Abstract
We have used proteomic technology to elucidate the complex cellular responses of Bacillus subtilis to antimicrobial compounds belonging to classical and emerging antibiotic classes. We established on two-dimensional gels a comprehensive database of cytoplasmic proteins with pIs covering a range of 4 to 7 that were synthesized during treatment with antibiotics or agents known to cause generalized cell damage. Although each antibiotic showed an individual protein expression profile, overlaps in the expression of marker proteins reflected similarities in molecular drug mechanisms, suggesting that novel compounds with unknown mechanisms of action may be classified. Indeed, one such substance, a structurally novel protein synthesis inhibitor (BAY 50-2369), could be classified as a peptidyltransferase inhibitor. These results suggest that this technique gives new insights into the bacterial response toward classical antibiotics and hints at modes of action of novel compounds. Such a method should prove useful in the process of antibiotic drug discovery.
Infectious diseases are the leading cause of death worldwide, and rapid resistance development is a growing threat even in developed countries (33). In addition to improved hygiene and prudent use of existing antibiotics, the development of novel antibacterial classes is a key to keeping pace with the remarkable adaptability of the bacteria. The search for such novel substances involves understanding of the molecular mechanism of action of the inhibitor and the related bacterial response. Today two major strategies are used in antibiotic drug discovery: (i) evaluation of the structural variations among existing antibiotic classes in order to find compounds which hit the same targets by similar molecular mechanisms yet avoid cross-resistance and (ii) evaluation of novel antibiotic substances arising either from high-throughput target-based assays or from screening for antibacterial activity. If, through its antibacterial activity alone, a novel compound class arouses interest, its molecular target needs to be identified so that undesirable side effects on eukaryotic cells can be minimized (target identification). In addition, for structurally modified antibiotics or compounds derived from the target-based assays it is necessary to prove that interaction with the cellular target is indeed the direct cause for bacterial cell death (target validation).
In this study we used the proteomic approach to study the responses of bacteria to antibacterial compounds. We show that proteome analysis is useful for both target identification and target validation. Proteomics facilitates a broad view on the cell's physiological state and, in combination with radioactive pulse-labeling, allows us to study the cellular response to any changes in growth conditions (32). Recently, proteome analysis has developed to a state which permits investigation of large numbers of different samples. Thus, we started to build a database from two-dimensional (2D) protein analysis of bacterial responses to antibiotic treatment considering all important established and emerging antibiotic classes as well as some substances causing generalized cell damage. We chose the gram-positive model organism Bacillus subtilis in order to profit from its fully sequenced genome (23) and earlier proteome studies focusing on the description of protein signatures of environmental stimuli (2, 5, 7) which are accessible in the Sub2D database (J. Bernhardt and H. Werner, http://microbio2.biologie.uni-greifswald.de:8880/sub2d.htm).
Here we present data for 30 antimicrobial compounds, most of which have been well characterized in terms of their mechanisms of action. By comparison with known antibiotics, we were able to predict the mode of action of the structurally new antibacterial BAY 50-2369. Another example is nitrofurantoin, which has been used for decades in the treatment of urinary tract infections, although its precise mode of action is still unclear (15). Nitrofurantoin led to a protein pattern very similar to that of diamide, suggesting that nitrofurantoin causes nonnative disulfide bonds in the bacterial cell.
Our results show that, by mirroring the complex molecular reactions of bacteria, proteomics may enlarge our view of known antibiotics and help us find new drugs.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
The MICs of the antimicrobial compounds for B. subtilis 168 (trpC2) (1) were determined in test tubes containing 5 ml of a previously described minimal medium (30) and appropriate concentrations of the respective compound inoculated with 105 cells/ml. The tubes were incubated at 37°C for 18 h. The MIC was defined as the lowest concentration that inhibited visible growth.
B. subtilis was cultivated at 37°C in minimal medium. For growth experiments different concentrations of inhibitors were added to exponentially growing cultures when the cultures reached an optical density at 500 nm of 0.4.
For quantitative 2D polyacrylamide gel electrophoresis (2D-PAGE) analysis, inhibitor concentrations that led to reduced growth rates were applied and incubation times at which the differences in the protein synthesis pattern compared to that for the control were most pronounced were chosen. Only in the cases of puromycin and 4-nitroquinoline-1-oxide were protein synthesis quantified at two different time points because the synthesis patterns changed over time. The following concentrations and incubation times were used for quantification: 25 μg of actinonin per ml, 60 min; 12 μg of bacitracin per ml, 10 min; 0.5 μg of BAY 50-2369 per ml, 10 min; 5 μg of cerulenin per ml, 60 min; 15 μg of chloramphenicol per ml, 10 min; 0.2 μg of ciprofloxacin per ml, 10 min; 170 μg of diamide per ml, 10 min; 75 μg of d-cycloserine per ml, 30 min; 0.5 μg of erythromycin per ml, 10 min; 1.6 μg of 5-fluorodeoxyuridine per ml, 60 min; 2 μg of 5-fluorouracil per ml, 10 min; 0.8 μg of fusidic acid per ml, 60 min; 0.25 μg of gentamicin per ml, 10 min; 0.025 μg of gramicidin A per ml, 10 min; 0.4 μg of gramicidin S per ml, 10 min; 3 μg of kanamycin per ml, 10 min; 0.07 μg of methicillin per ml, 10 min; 0.08 μg of mitomycin C per ml, 60 min; 3 μg of monensin per ml, 60 min; 0.06 μg of mupirocin per ml, 60 min; 2.5 μg of 4-nitroquinoline-1-oxide (4-NQO) per ml, 10 min (4-NQO-1) and 60 min (4-NQO-2); 50 μg of nitrofurantoin per ml, 10 min; 15 μg of novobiocin per ml, 10 min; 9 μg of puromycin (PUR) per ml, 10 min (PUR-1) and 40 min (PUR-2); 0.06 μg of rifampin per ml, 10 min; 90 μg of streptomycin per ml, 10 min; 17 μg of tetracycline per ml, 10 min; 0.09 μg of Triton X-100 per ml, 10 min; 10 μg of valinomycin per ml, 10 min; 1.5 μg of vancomycin per ml, 10 min.
