Abstract
Pseudomonas aeruginosa strains are less susceptible to tigecycline (previously GAR-936; MIC, 8 μg/ml) than many other bacteria (P. J. Petersen, N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa, Antimicrob. Agents Chemother. 43:738-744, 1999). To elucidate the mechanism of resistance to tigecycline, P. aeruginosa PAO1 strains defective in the MexAB-OprM and/or MexXY (OprM) efflux pumps were tested for susceptibility to tigecycline. Increased susceptibility to tigecycline (MIC, 0.5 to 1 μg/ml) was specifically associated with loss of MexXY. Transcription of mexX and mexY was also responsive to exposure of cells to tigecycline. To test for the emergence of compensatory efflux pumps in the absence of MexXY-OprM, mutants lacking MexXY-OprM were plated on medium containing tigecycline at 4 or 6 μg/ml. Resistant mutants were readily recovered, and these also had decreased susceptibility to several other antibiotics, suggesting efflux pump recruitment. One representative carbenicillin-resistant strain overexpressed OprM, the outer membrane channel component of the MexAB-OprM efflux pump. The mexAB-oprM repressor gene, mexR, from this strain contained a 15-bp in-frame deletion. Two representative chloramphenicol-resistant strains showed expression of an outer membrane protein slightly larger than OprM. The mexCD-OprJ repressor gene, nfxB, from these mutants contained a 327-bp in-frame deletion and an IS element insertion, respectively. Together, these data indicated drug efflux mediated by MexCD-OprJ. The MICs of the narrower-spectrum semisynthetic tetracyclines doxycycline and minocycline increased more substantially than did those of tigecycline and other glycylcyclines against the MexAB-OprM- and MexCD-OprJ-overexpressing mutant strains. This suggests that glycylcyclines, although they are subject to efflux from P. aeruginosa, are generally inferior substrates for P. aeruginosa efflux pumps than are narrower-spectrum tetracyclines.
Pseudomonas aeruginosa is a clinically important gram-negative opportunistic pathogen causing serious acute and chronic infections (8, 11). The exceptional array of intrinsic and acquired drug resistance mechanisms employed by P. aeruginosa renders antibiotic treatment of these infections problematic. One important resistance mechanism is efflux mediated by the so-called resistance nodulation division (RND) family of efflux pumps. Four RND pumps have been described in P. aeruginosa: MexAB-OprM (16), MexCD-OprJ (31), MexEF-OprN (14), and MexXY-OprM (1, 25, 40). RND pumps consist of an inner membrane transporter (MexB, MexD, MexF, and MexY), an outer membrane channel-forming component (OprM, OprJ, and OprN), and a membrane fusion protein (MexA, MexC, MexE, and MexX) (27). RND pumps show broad specificity, and their tripartite architecture allows extrusion of compounds directly from the cytoplasm to the external environment. Efflux pump action in P. aeruginosa leads to particularly high levels of drug resistance as a result of apparent synergism with the atypically impermeable outer membrane, which limits influx of antimicrobial agents (17).
The physiological function of efflux pumps and the regulation of efflux pump expression are not well understood. Indeed, in laboratory strain PAO1, the MexCD-OprJ and MexEF-OprN efflux pumps are significantly expressed under typical experimental growth conditions only as a result of mutations in regulatory loci (14, 15, 31). Conversely, MexAB-OprM is expressed but strains harboring mutations in the regulatory gene mexR (historically referred to as nalB strains) overproduce MexAB-OprM, with a corresponding increase in drug resistance (32, 34). Given the wide range of chemical structures recognized by RND family pumps, mutations in efflux pump regulatory genes selected by exposure to a single antibiotic lead to multidrug-resistant (MDR) strains. P. aeruginosa MDR clinical strains overexpressing MexAB-OprM (41) and expressing MexCD-OprJ (12) and MexEF-OprN (7) have been isolated. Furthermore, a P. aeruginosa MDR clinical isolate has recently been described that simultaneously expresses both the MexAB-OprM and MexEF-OprN efflux pumps (33). Multiple efflux pump expression by P. aeruginosa MDR veterinary isolates has also been reported (2). Exposure to the commonly used biocide triclosan can select for compensatory MexCD-OprJ expression in P. aeruginosa strains lacking the MexAB-OprM pump responsible for intrinsic triclosan resistance (3). Moreover, several additional putative RND pumps have been uncovered by searches of the PAO1 genome sequence (9, 39). This underscores the potential problems posed by efflux pumps in terms of both intrinsic and acquired drug resistance in P. aeruginosa and the likelihood that ever more resistant strains will emerge in response to antibiotic selection pressure.
