighbor-joining distance matrix method after alignment of the sequences with ClustalX (with manual editing of the alignment). All bootstrap values for the branching order (out of a total of 1,000 replicates) are over 700, with the exception of the branches shown with thinner lines. Branching order for these latter sequences cannot be determined with any degree of accuracy.
Initial screening of mini-Tn5 mutants.
Mini-Tn5 insertion mutants (H956-H969) in each of the genes encoding the novel OprM homologs (except opmQ) were provided by PathoGenesis Corporation (Chiron). Three of the mutants showed increased susceptibility to all four aminoglycoside antibiotics tested (Table 2). H958 (opmG) showed eightfold decreases in the MICs of kanamycin, gentamicin, and streptomycin, as well as a fourfold decrease in the MIC of tobramycin, compared to the wild-type H911 (PAK). H960 (opmI) showed a 16-fold decrease in the MIC of streptomycin, an eightfold decrease in the MIC of kanamycin, and fourfold decreases in the MICs of gentamicin and tobramycin. H966 (opmH) showed eightfold decreases in the MICs of kanamycin and tobramycin, a fourfold decrease in the MIC of streptomycin, and a twofold change in the MIC of gentamicin. Tetracycline resistance confirmed maintenance of the transposon insertion. Carbenicillin is not a substrate for MexX-MexY (12), and consistent with this, none of the three mutants showed a change in the MIC of this β-lactam compared to H911. The remaining 11 mutants showed no change in susceptibility to any of the four aminoglycosides tested, as shown for mutants in the relatively well-expressed outer membrane channels, opmA, opmD, and opmL. Similarly, deletion of OprM, which some authors have proposed to be the cognate outer membrane pore of MexXY (1, 11, 13), caused only a twofold change in aminoglycoside susceptibility (Table 3), which by convention is considered insignificant.
TABLE 2.
MICs of aminoglycosides, carbenicillin, and tetracycline for P. aeruginosa mutants lacking selected outer membrane channel proteins
| Strain | Phenotype | MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|---|
| Km | Gm | Sm | Tm | Cb | Tc | ||
| H911 | Parent | 100 | 0.8 | 8 | 0.25 | 50 | 1.3 |
| H958 | OpmG− | 13 | 0.1 | 1 | 0.063 | 50 | 25 |
| H966 | OpmH− | 13 | 0.4 | 2 | 0.031 | 50 | 25 |
| H960 | OpmI− | 13 | 0.2 | 0.5 | 0.063 | 100 | 13 |
| H957 | OpmA− | 100 | 0.8 | 8 | 0.25 | 100 | 13 |
| H963 | OpmD− | 100 | 0.8 | 4 | 0.25 | 100 | 25 |
| H962 | OpmL− | 50 | 0.8 | 8 | 0.5 | 100 | 25 |
Abbreviations: Km, kanamycin; Gm, gentamicin; Sm, streptomycin; Tm, tobramycin; Cb, carbenicillin; Tc, tetracycline.
TABLE 3.
Complementation of an OprM− mutant with OpmG and OpmH
| Strain | Phenotype of strain and plasmid | MIC (μg/ml)a
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km | Gm | Ery | Clin | Fus | Cm | Tc | Cb | Imi | Mer | Ctz | Nal | Nfx | Pmb | Acr | CV | ||
| H730 | Parent | 50 | 3.1 | 125 | >750 | 500 | 7.8 | 5 | 12.5 | >5 | 0.63 | 0.032 | 63 | 1.0 | 0.31 | 63 | 63 |
| H743 | OprM− | 25 | 1.6 | 7.8 | 23 | 16 | 1.0 | 0.63 | 0.78 | 5 | 0.63 | 0.02 | 16 | 1.0 | 0.31 | 7.8 | 63 |
| H743(pJJ106) | OprM− + OpmG | 25 | 0.8 | >500 | >750 | 250 | 16 | >20 | 0.78 | 5 | 0.63 | 0.04 | 250 | 64 | 0.31 | 31 | 31 |
| H743(pJJ105) | OprM− + OpmH | 50 | 0.8 | >500 | >750 | 250 | 31 | 10 | 0.78 | 5 | 0.63 | 0.04 | 250 | 64 | 0.16 | 63 | 63 |
Abbreviations: Km, kanamycin; Gm, gentamicin; Ery, erythromycin; Clin, clindamycin; Fus, fusidic acid; Cm, chloramphenicol; Tc, tetracycline; Cb, carbenicillin; Imi, imipenem; Mer, meropenem; Ctz, ceftazidime; Nal, nalidixic acid; Nfx, norfloxacin; Pmb, polymyxin B; Acr, acriflavin; CV, crystal violet.