Preparation of cytoplasmic l-[35S]methionine-labeled protein fraction.
Cells were labeled with 10 μCi of l-[35S]methionine per ml for 5 min at different time points after treatment with a growth inhibitor, as were untreated control cells when the cells reached an optical density at 500 nm of 0.4. l-[35S]methionine incorporation was stopped by the addition of 1 mg of chloramphenicol per ml and an excess of cold l-methionine (10 mM) while transferring the culture onto ice. The cells were disrupted by ultrasonic treatment, and the soluble cell fraction containing the soluble protein fraction was separated from the insoluble cell remnants by centrifugation. Incorporation of l-[35S]methionine into protein was measured by precipitation of aliquots of protein extracts with 10% trichloroacetic acid on filter papers, as described previously (3).
Analytical and preparative 2D-PAGE.
Analytical 2D-PAGE was performed by the immobilized pH gradient technique as described previously (5). In short, 50 μg of crude protein extract was loaded on immobilized pH gradient strips covering a pH range of 4 to 7 by in-strip rehydration (Amersham Pharmacia Biotech, Piscataway, N.J.). A Multiphor II apparatus (Amersham Pharmacia Biotech) was used for isoelectric focusing, while the second dimension was performed on an Investigator 2D electrophoresis running system (Genomics Solutions, Ann Arbor, Mich.). The gels were stained with silver nitrate (5). Afterwards the gels were dried on filter paper. Exposure to Phosphor screens (Molecular Dynamics, Sunnyvale, Calif.) was followed by detection with a Phosphor Imager SI instrument (Molecular Dynamics). For identification of the proteins by mass spectrometry, nonradioactive protein samples of 200 μg were separated by preparative 2D-PAGE, and the gels were stained with SYPRO Ruby (Molecular Probes, Eugene, Oreg.). SYPRO Ruby-stained gels were detected with a Storm 840 fluorescence scanner (Molecular Dynamics).
Peptide mass fingerprinting.
Tryptic in-gel digestion of protein spots and matrix-assisted laser desorption ionization-time of flight mass spectrometry were performed as described previously (4). Peptide mass fingerprints were analyzed by using MS-Fit software (P. R. Baker and K. R. Clausner, http://prospector.ucsf.edu).
Image analysis and spot quantification.
Delta2D software (Decodon GmbH, Greifswald, Germany) was used to warp autoradiographs of treated cells onto the corresponding control and to quantify the protein spots. Proteins comprising at least 0.05% of the protein synthesized during the l-[35S]methionine pulse and showing an induction of at least twofold in two independent experiments were designated marker proteins. The names of unidentified markers were assigned an abbreviation consisting of the antibiotic by which they were induced and a number.
RESULTS
Setup of a comprehensive database from 2D analysis.
Reproducible growth conditions as well as suitable antibiotic concentrations and incubation times are critical for obtaining high-quality results in protein synthesis pattern analysis. B. subtilis was grown in a defined synthetic medium, and the optical density was measured. During the exponential growth phase different concentrations of the antimicrobial compound under investigation were applied to culture aliquots, and growth was monitored until the untreated control reached stationary phase. For proteome analysis samples were collected at time points from 10 to 60 min after treatment with antibiotics at concentrations that during the time monitored caused a reduction in the growth rate but did not inhibit growth completely. This was difficult only in those cases in which growth was unaffected up to a critical concentration which, once reached, caused rapid cell lysis, as seen with gramicidin S.
2D-PAGE separates cytoplasmic proteins from crude protein extracts in a first step by their pIs and in a second step by their Mrs (28). The theoretical 2D protein map of B. subtilis (7) shows that the pIs of the majority of cytoplasmic proteins are in the range of 4 to 7. In the present study we concentrated on this pI range as it also gives a good spot resolution. Pulse-labeling with l-[35S]methionine enabled us to specifically detect the protein fraction newly synthesized in response to antibiotic treatment on autoradiographs of the dried 2D gels. Antibiotic treatment resulted in a reduction in l-[35S]methionine incorporation of up to 3 orders of magnitude, which led us to apply onto the 2D gels equal amounts of protein extract rather than radioactivity. The times of exposure of the dried 2D gels to Phosphor screens were adjusted in order to guarantee full exploitation of the screens' dynamic range by the strongest signal. For quantification we calculated the percentage of radioactivity in the protein spot of interest in relation to the total radioactive signal generated by the sum of all protein spots on the gel. Thus, it is important to bear in mind that such relative synthesis rates do not allow conclusions to be drawn about the absolute amount of protein synthesized during the pulse under the various conditions.