Tigecycline, an analog of the semisynthetic tetracycline derivative minocycline, shows broad-spectrum antimicrobial activity that is not significantly affected by either of the widely distributed tetracycline resistance determinants ribosomal protection [e.g., tet(M)] and tetracycline-specific efflux [e.g., tet(A)] (30). Tetracycline-specific efflux pumps differ from RND pumps, being highly specific, single-component, major-facilitator-type efflux pumps that are common in both gram-positive and gram-negative bacteria (29). Despite the ability of tigecycline to circumvent these resistance mechanisms, attributed to the 9-t-butylglycylamido substitution of minocycline, P. aeruginosa clinical strains (30) and laboratory strain PAO1 exhibited reduced susceptibility to tigecycline. Since both MexAB-OprM and MexCD-OprJ have been shown to mediate resistance to tetracycline in P. aeruginosa (23), it was of interest to investigate the involvement of RND-type efflux in resistance to tigecycline. Therefore, an investigation of the mechanism underlying reduced susceptibility to tigecycline in P. aeruginosa was undertaken.
MATERIALS AND METHODS
Bacterial strains, medium, and culture conditions.
The bacterial strains used in this study are described in Table 1. L broth (5) was used for routine cultivation of P. aeruginosa. Solid medium was obtained by addition of 1.5% (wt/vol) Bacto Agar (Difco).
TABLE 1.
Bacterial strains used in this study
| P. aeruginosa strain | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| K767 | Prototroph | 22 |
| K1523 | K767 mexB; unmarked in-frame deletion | 10 |
| K1525 | K767 mexXY; unmarked in-frame deletion | 10 |
| XY1 | Spontaneous tigecycline-resistant derivative of K1525 | This study |
| XY3 | Spontaneous tigecycline-resistant derivative of K1525 | This study |
| K1542 | K767 mexB/mexXY; unmarked in-frame deletions | K. Poolea |
| BXY1 | Spontaneous tigecycline-resistant derivative of K1542 | This study |
| BXY4 | Spontaneous tigecycline-resistant derivative of K1542 | This study |
| K1119 | K797 mexAB-oprM; unmarked in-frame deletion | 18 |
| M1 | Spontaneous tigecycline-resistant derivative of K1119 | This study |
| M9 | Spontaneous tigecycline-resistant derivative of K1119 | This study |
| K1455 | K767 nalB; MexAB-OprM-overproducing strain | 38 |
Deletions constructed as for strains K1523 and K1525 (K. Poole, personal communication).
Antimicrobial sensitivity testing.
Antibiotic and substrate MICs were determined by broth microdilution using twofold dilution in Mueller-Hinton II broth (BBL, Cockeysville, Md.) in accordance with the procedures established by the NCCLS (26). All of the substrates and antibiotics tested were prepared fresh on the day of testing. Tigecycline, minocycline, doxycycline, PAM-MINO, DMG-MINO, and DMG-DMDOT were synthesized at Wyeth Research, Pearl River, N.Y. Trimethoprim, carbenicillin, and chloramphenicol were obtained from Sigma Chemical Co. (St. Louis, Mo.). Cefepime was obtained from Bristol-Myers Squibb (Hounslow, United Kingdom)
DNA manipulations.