The opmG and opmH genes were PCR amplified from PAO1 genomic DNA and cloned. The identities of opmG and opmH were confirmed by sequencing. However, this strategy was unsuccessful in our hands for cloning the opmI gene, so only the roles of OpmG and OpmH were explored in greater detail.
Complementation of an oprM mutant susceptibility phenotype.
If OpmG and OpmH were indeed channel-forming outer membrane proteins, then they might be able to complement an oprM deletion mutation. Mutant H743, deficient in OprM, was confirmed to be more susceptible than parent strain H730 to nalidixic acid, carbenicillin, erythromycin, clindamycin, chloramphenicol, and tetracycline, in agreement with previous studies (11). In addition, we observed supersusceptibility to fusidic acid and acriflavin. As noted above, aminoglycoside susceptibility was not significantly changed, nor was susceptibility to the carbapenems, ceftazidime and polymyxin B.
When multicopy plasmids bearing opmG (pJJ106) or opmH (pJJ105) cloned behind the lac promoter in pUCP27 were introduced into strain H743, antibiotic resistance was restored, as indicated by the increased MICs of erythromycin, clindamycin, fusidic acid, chloramphenicol, nalidixic acid, and acriflavin. The MICs of erythromycin, chloramphenicol, and nalidixic acid were indeed higher for the complemented strains than for the parent strain H730. The increased MIC of tetracycline was due to the presence of a tetracycline resistance gene present in the vector pUCP27. No increase in resistance to the aminoglycosides, carbapenems, or polymyxin B was observed, however.
Complementation of opmG and opmH mutant susceptibility phenotypes.
The opmG and opmH genes were cloned behind the lac promoter in pUCP21 to create pJJ107 and pJJ109 and introduced into strains H958 and H966, respectively, by electroporation. The results are shown in Table 4. Strain H958 (opmG::miniTn5-Tcr) showed an eightfold decrease in the MICs of the three aminoglycosides kanamycin, gentamicin, and streptomycin, as well as a fourfold decrease in the MIC of fusidic acid, compared to the parent wild-type strain H911. There were no other significant changes in MICs as a result of insertional inactivation of the opmG gene. Introduction of pJJ107 into strain H958 resulted in full complementation of the MICs of gentamicin, streptomycin, and fusidic acid and partial complementation of the MIC of kanamycin. Therefore, OpmG apparently played a role in aminoglycoside (and fusidic acid) efflux. The increase in carbenicillin resistance upon introduction of pJJ107 was due to the β-lactamase marker on pUCP21.
TABLE 4.
Complementation of OpmG− and OpmH− mutants with OpmG and OpmH
| Strain | Phenotype of strain and plasmid | MIC (μg/ml)a
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km | Gm | Sm | Ery | Clin | Fus | Cm | Tc | Cb | Imi | Mer | Ctz | Nal | Nfx | Pmb | Acr | CV | ||
| H911 | PAK wild type | 100 | 0.8 | 8 | 125 | >750 | 16 | 125 | 1.3 | 50 | 1.6 | 0.63 | 0.08 | 31 | 0.25 | 0.16 | 32 | 125 |
| H958 | OpmG− | 13 | 0.1 | 1 | 63 | 375 | 4 | 125 | >20 | 50 | 1.6 | 0.63 | 0.08 | 31 | 0.25 | 0.16 | 32 | 125 |
| H958 (pJJ107) | OpmG− + OpmG | 25 | 0.8 | 8 | 250 | >750 | 16 | 125 | >20 | 800 | 1.6 | 0.63 | 0.08 | 31 | 0.25 | 0.08 | 32 | 125 |
| H966 | OpmH− | 13 | 0.4 | 2 | 63 | 375 | 2 | 125 | >20 | 50 | 0.8 | 0.15 | 0.08 | 31 | 0.25 | 0.16 | 4 | 16 |
| H743 (pJJ109) | OpmH− + OpmH | 25 | 0.8 | 4 | 125 | >750 | 8 | 250 | >20 | 800 | 1.6 | 0.15 | 0.08 | 31 | 0.25 | 0.3 | 16 | 63 |
Abbreviations: Km, kanamycin; Gm, gentamicin; Sm, streptomycin; Ery, erythromycin; Clin, clindamycin; Fus, fusidic acid; Cm, chloramphenicol; Tc, tetracycline; Cb, carbenicillin; Imi, imipenem; Mer, meropenem; Ctz, ceftazidime; Nal, nalidixic acid; Nfx, norfloxacin; Pmb, polymyxin B; Acr, acriflavin; CV, crystal violet.