Image analysis software assists with the warping of one gel image so that it fits onto another, which allowed us to directly compare the protein synthesis patterns of antibiotic-treated and untreated cells. The gel image derived from antibiotic-treated cells was assigned the false color red and warped to fit onto a green control image. Thus, antibiotic-induced proteins appear red, repressed proteins appear green, and proteins synthesized under both conditions appear yellow. Among all concentrations and time points tested for each antibiotic, in this visual assessment of the differences, the image that we used for quantification was the one that showed the greatest changes in the protein synthesis pattern compared to the control pattern. To facilitate the comparison of protein expression profiles, for each compound we selected marker proteins which in two independent experiments had relative synthesis rates of at least 0.05% and showed at least a twofold induction. The identities of the marker proteins, which accumulated to an extent that allowed analyses by mass spectrometry, were confirmed. About 50 of the marker proteins were identified for the first time after treatment with the antimicrobial agents and could be added to the cytosolic 2D protein map of B. subtilis (7).
To build a comprehensive database with data from 2D analysis, antibiotics from all important classical and emerging classes were tested for their influence on the B. subtilis proteome. These included all important classes of protein synthesis inhibitors (mupirocin, tetracycline, chloramphenicol, erythromycin, fusidic acid, puromycin, gentamicin, streptomycin, kanamycin) and representatives of important inhibitors of cell wall biosynthesis (d-cycloserine, bacitracin, vancomycin, methicillin). Furthermore, the effects of the base analogues 5-fluorouracil and 5-fluorodeoxyuridine were investigated, RNA biosynthesis was inhibited by rifampin, and DNA gyrase function was addressed by using novobiocin and ciprofloxacin. Covalent modification of DNA was studied by using mitomycin C and 4-nitroquinoline-1-oxide. Novel target areas such as fatty acid synthesis and peptide deformylase were studied by using cerulenin and actinonin, respectively. Less specific interactions with the integrity of the cytoplasmic membrane were addressed by gramicidin S and gramicidin A, monensin, valinomycin, and Triton X-100. In addition, we tested nitrofurantoin, for which contradictory mechanisms of action have been described. Finally, we included the structurally novel antibiotic agent BAY 50-2369, the molecular mechanism of action of which was not known.
Consistency check: do the 2D patterns reflect the antimicrobial mode of action?
The cellular response to antibiotic treatment can be expected to be of different complexities depending on the mode of action of the antibiotic compound and on the cells' sensory capabilities and responsiveness. Changes in the protein synthesis pattern mirror the global response to the damage caused in the cell. This might explain why each of the compounds tested showed an individual protein expression profile which is also reflected by a distinct set of marker proteins. We found this high selectivity of 2D protein analysis to be rather unexpected.
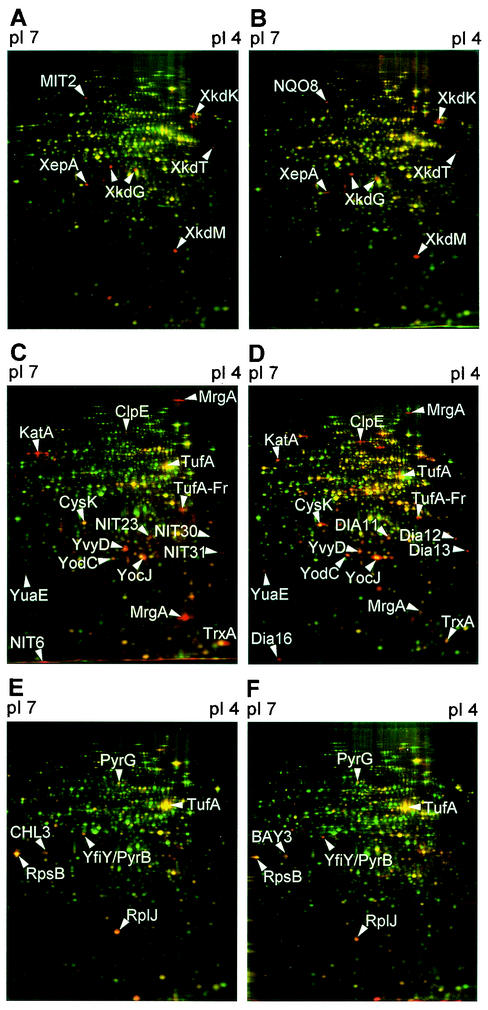
Many marker proteins can be explained by the regulatory phenomena described in the literature. Mitomycin C and 4-nitroquinoline-1-oxide, known to disturb the DNA structure by covalent binding, induce the SOS response in B. subtilis (12). Consistent with this report, proteins of the prophage PBSX, whose induction is RecA dependent, constitute marker proteins for both substances (Fig. 1A and B). A distinction between both substances was still possible. 4-Nitroquinoline-1-oxide only transiently induced RecA and part of the well-known oxidative stress response (2) 10 min after treatment. In the case of mitomycin C, RecA was continuously induced for 60 min, and DinB, another protein belonging to the SOS response, was also among the markers. When RNA synthesis was inhibited by rifampin for 10 min, only a few of the housekeeping proteins were still detectable (3); those proteins were probably translated from relatively stable mRNAs. For mupirocin, which inhibits Ile-tRNA synthetase, the proteome signature was indicative of the classical stringent response (10, 11) and included the induction of the gene products of the ilv-leu operon. Antibiotics that inhibited protein biosynthesis by interfering with ribosomal translation accuracy, such as gentamicin, kanamycin, and streptomycin (24), as well as puromycin, which causes abortive translation (20), induced GroES/L (representing class I heat shock proteins). This chaperon machinery is induced by misfolded proteins appearing due to mistranslation. As observed earlier for Escherichia coli (31) and Haemophilus influenzae (9), the group of inhibitors of translation elongation (tetracycline, erythromycin, chloramphenicol, fusidic acid) shared marker proteins that belong to the translation machinery: ribosomal proteins and elongation factors.