P. aeruginosa genomic DNA was isolated by using the Puregene tissue kit (Gentra Systems Inc, Minneapolis, Minn.) in accordance with the manufacturer's instructions. Oligonucleotides for PCR and sequencing were obtained from GeneLink (Hawthorne, N.Y.). PCR amplifications were performed by using the Easystart mix-in-a-tube system (Molecular Bio-Products Inc., San Diego, Calif.) and a Perkin-Elmer GeneAmp PCR system 2400 thermocycler in accordance with the manufacturers' instructions. DNA fragments were isolated following agarose gel electrophoresis by using the QIAquick gel extraction kit (Qiagen, Inc., Valencia, Calif.) as specified by the manufacturer. The oligonucleotides used for PCR of mexR were P5for (5′-CATGGCCCATATTCAGAACC-3′) and P5rev (5′-CCAGTAAGCGGATACCTG-3′) (34), and those used for PCR of nfxB were NFXB1 (5′-CGATCCTTCCTATTGCACG-3′) and NFXB2 (5′-GCCAAGTGCCAGTATCG-3′) (31).
Outer membrane isolation.
Outer membranes were prepared by the method of Masuda et al. (23). Briefly, P. aeruginosa strains were grown to early stationary phase at 37°C in Mueller-Hinton broth. Twenty milliliters of culture was recovered by centrifugation and frozen at −20°C. The pellets were thawed on ice, suspended in 30 mM Tris-HCl (pH 8.0), and passed through a French pressure cell. Unbroken cells were removed by low-speed centrifugation, and whole membranes were isolated from the supernatant by centrifugation at 100,000 × g for 1 h. The membrane pellet was extracted with 30 mM Tris-HCl containing 1% Sarkosyl for 30 min at 30°C. Outer membranes were then isolated by centrifugation at 100,000 × g for 1 h and resuspended in water.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS PAGE) and Western immunoblotting.
Outer membranes were solubilized in Laemmli running buffer (Bio-Rad Laboratories, Hercules, Calif.) with 5% β-mercaptoethanol at 95°C for 5 min and separated on Criterion precast 10% polyacrylamide gels (Bio-Rad Laboratories). Proteins were transferred to nitrocellulose membrane (Bio-Rad Laboratories) by using a Trans-Blot semidry electrophoretic transfer apparatus (Bio-Rad Laboratories) in accordance with the manufacturer's instructions. Western immunoblot assays were performed as previously described (4), except that Tris-buffered saline replaced phosphate-buffered saline. The primary antibody used was polyclonal antiserum specific for OprM (kindly provided by K. Poole). Blots were developed at room temperature with anti-rabbit immunoglobulin G-alkaline phosphatase (Bio-Rad Laboratories) by using the Bio-Rad alkaline phosphatase substrate kit in accordance with the manufacturer's instructions.
Transcriptional profiling.
Overnight cultures of P. aeruginosa grown in L broth were diluted to an optical density at 600 nm (OD600) of 0.025 in the same medium and incubated with shaking at 37°C until they reached an OD600 of approximately 0.1. The cultures were then supplemented with 0, 1, 2, or 4 μg of tigecycline per ml or 4 μg of tetracycline per ml and allowed to grow to an OD600 of approximately 0.9. Cells were harvested by centrifugation and frozen overnight at −80°C. Total RNA was isolated from thawed cell pellets by using the Purescript tissue kit (Gentra Systems Inc.) in accordance with the manufacturer's instructions. To remove any contaminating genomic DNA, approximately 125 μg of RNA was treated with 20 U of DNase I (Amersham Biosciences, Piscataway, N.J.) at 37°C for 45 min. The RNA was then purified and size fractionated with an RNeasy mini column (Qiagen). Reverse transcription-PCR, cDNA fragmentation, cDNA terminal labeling, and hybridization of approximately 1.6 μg of labeled cDNA to P. aeruginosa gene chips were carried out in accordance with the manufacturer's (Affymetrix Inc., Santa Clara, Calif.) instructions. GeneChip arrays were scanned by using the Agilent GeneArray laser scanner (Agilent Technologies, Palo Alto, Calif.). GeneChip scan data for biological duplicates were normalized and analyzed by using the GeneSpring gene expression software package (Silicon Genetics, Redwood City, Calif.).