Strain H966 (opmH::miniTn5-Tcr) showed less dramatic decreases in the MICs of the three aminoglycosides. There was an eightfold reduction in the MIC of kanamycin, but only two- and fourfold reductions in the MICs of gentamicin and streptomycin, respectively. However, there were also eightfold reductions in the MICs of the dyes acriflavin and crystal violet. Reintroduction of the opmH gene on pJJ109 resulted in only twofold increases in the MICs of the three aminoglycosides, but also a fourfold restoration of the MICs of acriflavin and crystal violet.
The introduction of control pUCP21 and pUCP27 plasmids lacking cloned OpmH or OpmG did not result in any significant alterations to antibiotic resistance except for that conferred by plasmid-borne resistance markers.
Use of DNA mini-microarrays to assess compensation.
DNA mini-microarrays were used to assess the possibility that the antibiotic supersusceptibility phenotypes of opmG and opmH knockout strains were due to a compensatory alteration in expression of another outer membrane channel. Internal 600-bp fragments of each OprM homolog, and the uvrD gene, were PCR amplified from PAO1 genomic DNA. The DNA repair enzyme uvrD was previously determined to be constitutively expressed under all tested conditions (Brazas, unpublished data) and was used as an internal standard. RNA was isolated from H911, H958, and H966 and reverse transcribed to cDNA by using a 5′ primer pool consisting of a mixture of the 5′ primers of each amplicon. cDNA was used as template for low-cycle PCR during which [α-32P]dCTP was incorporated. Due to the relatively low levels of most of the starting mRNAs, this labeling method did not skew the relative transcription levels of individual genes, as confirmed in part by direct assessment of specific DNA hybridization to custom microarrays (data not shown). The resultant radioactive DNA, reflecting the amount of original mRNA, was used to probe the purified PCR amplicons that were spotted onto positively charged nylon membranes by using a replicator.
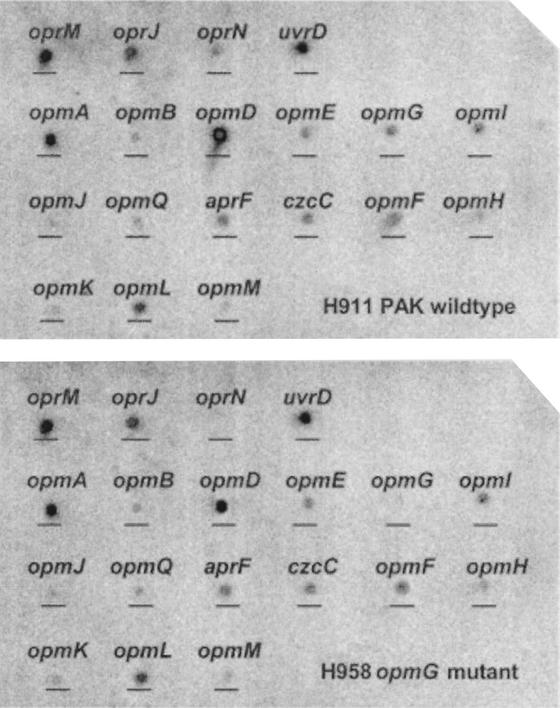
Figure 2 indicates the hybridization pattern for the opmG mutant H958 (opmG::Tn5) compared to that of the parent strain H911. When results were standardized to the level of expression of uvrD, the only significant changes were in the levels of opmG (as anticipated) and oprN message, which were not measurably expressed in the mutant H958. Since it is known that OprN does not contribute to aminoglycoside resistance (10, 15, 20), it is unlikely that the downregulation of OprN would have an effect on the aminoglycoside resistance phenotype of strain H958. Similar expression studies were performed on the mutant H966 (opmH::miniTn5-Tcr) and the parent strain H911. As expected, opmH was not expressed in the mutant, but there were modest decreases in opmG (1.9-fold) and opmI (2-fold) that may have contributed to the H966 resistance phenotype. No alteration in the expression of oprM was observed in either H958 or H966.
FIG. 2.