FIG. 1.
The protein expression profiles of B. subtilis 168 60 to 65 min after treatment with mitomycin C at 0.08 μg/ml (the MIC) (A) and 4-nitroquinoline-1-oxide at 2.5 μg/ml (five times the MIC) (B) share marker proteins. Marker proteins overlap strongly for the protein expression profiles obtained 10 to 15 min after treatment with nitrofurantoin at 50 μg/ml (five times the MIC) (C) and diamide at 170 μg/ml (two times the MIC) (D). Also, the protein expression profiles obtained 10 to 15 min after treatment with chloramphenicol at 15 μg/ml (five times the MIC) (E) and BAY 50-2369 at 0.5 μg/ml (the MIC) (F) have marker proteins in common. 2D-PAGE was used to separate pulse-labeled cytoplasmic proteins in the pI range of 4 to 7 according to their pIs and Mrs. Red false-color images of autoradiographs of antibiotic-treated cells are warped to fit onto the corresponding green control images. Proteins induced by the antibiotic appear red, repressed proteins appear green, and proteins which are synthesized in the control as well as after antibiotic treatment appear yellow. Arrowheads indicate shared marker proteins (for a full list of marker proteins, see supplementary material at http://microbio2.biologie.uni-greifswald.de:8880/antibiotics/).
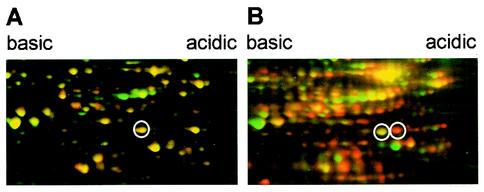
The protein pattern for the peptide deformylase inhibitor actinonin was unique among the compounds tested. It was impossible to directly warp the antibiotic gel image so that it fit onto the control because the positions of many spots had greatly shifted to a more acidic pI. Therefore, as a variation of the procedure described above, dual-channel imaging (5) was used to fit the autoradiographs for the protein fraction newly synthesized during actinonin treatment onto the corresponding SYPRO Ruby-stained images for the total cellular protein (Fig. 2). We were able to demonstrate that the pI shift is caused by the formyl residue still attached to the N terminus due to peptide deformylase inhibition (4). This pI shift makes the actinonin signature characteristic, although it was impossible to define marker proteins without verifying the identity of each protein spot on the gel.
FIG. 2.
Treatment of B. subtilis with actinonin leads to a shift in the pI of the newly synthesized protein fraction. The newly synthesized pulse-labeled protein fraction was set against SYPRO Ruby-stained total protein. The details of 2D images show red false-color images of autoradiographs of untreated control cells (A) and cells treated with actinonin at 125 μg/ml (five times the MIC) for 10 min (B) that are warped to fit onto the corresponding green images of the SYPRO Ruby-stained gels. Proteins that had already accumulated before actinonin treatment and that are still being synthesized during the l-[35S]methionine pulse appear yellow. Accumulated proteins no longer synthesized appear green, and proteins newly induced in response to actinonin treatment appear red. Circles exemplify one protein which shifts to a more acidic pI after actinonin treatment.
Little is known about the global cellular reaction to membrane disruption. The pattern of gramicidin S resembled that of the untreated control probably because there is no intermediate stage between cells being either unaffected or rapidly killed by gramicidin S. This was supported by growth experiments in which different concentrations of gramicidin S were added to exponentially growing B. subtilis cultures, in which cells were unaffected by gramicidin S at twice the MIC but lysed shortly after addition of gramicidin S at four times the MIC. Gramicidin A, valinomycin, and Triton X-100 all induced NH4-dependent NAD+ synthase and shared other marker proteins of mostly unknown function. Cerulenin, an inhibitor of fatty acid synthesis, induced proteins involved in fatty acid synthesis and, moreover, shared some additional marker proteins with monensin, suggesting that fatty acid depletion leads to membrane destabilization.
These examples demonstrate that many details of the bacterial proteomic response are consistent with preexisting knowledge on the antibiotic mechanisms of action. In addition, many of the protein expression profiles showed reactions not anticipated before, adding novel pieces to the puzzle of understanding antibiotic action.