Nucleotide sequence accession number.
The complete nucleotide sequence of the region described in the legend to Fig. 3 has been submitted to the GenBank database and assigned accession no. AY180395.
FIG. 3.
Genetic rearrangement within the nfxB gene from strain BXY1. The IS element region is delimited by a 10-bp inverted repeat of the sequence TGTAGTGGTC.
RESULTS AND DISCUSSION
Intrinsic resistance to tigecycline is mediated by MexXY-OprM.
Despite the broad-spectrum activity of the novel glycylcycline antibiotic tigecycline (GAR-936), P. aeruginosa clinical strains are less susceptible to this antibiotic than are most other bacteria (30). In this study, we found that laboratory strain PAO1 (K767) also exhibits less susceptibility to tigecycline (MIC, 8 μg/ml; Table 2). In P. aeruginosa PAO1, MexCD-OprJ and MexEF-OprN are not typically expressed under laboratory growth conditions (28, 36) and were therefore less likely to play a role in reducing tigecycline susceptibility. MexAB-OprM is expressed under laboratory growth conditions; however, PAO1 lacking the mexB gene (K1523), which encodes the inner membrane transporter component of the MexAB-OprM efflux pump, showed no alteration in susceptibility to tigecycline (Table 2). The MexXY efflux pump has been shown to require induction by exposure of cells to various antibiotics, including tetracycline (24). PAO1 lacking mexXY (K1525) exhibited significantly increased susceptibility to tigecycline (Table 2), indicating that efflux by MexXY mediates a significant reduction in susceptibility to tigecycline in PAO1.
TABLE 2.
Contributions of RND efflux pumps to tigecycline resistance
| Strain | Deletion(s) | MIC (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|---|
| TMP | CHL | GEN | CIP | CAR | FEP | TGC | ||
| K767 | None (wild type) | 64 | 250 | 1 | 0.06 | 160 | ≤0.625 | 8 |
| K1523 | mexB | 16 | ≤8 | 2 | 0.06 | 20 | ≤0.625 | 8 |
| K1525 | mexXY | 125 | 125 | ≤0.125 | 0.06 | 160 | 1.25 | 0.5 |
| XY1 | 500 | 1,000 | ≤0.125 | 2 | 20 | 5 | 4 | |
| XY3 | 500 | 188 | ≤0.125 | 0.5 | 640 | 5 | 4 | |
| K1542 | mexB/mexXY | 4 | 125 | ≤0.125 | ≤0.03 | 1.25 | ≤0.625 | 0.25 |
| BXY1 | 250 | 1,000 | 0.25 | 2 | 1.25 | 5 | 8 | |
| BXY4 | 250 | 1,000 | ≤0.125 | 2 | 1.25 | 5 | 4 | |
| K1119 | mexAB-oprM | 4 | 32 | ≤0.125 | ≤0.03 | 1.25 | ≤0.625 | 0.25 |
| M1 | 500 | 1,000 | ≤0.125 | 2 | 1.25 | 5 | 8 | |
| M9 | 500 | 1,000 | ≤0.125 | 2 | 1.25 | 5 | 8 | |
Abbreviations: CAR, carbenicillin; CHL, chloramphenicol; CIP, ciprofloxacin; FEP, cefepime; GEN, gentamicin; CIP, ciprofloxacin; TGC, tigecycline; TMP, trimethoprim.