Comparison of expression of OprM homolog genes in wild-type PAK (H911) (top panel) and the opmG mutant (H958) (bottom panel) by using mini-microarrays. RNA isolated from each strain was reverse transcribed to cDNA, radiolabeled with [α-32P]dCTP, and hybridized to a nylon membrane containing 10-ng spots of 600-bp internal fragments corresponding to each of the OprM homologs. Spot densitometry revealed an absence of oprN expression as well as the expected absence of opmG expression.
DISCUSSION
P. aeruginosa has the largest bacterial genome comprising a single circular chromosome (6.3 Mbp) that has been sequenced to date, and three apparent large families of outer membrane proteins have been identified through genome sequence analysis (21). The 18 members homologous to OprM have been implicated in forming the outer membrane repertoire of proteins required for efflux of toxic compounds and ions and for secretion of extracellular proteins. Eleven of these members appear to form a tight subfamily, according to phylogenetic analysis, and we have termed them the OprM subfamily. Functional similarities within this subfamily appear likely. Of the 18 members of the larger family, 11 (OprM, -J, and -N and OpmA, -B, -D, -E, -G, -I, -J, and -N) are encoded as part of an efflux operon containing genes for an inner membrane RND or MFS efflux pump and a periplasmic linker protein, which are characteristics of the tripartite efflux secretion systems used to export compounds across the gram-negative double membrane. Another six (AprF and OpmQ, -F, -K, -L, and -M) are encoded in operons with genes for ABC-type transporters as well as those for periplasmic linkers proteins. Consequently, of the 18 members of the extended OprM family, only opmH, the proposed ortholog of the E. coli efflux outer membrane channel TolC, is encoded without genes encoding a cognate transporter and linker.
Aminoglycosides are a class of highly cationic amphipathic antibiotics that only recently have been found to be a substrate for multidrug efflux pumps. Of the four characterized antibiotic efflux systems of P. aeruginosa, only MexX-MexY has been implicated in conferring resistance to these drugs. Notably, MexX-MexY is not only constitutively expressed but also inducible by subinhibitory concentrations of some of its substrates (11). However, despite strong evidence about the role of MexX-MexY in intrinsic aminoglycoside resistance (1, 11, 13, 22), data regarding the identity of the native outer membrane component for this tripartite pump are less conclusive (22). It was suggested in some studies that the native outer membrane channel was OprM (11, 12, 13), but this was contradicted by the results of another study (22) and also by this investigation.
Given the number of OprM homologs that have yet to be studied, it seemed possible that another candidate outer membrane pore could be found from within the OprM family. By screening mini-Tn5 insertion mutants in 14 of the uncharacterized OprM homologs, we managed to find three proteins, OpmG, OpmH, and OpmI, that influenced aminoglycoside susceptibility when deleted (Table 2). Significant decreases in the MICs (4- to 16-fold) of kanamycin, gentamicin, streptomycin, and tobramycin were recorded for strains H958, H960, and H966 compared to that of the wild-type strain H911; however, each strain had a different profile with respect to the fold decrease for each individual antibiotic. For example, OpmG disruption had a larger effect on gentamicin susceptibility, while OpmI disruption had a larger effect on streptomycin susceptibility, and knockout of OpmH had a lesser effect on gentamicin and streptomycin susceptibility. It appeared, therefore, that each protein had different, though overlapping, functional specificity for aminoglycosides.
Phylogenetic analysis indicates that OpmG and OpmI are more closely related to each other than to any of the other OprM homologs, reflecting a more recent gene duplication leading to their formation (Fig. 1); however, they share apparent orthologous relationships with putative outer membrane channels from other organisms, so this gene duplication is still ancient. Because of this close paralogous relationship, OpmG and OpmI would be predicted to share a more notable degree of functional similarity with each other than with other OprM family members. However, OpmG and OpmI would not be predicted to share identical functionality, since it is not cost effective for a bacterium to maintain two copies of a gene with identical function. Based on our results obtained to date, we propose that OpmG and OpmI may share some functional similarity in terms of aminoglycoside efflux, though their specificity for particular aminoglycosides may differ. Although inner membrane efflux pumps are thought to be responsible for determining substrate specificity (20), it is possible that such channel-forming outer membrane proteins may have preferred interactions with particular pump-linker components or possibly even a preference for certain general classes of compounds.