New view on well-known antibiotics.
Three different molecular mechanisms contribute to the inhibitory activity of 5-fluorouracil: incorporation into RNA (16, 18, 22), inhibition of tRNA-uracil methyltransferase (18), and inhibition of thymidylate synthetase after conversion to 5-fluorodeoxyuridine monophosphate by uracil phosphoribosyltransferase (22, 27). 5-Fluorodeoxyuridine is converted to 5-fluorodeoxyuridine monophosphate by thymidine kinase (26). In contrast to rifampin, in our experiments neither 5-fluorouracil nor 5-fluorodeoxyuridine led to an immediate decrease in protein synthesis, nor did we observe an induction of the SOS response as described for E. coli (27). Furthermore, the two compounds did not show overlapping protein signatures under the conditions tested, contrary to what might have been expected from the overlapping mode of action. Interestingly, antitermination-regulated aminoacyl-tRNA synthetases (25) are repressed after 5-fluorouracil treatment. The interaction of uncharged tRNA with the leader RNA is probably prevented by changes in secondary structure due to 5-fluorouracil incorporation into mRNA or tRNA. The first two enzymes involved in pyrimidine synthesis were repressed by 5-fluorouracil, while CTP synthetase was induced. This deregulation of pyrimidine synthesis seems to be the major effect of 5-fluorouracil on B. subtilis. This interpretation was supported by subsequent MIC tests. The MIC of 5-fluorouracil was found to be 0.039 μg/ml; and supplementation with uracil, uridine, and cytidine (MIC > 10 μg/ml) but not with thymidine was successful. 5-Fluorodeoxyuridine treatment (MIC = 0.0098 μg/ml) was complemented most efficiently by thymidine (MIC = 2.5 μg/ml), confirming that 5-fluorodeoxyuridine is a more potent inhibitor of thymidylate synthetase than 5-fluorouracil (6).
As with E. coli (31) and H. influenzae (9), the inhibitors of translation elongation, tetracycline, chloramphenicol, erythromycin, and fusidic acid, led to induction of stringently controlled ribosomal proteins and translation factors. However, in contrast to the response in E. coli, in which ribosomes were suggested to be the sensors of heat and cold shock (31), the response to these antibiotics in B. subtilis did not overlap with the cold shock response of B. subtilis, which involves induction of cold shock-specific proteins CspB, CspC, and CspD (19).
Different ideas on the antibacterial mechanism of nitrofurantoin were published during the last 5 decades. These include inhibition of the bacterial enzymes involved in DNA and RNA synthesis and carbohydrate metabolism and other metabolic enzymes (15). The toxic side effects on eukaryotic cells were attributed to a rapid formation of glutathione-glutathione disulfides, mixed glutathione-protein disulfides, and protein-protein disulfides (17, 29). Surprisingly, this oxidative protein damage has never been discussed to play a role in antimicrobial activity. We happened to test diamide, an agent used to cause oxidative damage by inducing nonnative disulfide bonds (21). The protein signatures of nitrofurantoin and diamide, although distinct in detail, overlap to a great extent (Fig. 1C and D). On the basis of the data from 2D-PAGE, we therefore suggest that protein inhibition by nonnative disulfide bonds may be the primary mode of action of nitrofurantoin in prokaryotes as well as in eukaryotes. The inactivation of cysteines in the catalytic centers of enzymes together with the structural distortions caused by nonnative disulfide bonds could present a straightforward explanation for the low level of evolutionary resistance development and the pleiotropic effects on different metabolic pathways. We observed the formation of glutathione-glutathione disulfides in vitro after diamide incubation but not after nitrofurantoin incubation, which is in accordance with the previous observation that nitrofurantoin needs to be activated in vivo (15).
Does clustering by marker proteins reflect molecular drug mechanisms?
In light of the complexity of the proteomic responses to the antibiotics, a simplified tool used independently of preexisting knowledge on a specific mode of action would be instrumental in the classification of novel compounds. To this end, we intentionally left the level of physiological interpretation of the protein expression profiles and tried to group antibiotics solely by the number of shared marker proteins (Table 1) antibiotic varied from 0 to 34, with an average of 13.3. The highest number of common marker proteins shared by two individual antibiotics was 12. This maximal overlap was observed for nitrofurantoin and diamide, for which we propose a common mode of action.
TABLE 1.