Unlike mexAB-oprM, mexCD-oprJ, and mexEF-oprN, the mexXY locus does not have an associated gene encoding an outer membrane component. This is similar to the acrAB multidrug efflux operon of Escherichia coli, which also lacks an associated outer membrane component. In the case of AcrAB, the outer membrane function is provided by the outer membrane component TolC (6), which is encoded elsewhere on the chromosome. Several lines of evidence indicate that the OprM component of the constitutively expressed MexAB-OprM efflux pump provides the outer membrane channel function for MexXY in P. aeruginosa (1, 24). Consistent with this, PAO1 lacking mexAB-oprM (K1119) exhibits the same susceptibility to tigecycline as the mexXY strain (K1525) while disruption of the MexAB-OprM pump by loss of the mexB gene alone (K1523) (Table 2) has no discernible effect. The specific association of oprM with MexXY-mediated resistance to tigecycline reported here strongly supports the notion that OprM is the outer membrane channel component that functions with MexXY.
Tigecycline induces mexZ, mexX, and mexY transcription.
P. aeruginosa lacking MexZ has been shown to exhibit increased transcription of mexXY, suggesting that MexZ acts as a repressor (40). Previously, MexX protein production in P. aeruginosa, as determined by Western blot analysis, was shown to be inducible by various antibiotics, including tetracycline (24). Therefore, efflux of tigecycline by MexXY resulted from efficient efflux by uninduced levels of MexXY or, alternatively, tigecycline induces expression of the efflux pump. To investigate this, P. aeruginosa PAO1 GeneChip arrays were employed to determine the transcriptional status of efflux pump genes in cells treated with tigecycline and tetracycline. Tigecycline at 1, 2, or 4 μg/ml strongly induced transcription of mexX (13.4-fold at 1 μg/ml) and mexY (12.8-fold at 1 μg/ml) (Table 3), as did tetracycline at 4 μg/ml, indicating that despite their structural differences, both compounds are strong mexXY inducers. This also shows that induction of MexXY efflux pump expression by tetracycline and its derivatives is mediated at the transcriptional level.
TABLE 3.
Induction of efflux genes by tigecycline and tetracyclinea
| Treatment | Fold change
|
|||
|---|---|---|---|---|
| mexZ | mexX | mexY | oprM | |
| TGC, 1 μg/ml | 4.52 | 13.43 | 12.85 | 0.9 |
| TGC, 2 μg/ml | 5.28 | 15.48 | 13.34 | 0.63 |
| TGC, 4 μg/ml | 10.92 | 14.85 | 11.45 | 0.47 |
| TET, 4 μg/ml | 4.93 | 15.81 | 14 | 0.78 |
Values represent fold changes in transcript levels detected compared to untreated control samples and are averages of two biological replicates. TGC, tigecycline; TET, tetracycline.
Detection of mexY was lower in our system than that of mexX, resulting in mexY mRNA being considered marginal or absent in untreated (uninduced) cells while mexX mRNA was considered present. Therefore, the specific fold change in mexY induction is only an estimate and could be greater than that reported here. However, as calculated, it closely follows the fold induction of mexX (Table 3), consistent with these two genes forming an operon, and therefore likely represents the true level of induction.
Expression of mexZ, the putative repressor of mexXY, was also significantly induced by both tigecycline and tetracycline (4.5-fold at a tigecycline concentration of 1 μg/ml). This indicates that mexZ expression is probably negatively autoregulated. What is less clear is what constitutes the specific signal recognized by MexZ. While tetracycline and tigecycline have structural differences, they also have core structural similarities that could be recognized by MexZ. However, given that MexXY-OprM function is not specific to tetracycline and MexX induction is responsive to erythromycin and gentamicin, as well as tetracycline (24), it is possible that MexZ responds to a signal generated within the cell as a result of antibiotic activity. Alternatively, certain efflux pump regulatory proteins, such as MarR, BmrR, and QacR, exhibit multidrug binding (35), suggesting that MexZ may also directly bind structurally diverse antibiotics.