P. aeruginosa OpmH is the apparent ortholog of E. coli TolC, a multifunctional efflux channel that has several roles, including hemolysin secretion, colicin V secretion, and multiple antibiotic efflux (11). OpmH is also of particular interest since it is the only member of the extended OprM family that is not encoded as part of an operon containing a periplasmic linker and/or some class of inner membrane transporter. Presumably, the native systems for the other OprM homologs are the systems with which they are each encoded, although this does not rule out the possibility that one protein might function as the native outer membrane component for two (or more) efflux systems. However, for OpmH, the apparent lack of other cotranscribed components of efflux/secretion machinery suggests that, like TolC, it may play a more general role, interacting with more than one efflux system in a nonspecific fashion as required.
To investigate general or specific roles more fully, the genes encoding opmG and opmH were cloned behind the lac promoter of the pUCP27 vector to create the two recombinant vectors pJJ106 and pJJ105. Both the OpmG and OpmH proteins were able to complement the OprM deficiency in strain H743. This ability to complement an OprM deficiency, restoring the substrate specificity of the MexA-MexB-OprM system, suggests a number of important points, namely: (i) both OpmG and OpmH are channel-forming outer membrane proteins capable of mediating efflux of antibiotics from the cell, (ii) substrate specificity for MexA-MexB-OprM is indeed largely determined by the inner membrane components of the efflux pumps, as has been noted by several separate research groups (5, 20, 23), (iii) the ability of channel-forming components to function with alternate pump and linker components (5, 23) is a common feature of this family, and (iv) the outer membrane channels, despite having modest sequence identity (3), still share the same structure and mechanism of action of gating necessary to function in RND efflux.
Cloned OpmG was also able to complement the opmG::miniTn5-Tcr mutant H958, a finding that supports the hypothesis that OpmG plays a role in aminoglycoside resistance. In contrast, OpmH only partly complemented the opmH::miniTn5-Tcr mutant H966. In addition to an aminoglycoside susceptibility phenotype, strain H966 showed decreased MICs for the dyes acriflavin and crystal violet, and cloned OpmH was able to increase the MICs of each dye fourfold, which is a notable finding, since an ortholog of OpmH, TolC of E. coli, was first implicated in the efflux of acriflavin.
MexC-MexD-OprJ and MexE-MexF-OprN have been shown to be upregulated in response to the loss of MexA-MexB-OprM by an unknown mechanism (10). We therefore considered the possibility that a mini-Tn5 insertion mutation in these OprM-family homologs could result in compensatory upregulation or downregulation of other homologs that would then prove problematic in assessing the roles of OpmG and OpmH in aminoglycoside resistance. Mini-microarrays were constructed to monitor expression of all members of this family in the wild-type and mutant strains. It should be noted that microarray analysis measures the amount of transcript as opposed to the amount of protein.
By comparing the expression of the 18 OprM homologs in the parent strain H911 (Fig. 2, top panel) and the opmG mutant H958 (Fig. 2, bottom panel), we confirmed the expected loss of opmG expression as well as the loss of OprN expression. Since OprN does not appear to play a role in aminoglycoside resistance (10, 15, 20), it is unlikely that this change in OprN expression would influence aminoglycoside resistance in strain H958. Conversely, by comparing the expression of the 18 OprM homologs in the parent strain H911 (Fig. 3, top panel) and the opmH mutant H966 (Fig. 3, bottom panel), we showed the expected loss of opmH expression, but in addition, there were modest decreases in opmG (1.9-fold) and opmI (2-fold) expression in all three independent trials of OpmH action. Therefore, it seems possible that the effect of opmH mutations on aminoglycoside resistance was exaggerated by the downregulation of two other outer membrane proteins which appeared to play a role in aminoglycoside resistance. Consistent with this interpretation, cloned OpmH only partially restored aminoglycoside susceptibility.
FIG. 3.
Comparison of expression of OprM homolog genes in wild-type PAK (H911) (top panel) and the opmH mutant (H966) (bottom panel) by using mini-microarrays. RNA isolated from each strain was reverse transcribed to cDNA, radiolabeled with [α-32P]dCTP, and hybridized to a nylon membrane containing 10-ng spots of 600-bp internal fragments corresponding to each of the OprM homologs. Spot densitometry revealed the expected absence of opmH expression as well as 1.9- and 2-fold downregulation of opmG and opmI expression, respectively.