Total and shared marker proteinsa
| Overlapping marker proteins | No. of shared marker proteins
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Actin- onin | Erythro- mycin | BAY502369 | Chloram- phenicol | Tetra- cycline | Fusidic acid | PUR-2 | Diamide | Nitrofur- antoin | 4-NQO-1 | PUR-1 | Genta- mycin | Kana- mycin | Strepto- mycin | 4-NQO-2 | Mito- mycin | |
| Actinonin | ||||||||||||||||
| Erythromycin | ||||||||||||||||
| BAY 50-2369 | 4 | |||||||||||||||
| Chloramphenicol | 4 | 6 | ||||||||||||||
| Tetracycline | 3 | 6 | 7 | |||||||||||||
| Fusidic acid | 5 | 4 | 6 | 6 | ||||||||||||
| PUR-2 | 1 | 2 | 2 | 2 | 3 | |||||||||||
| Diamide | 1 | 4 | ||||||||||||||
| Nitrofurantoin | 1 | 2 | 2 | 2 | 3 | 12 | ||||||||||
| 4-NQO-1 | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 4 | ||||||||
| PUR-1 | 3 | 3 | 1 | 2 | ||||||||||||
| Gentamicin | 1 | 1 | 1 | 3 | ||||||||||||
| Kanamycin | 1 | 2 | 1 | 3 | 2 | |||||||||||
| Streptomycin | 1 | 2 | 1 | 2 | 1 | 2 | ||||||||||
| 4-NQO-2 | ||||||||||||||||
| Mitomycin | 1 | 1 | 4 | |||||||||||||
| Gramicidin A | 1 | 1 | 1 | |||||||||||||
| Triton X-100 | 1 | 1 | 1 | 1 | ||||||||||||
| Valinomycin | 1 | 1 | 1 | |||||||||||||
| d-Cycloserine | 1 | 1 | ||||||||||||||
| Mupirocin | 1 | 1 | 1 | 1 | ||||||||||||
| Cerulenin | ||||||||||||||||
| Monensin | 2 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| Rifampin | 1 | 1 | 1 | 1 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | 1 | ||||
| 5-Fluorouracil | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | ||||||||
| Gramicidin S | 1 | |||||||||||||||
| Vancomycin | 1 | 1 | ||||||||||||||
| Bacitracin | 1 | 1 | 1 | 2 | ||||||||||||
| Novobiocin | 1 | 1 | 1 | 2 | 2 | 1 | ||||||||||
| Ciprofloxacin | 1 | |||||||||||||||
| 5-Fluorodeoxyuridine | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | ||||||
| Methicillin | ||||||||||||||||
| Total marker proteins | 0 | 13 | 12 | 14 | 10 | 20 | 24 | 29 | 34 | 10 | 7 | 4 | 4 | 4 | 7 | 8/PICK> |
| 4 | ||||||||||||||||
| 2 | 6 | |||||||||||||||
| 2 | 4 | |||||||||||||||
| 3 | 1 | 6 | ||||||||||||||
| 1 | 2 | 2 | 3 | 5 | ||||||||||||
| 1 | 1 | 1 | 2 | 2 | 5 | |||||||||||
| 1 | ||||||||||||||||
| 1 | ||||||||||||||||
| 1 | 1 | 1 | ||||||||||||||
| 1 | 2 | 1 | 1 | 1 | 2 | |||||||||||
| 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||
| 1 | 1 | 1 | 1 | 1 | ||||||||||||
| 5 | 26 | 26 | 6 | 20 | 19 | 20 | 32 | 10 | 5 | 5 | 11 | 21 | 10 | 8 | 0 | |
Relatively few marker proteins were obtained for cell wall synthesis inhibitors, with the tendency that the later in the process the inhibition takes place, the less the cells are able to react to it. Thus, d-cycloserine, which inhibits cytosolic cell wall biosynthesis steps, yielded six marker proteins; vancomycin, which binds to a membrane-associated peptidoglycan precursor, resulted in five marker proteins; and the β-lactam methicillin, which acts at the final transpeptidation step outside the cytoplasmic membrane, showed no marker protein. The last finding is in accordance with the view that β-lactam-treated cells die as they continue to initiate cell division because they do not sense the absence of the cross wall (13). Bacitracin, which traps the universal lipid carrier undecaprenyl-pyrophosphate and effects not only peptidoglycan biosynthesis but also teichoic, teichuronic, and lipoteichoic acid biosynthesis, yielded 12 marker proteins.
4-Nitroquinoline-1-oxide and mitomycin C, two DNA-damaging agents, clustered together and shared marker proteins that belong to the SOS response of B. subtilis.
The membrane-damaging agents monensin, gramicidins A and S, valinomycin, and Triton X-100 did not fall into a uniform cluster, demonstrating that each of them has a distinct mode of action and leads to a different cellular response. Nevertheless, subsets of Triton X-100 marker proteins overlap with those of valinomycin and gramicidin A, implying common characteristics of the bacterial response in disturbance of the membrane structure. It is noteworthy that cerulenin shared marker proteins with monensin, suggesting that the cerulenin-induced inhibition of fatty acid biosynthesis eventually leads to a loss of membrane integrity.
Inhibitors of protein synthesis could be roughly divided into two groups. The first comprised the aminoglycosides kanamycin, gentamicin, and streptomycin, which interfere with translation accuracy, and puromycin, which leads to abortive translation. The translation elongation inhibitors chloramphenicol, erythromycin, tetracycline, and fusidic acid represented the second group and shared an average of 5.2 marker proteins. Interestingly, the Ile-tRNA synthetase inhibitor mupirocin did not fall into one of these clusters but shared marker proteins with rifampin, monensin, cerulenin, and d-cycloserine. This result might reflect the multiple aspects of the stringent response including down-regulation of mRNA, membrane, and cell wall biosynthesis (8).