Transcription of oprM, which encodes the outer membrane channel component of the MexXY-OprM pump, was not induced by tigecycline or tetracycline (Table 3), instead showing a possible slight decline, as did that of mexA and mexB (data not shown). Therefore, the OprM component is not coregulated with its MexXY partners, save for a possible slight negative impact of mexXY expression on mexAB-oprM expression. This suggests that the constitutive level of OprM produced in wild type strains is sufficient to function with induced MexXY and MexAB, at least under these growth conditions.
Acquired resistance to tigecycline in P. aeruginosa.
Given the potential complications of antibiotic treatment of P. aeruginosa infections posed by efflux pumps, attention has recently focused on inhibition of efflux pump activity as a way to increase and extend the activity of currently available antibiotics (19, 20). Inhibition of efflux pumps in P. aeruginosa and other microorganisms is complicated by a combination of the multiplicity of pumps and their broad and often overlapping specificities. Therefore, it is likely that efflux pump inhibitors would ideally need to be broadly active. However, there is some pump specificity for classes of compounds. For example, high-level efflux of several β-lactams is mediated primarily by MexAB-OprM (37) and the MexXY-OprM efflux pump shows specificity for aminoglycosides (1, 40). Moreover, Mao et al. (21) have shown that loss of MexXY-OprM in P. aeruginosa eliminates the phenomenon of divalent cation antagonism of aminoglycosides that negatively affects their use in the treatment of P. aeruginosa infections. This suggests that specific inhibition of pumps may have utility in extending the activity of certain classes of preexisting drugs, assuming that additional pump expression replacing the inhibited pump does not occur.
To test the possibility that specific inhibition of MexXY might be useful in extending tigecycline activity to include P. aeruginosa, the PAO1 mexXY (K1525), mexB/mexXY (K1542), and mexAB-oprM (K1119) strains, which mimic complete inhibition of MexXY-OprM, were subjected to selection on L-agar plates containing 4 or 6 μg of tigecycline per ml. For each strain, spontaneous mutants resistant to this level of tigecycline were readily obtained (average frequency, 10−7 to 10−8). Susceptibility profiles of two representative mutants derived from each parent strain (XY1 and XY3 from the mexXY mutant strain, BXY1 and BXY4 from the mexB/mexXY mutant strain, and M1 and M9 from the mexAB-oprM mutant strain) are shown in Table 2. For each mutant, susceptibility to tigecycline was reduced to the intrinsic level of PAO1. Each mutant also exhibited a multidrug resistance phenotype, indicating the likelihood of efflux pump involvement in the acquired resistance. The rapid appearance of alternative resistance mechanisms upon exposure to tigecycline, in the absence of MexXY-OprM, indicates that specific inhibition of MexXY would not be a successful long-term strategy by which to improve the activity of tigecycline against P. aeruginosa.
Tigecycline is a substrate for MexAB-OprM.
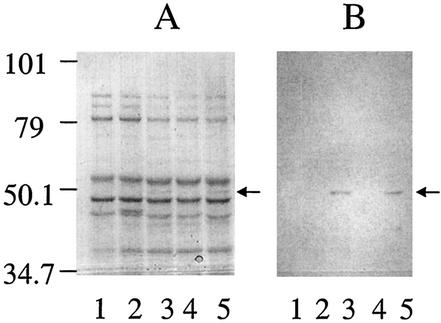
Strain XY3 exhibits resistance to carbenicillin at levels typical of so-called nalB mutant strains, which overexpress MexAB-OprM (Table 2). Several other mutants derived from the mexXY mutant strain also exhibited this resistance profile (data not shown). Carbenicillin resistance in nalB mutant strains is associated with obvious overexpression of the outer membrane component OprM. An examination of outer membrane proteins prepared from strain XY3 revealed overexpression of an outer membrane protein of approximately 50 kDa, the same size as the protein overexpressed by a previously characterized nalB mutant (Fig. 1A, lanes 3 and 5). Moreover, this band was detected by polyclonal antiserum specific for OprM (Fig. 1B), suggesting that strain XY3 is a nalB mutant strain. Typically, nalB mutant strains harbor mutations in the putative repressor gene mexR, which is located immediately upstream of the mexAB-oprM operon. Consistent with this, sequence analysis of the mexR gene from XY3 revealed a 15-bp in-frame deletion of nucleotides 299 to 313.