An interesting observation is that the wild-type strain H911 expressed measurable levels of mRNA to all 18 OprM family outer membrane proteins. Some of these, including OpmA, OpmD, and OpmL, were expressed almost as strongly as OprM, while others, including OprJ, OprN, OpmB, OpmE, OpmJ, OpmQ, OpmH, OpmK, and OpmM, were very poorly expressed under the conditions investigated. Previous studies have suggested that OprJ and OprN are not expressed under normal growth conditions, but it seems possible from these studies that they are instead very weakly expressed. Note that this result is not likely due to cross-hybridization between different family members, since no expression signal was ever observed for a gene from this family when it was knocked out. It is obvious that there is much we do not yet understand about efflux systems, their interrelationships with each other, and the roles they play in survival of P. aeruginosa. Nevertheless, the ability to export harmful substances probably plays an important role in the ability of this organism to successfully colonize so many differing environments. Similarly, the intrinsic resistance to antibiotic therapy afforded by these efflux systems plays a large role in the success of P. aeruginosa in producing chronic infections in immunocompromised patients. In particular, the efflux of aminoglycoside antibiotics is of particular interest, since they are a common therapeutic drug for recalcitrant Pseudomonas infections (4) and because it was believed until quite recently that highly charged hydrophilic molecules were not substrates for efflux (15).
This study has identified three possible candidate outer membrane proteins, OpmG, OpmI, and OpmH, that may function in conjunction with the MexY inner membrane RND efflux pump and the MexX linker in aminoglycoside efflux. More detailed studies are consistent with the suggestion that OpmG is a major outer membrane efflux channel in P. aeruginosa PAK, with its relatively closely related paralog OpmI also having a probable overlapping functional role. A lesser role for OpmH may be possible; however, indirect effects on OpmG and OpmI make it possible that OpmH does not play a role or plays a more general, nonspecific role in the cell. Further studies into the regulation of these outer membrane proteins will be required to evaluate their roles in determining resistance in different P. aeruginosa isolates.
Acknowledgments
The financial assistance of the Canadian Cystic Fibrosis Foundation (CCFF), U. S. Cystic Fibrosis Foundation, and Canadian Institutes of Health Research is gratefully acknowledged. J.J. was the recipient of a CCFF studentship, and R.E.W.H. was a Canada Research Chair holder.
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amsterdam, D. 1991. Susceptibility testing of antimicrobials in liquid media, p. 72-78. In V. Lorian (ed.), Antibiotics in laboratory medicine, 3rd ed. Williams and Wilkins, Baltimore, Md.
- 3.Andersen, C., C. Hughes, and V. Koronakis. 2001. Protein export and drug efflux through bacterial channel-tunnels. Curr. Opin. Cell Biol. 13:412-416. [DOI] [PubMed] [Google Scholar]
- 4.Davies, J., and G. D. Wright. 1997. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 5:234-240. [DOI] [PubMed] [Google Scholar]
- 5.Gotoh, N., H. Tsujimoto, A. Nomura, K. Okamoto, M. Tsuda, and T. Nishino. 1998. Functional replacement of OprJ by OprM in the MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 165:21-27. [DOI] [PubMed] [Google Scholar]
- 6.Hancock, R. E. W., and J. S. Lam. 1998. Pseudomonas aeruginosa: infection and immunity, p. 2042-2045. In P. J. Delves (ed.), Encyclopedia of immunology, vol. 1. Academic Press, London, United Kingdom.
- 7.Hancock, R. E. W., and D. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updates 3:247-255. [DOI] [PubMed] [Google Scholar]
- 8.Kohler, T., M. Michea-Hamzepour, U. Henze, N. Gotoh, L. K. Curty, and J. C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 9.Li, X., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, X., N. Barre, and K. Poole. 2000. Influence of the MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and Mex-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 46:885-893. [DOI] [PubMed] [Google Scholar]
- 11.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, T. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, T. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. Yamagachi, X. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 17.Postle, K., and H. Vakharia. 2000. TolC, a macromolecular periplasmic ‘chunnel’. Nat. Struct. Biol. 7:527-530. [DOI] [PubMed] [Google Scholar]
- 18.Rosenberg, E. Y., D. Ma, and H. Nikaido. 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182:1754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Srikumar, R., X. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, J. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, A. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:954-960. [DOI] [PubMed] [Google Scholar]
- 22.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoneyama, H., A. Ocaktan, N. Gotoh, T. Nishino, and T. Nakae. 1998. Subunit swapping in the Mex-extrusion pumps in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 244:898-902. [DOI] [PubMed] [Google Scholar]
- 24.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]