Several of the antibiotics tested did not share marker proteins with any of the other compounds, indicative of their unique modes of action. Examples include novobiocin and ciprofloxacin, which possess distinct molecular mechanisms, although they both interfere with the activity of DNA gyrase. Similarly, among the inhibitors of cell wall synthesis the maximum overlap was two marker proteins, again showing their diverse modes of action.
Finally, we applied the concept of marker proteins to the novel pyrimidinone antibiotic BAY 50-2369, which is structurally related to the natural compound TAN 1057 A/B. BAY 50-2369 clustered along with the translation elongation inhibitors (Table 1), and its protein expression profile closely resembled those of the other members of the group, especially chloramphenicol and tetracycline (Fig. 1E and F). This result leads us to propose that BAY 50-2369 acts by inhibition of the peptidyltransferase reaction, an interpretation that is supported by independent studies performed in the meantime with TAN 1057 A/B (N. Boddecker, G. Bahador, E. Mabery, J. J. Wolf, L. Xu, and J. C. Watson, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., p. 245, 2001).
DISCUSSION
Proteomic technologies have been greatly refined during the past decade and have been applied to investigate differences in the protein expression profiles of cells grown under a broad spectrum of growth conditions and with different stress factors including some antibiotics inhibiting protein synthesis or gyrase function (9, 14, 31). We envisioned a further application for proteomics (9, 31): study of the complex cellular responses to antibiotics from all modern classes should give us new insights into antibiotic action and should provide us with a large set of data to which data for novel antibacterial compounds could be compared. The cellular response manifest in the protein expression profiles could be a new means of antibiotic classification.
To this end, we established a database of bacterial responses to a comprehensive set of antibiotics covering well-known and emerging target areas. Each of the 30 compounds tested revealed a complex but characteristic protein expression profile. Many changes in the protein synthesis patterns in response to the antibiotics were consistent with existing knowledge on the modes of action and on the cellular responses to changes in environmental conditions. Not surprisingly, for instance, inhibition of Ile-tRNA synthetase induced the stringent response, and protein synthesis inhibitors that interfere with translation accuracy induced class I heat shock proteins known to be induced by misfolded proteins. Each response, however, also yielded new information, be it, for example, the expression of proteins with unknown function, a shift in the pIs of proteins newly synthesized after actinonin treatment, or the good correlation of the protein expression profiles of nitrofurantoin and diamide. Such examples expose our still limited understanding of the mechanisms of action of even long-established antibiotics.
We were able to demonstrate that similarities in molecular drug mechanisms resulted in a good correlation of protein expression profiles. This enabled us to define 122 marker proteins (a maximum of 34 per antibiotic), the expression of which was induced at least twofold compared to that for the untreated control. The marker protein concept was then successfully applied to BAY 50-2369. This novel compound was classified by this method as an inhibitor of the peptidyltransferase step of protein synthesis, a result that was independently confirmed by the activity of the structurally related compound TAN 1057 A/B in cell-free translation systems (Boddecker et al., 41st ICAAC). Another example for target identification is provided by the good correlation between the protein expression profile of nitrofurantoin, the mechanism of action of which is still puzzling, despite 50 years of therapeutic application (15), and diamide, an agent known to cause protein damage by induction of nonnative disulfide bonds (21).
Target identification will critically rely either on the availability of similar protein expression profiles for comparison or on the detailed investigation of proteome signatures induced by the compound tested. For target validation, proteome analysis of mutants may be helpful if an inhibitor of a novel target of interest is not yet available. For certain groups of inhibitors it might be beneficial to extend the analysis to different pI ranges or to include different protein fractions to increase the number of marker proteins. For instance, it would be interesting to identify marker proteins in the membrane fraction that help differentiate between membrane-active antibacterial compounds. There are limitations to using 2D-PAGE to separate membrane proteins, however, as this method crucially relies on protein solubilization. RNA profiling, which is capable of addressing the expression of all genes in an organism, can be used to complement proteome analysis. It will be crucial to gather in one database all available information on protein expression profiles, including protein modifications that have been detected, and supplement this data with RNA profiling data for as many conditions as possible. A database of this type will be able to facilitate the identification of more comprehensive signatures for treatment with antimicrobial agents and support functional analysis by combined protein and RNA profiling.
Acknowledgments
We thank Karin Binder for excellent support with 2D-PAGE and Doreen Kliewe, Thomas Kretschmann, Anita John, Dorian Schönfeld, Katy Raddatz, and Falko Hochgräfe for assisting in the identification of marker proteins. We thank Ben Newton for carefully reading and improving the manuscript.