FIG. 1.
Overexpression of OprM determined by SDS-PAGE (A) and Western immunoblot analysis (B) of outer membranes prepared from P. aeruginosa strains. Lanes: 1, K767; 2, K1119; 3, K1455; 4, K1525; 5, XY3. The antibody is polyclonal antiserum specific for OprM. The positions of molecular size markers (in kilodaltons on the left) and OprM (arrows) are indicated.
The decrease in susceptibility to tigecycline conferred by overexpressed MexAB-OprM in the absence of MexXY in strain XY3 indicates that tigecycline is also a substrate for MexAB-OprM. This is consistent with the finding that MexAB-OprM mediates resistance to tetracycline (23). However, overexpression of the pump is required to yield only the wild-type PAO1 levels of resistance normally conferred by MexXY-OprM and significant efflux of tigecycline by MexAB-OprM seemingly occurs only in the absence of MexXY, implying that tigecycline is preferentially recognized by MexXY-OprM when both pumps are present.
Tigecycline is a substrate for MexCD-OprJ.
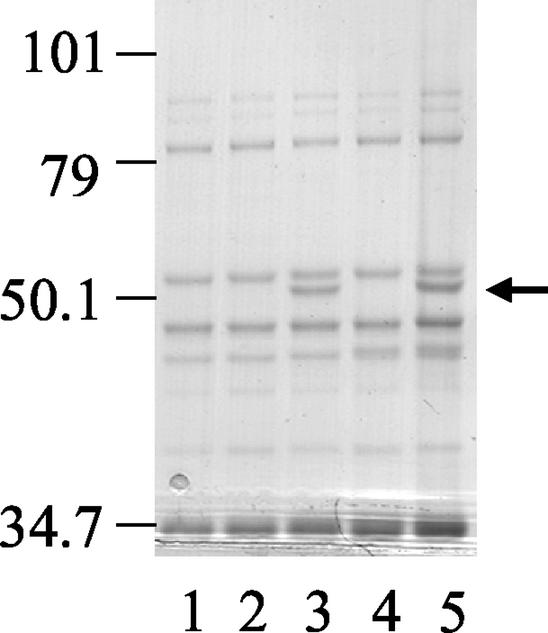
Mutant strain XY1, derived from PAO1 mexX (K1525), differs from mutant strain XY3 (derived from the same parent strain) in that it does not exhibit carbenicillin resistance (Table 2). Overall, the resistant mutants derived from PAO1 mexXY were evenly divided between these two types. Mutants BXY1, BXY4, M1, and M9 (Table 2) all appear to fall into the same class as XY1 with respect to their drug resistance profiles. Very high resistance to chloramphenicol and cefepime, and increased susceptibility to carbenicillin in the case of XY1, suggested MexCD-OprJ-mediated efflux. Consistent with this, analysis of outer membrane proteins prepared from representative strains BXY1 and M1 revealed the appearance of a highly expressed protein of ca. 54 kDa, the reported size of OprJ (Fig. 2, lanes 3 and 5). Expression of OprJ is typically associated with mutation of mexCD-OprJ repressor gene nfxB, which is located immediately upstream of and in the opposite orientation to the efflux operon. Analysis of nfxB gene products generated by PCR from several resistant mutants showed that nfxB from M1 was smaller than predicted while nfxB from BXY1 was larger. This indicated that more significant genetic rearrangement had occurred in these nfxB genes than the point mutations typically described in repressor genes from efflux pump-expressing strains. Nucleotide sequence analysis of these nfxB gene products revealed a 327-bp in-frame deletion of nucleotides 71 to 397 from nfxB in M1, while the nfxB gene from BXY1 was interrupted by insertion of an IS element (Fig. 3). Taken together, these data indicate that the reduced susceptibility to tigecycline exhibited by BXY1 and M1 is mediated by MexCD-OprJ.