This work was supported by a grant from Bayer AG, Wuppertal, Germany.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., J. Bernhardt, R. Schmid, H. Mach, U. Volker, and M. Hecker. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18:1451-1463. [DOI] [PubMed] [Google Scholar]
- 3.Bandow, J. E., H. Brötz, and M. Hecker. 2002. Bacillus subtilis tolerance of moderate concentrations of rifampin involves the sigma(B)-dependent general and multiple stress response. J. Bacteriol. 184:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandow, J. E., D. Becher, K. Büttner, F. Hochgräfe, C. Freiberg, H. Brötz, and M. Hecker. The role of peptide deformylase in protein biosynthesis-a proteomic study. Proteomics, in press. [DOI] [PubMed]
- 5.Bernhardt, J., K. Buttner, C. Scharf, and M. Hecker. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225-2240. [DOI] [PubMed] [Google Scholar]
- 6.Bosch, L., E. Harbers, and C. Heidelberger. 1958. Studies on fluorinated pyrimidines. V. Effects on nucleic acid metabolism in vitro. Cancer Res. 18:335-343. [PubMed] [Google Scholar]
- 7.Buttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Volker, U. Volker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 8.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 9.Evers, S., K. Di Padova, M. Meyer, H. Langen, M. Fountoulakis, W. Keck, and C. P. Gray. 2001. Mechanism-related changes in the gene transcription and protein synthesis patterns of Haemophilus influenzae after treatment with transcriptional and translational inhibitors. Proteomics 1:522-544. [DOI] [PubMed] [Google Scholar]
- 10.Eymann, C., and M. Hecker. 2001. Induction of sigma(B)-dependent general stress genes by amino acid starvation in a spo0H mutant of Bacillus subtilis. FEMS Microbiol. Lett. 199:221-227. [DOI] [PubMed] [Google Scholar]
- 11.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman, B. M., and R. E. Yasbin. 1983. The genetics and specificity of the constitutive excision repair system of Bacillus subtilis. Mol. Gen. Genet. 190:481-486. [DOI] [PubMed] [Google Scholar]
- 13.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62:1371-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gmuender, H., K. Kuratli, K. Di Padova, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guay, D. R. 2001. An update on the role of nitrofurans in the management of urinary tract infections. Drugs 61:353-364. [DOI] [PubMed] [Google Scholar]
- 16.Heidelberger, C. 1965. Fluorinated pyrimidines. Prog. Nucleic Acid Res. Mol. Biol. 4:1-50. [DOI] [PubMed] [Google Scholar]
- 17.Hoener, B., A. Noach, M. Andrup, and T. S. Yen. 1989. Nitrofurantoin produces oxidative stress and loss of glutathione and protein thiols in the isolated perfused rat liver. Pharmacology 38:363-373. [DOI] [PubMed] [Google Scholar]
- 18.Ivanetich, K. M., and D. V. Santi. 1988. Thymidylate synthase and fluorouracil. Adv. Exp. Med. Biol. 244:113-124. [DOI] [PubMed] [Google Scholar]
- 19.Kaan, T., G. Homuth, U. Mader, J. Bandow, and T. Schweder. 2002. Genome-wide transcriptional profiling of Bacillus subtilis cold-shock response. Microbiology 148:3441-3455. [DOI] [PubMed] [Google Scholar]
- 20.Kirillov, S., B. T. Porse, B. Vester, P. Woolley, and R. A. Garrett. 1997. Movement of the 3′-end of tRNA through the peptidyl transferase centre and its inhibition by antibiotics. FEBS Lett. 406:223-233. [DOI] [PubMed] [Google Scholar]
- 21.Kosower, N. S., and E. M. Kosower. 1995. Diamide: an oxidant probe for thiols. Methods Enzymol. 251:123-133. [DOI] [PubMed] [Google Scholar]
- 22.Koyama, F., H. Sawada, H. Fuji, H. Hamada, T. Hirao, M. Ueno, and H. Nakano. 2000. Adenoviral-mediated transfer of Escherichia coli uracil phosphoribosyltransferase (UPRT) gene to modulate the sensitivity of the human colon cancer cells to 5-fluorouracil. Eur. J. Cancer 36:2403-2410. [DOI] [PubMed] [Google Scholar]
- 23.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 24.Lafontaine, D. L., and D. Tollervey. 2001. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2:514-520. [DOI] [PubMed] [Google Scholar]
- 25.Luo, D., J. Leautey, M. Grunberg-Manago, and H. Putzer. 1997. Structure and regulation of expression of the Bacillus subtilis valyl-tRNA synthetase gene. J. Bacteriol. 179:2472-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyata, S., H. Mikami, M. Tai, T. Hori, and H. Fujita. 1991. Competitive binding radioassay for the determination of 5-fluorodeoxyuridine and 5-fluorodeoxyuridine-5′-monophosphate levels in plasma and tumor tissue. Jpn. J. Cancer Res. 82:742-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oda, Y. 1987. Induction of SOS responses in Escherichia coli by 5-fluorouracil. Mutat. Res. 183:103-108. [DOI] [PubMed] [Google Scholar]
- 28.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 29.Silva, J. M., S. Khan, and P. J. O'Brien. 1993. Molecular mechanisms of nitrofurantoin-induced hepatocyte toxicity in aerobic versus hypoxic conditions. Arch. Biochem. Biophys. 305:362-369. [DOI] [PubMed] [Google Scholar]
- 30.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041-2045. [DOI] [PubMed] [Google Scholar]
- 31.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VanBogelen, R. A., E. E. Schiller, J. D. Thomas, and F. C. Neidhardt. 1999. Diagnosis of cellular states of microbial organisms using proteomics. Electrophoresis 20:2149-2159. [DOI] [PubMed] [Google Scholar]
- 33.World Health Organization. 2001. WHO global strategy for containment of antimicrobial resistance. Report W.H.O./CDS/CSR/DRS/2001.2. World Health Organization, Geneva, Switzerland.