FIG. 2.
Expression of OprJ determined by SDS-PAGE analysis of outer membranes prepared from P. aeruginosa strains. Lanes: 1, PAO1; 2, K1542; 3, BXY1; 4, K1119; 5, M1. The positions of molecular size markers (in kilodaltons on the left) and OprJ (arrow) are indicated.
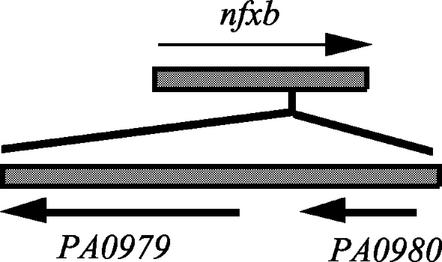
BLAST analysis of the IS element residing in nfxB from BXY1 (Fig. 2) against the P. aeruginosa PAO1 genome sequence (The Institute for Genomic Research database) revealed near identity to two separate regions encompassing open reading frames PA0979 and PA0980 in the first case and PA1937 and PA1938 in the second. These regions appear to be essentially two copies of an IS element, with the predicted proteins from both having extensive homology to numerous transposases. The involvement of an IS element in conversion to an MDR phenotype mirrors the IS186- and IS2-mediated conversion to AcrAB and AcrEF efflux pump expression, respectively, observed when E. coli strains defective in the acrAB regulatory locus marRAB, or in acrAB, were exposed to fluoroquinolones (13). The observations presented here for P. aeruginosa further support the notion that larger genetic deletions and movement of mobile genetic elements can come into play in the conversion to an efflux-mediated MDR phenotype under selective conditions.
The genetic rearrangements disrupting nfxB in BXY1 and M1 likely completely abolish any potential NfxB repressor function. This is consistent with the very high level of expression of OprJ observed in the outer membranes of BXY1 and M1 (Fig. 2), which might be expected in the absence of any repression. This high level of expression, which presumably also reflects cellular levels of the whole MexCD-OprJ pump complex, does not reduce susceptibility to tigecycline beyond the intrinsic resistance level conferred by MexXY-OprM. This suggests that MexCD-OprJ may be less efficient than MexXY-OprM at pumping tigecycline.
Efflux of glycylcycline derivatives.
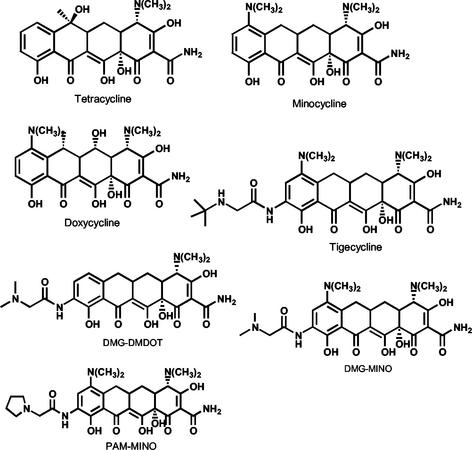
Since the 9-t-butylglycylamido substitution of tigecycline prevents efflux by tetracycline-specific major facilitator pumps, it was of interest to examine the relationship, if any, between glycylcycline structures and RND pump-mediated efflux. The MICs of several glycylcycline derivatives (Fig. 4) for various strains used in this study were compared to those of the narrower-spectrum semisynthetic tetracyclines minocycline and doxycycline and those of tetracycline (Table 4). PAO1 with a deletion in mexAB-oprM (K1119), and therefore lacking both MexAB-OprM and MexXY-OprM functions, was much more susceptible to all of the tetracyclines tested (MIC, ≤0.5 μg/ml in all cases) than the parent strain, K767 (Table 4). However, specific loss of mexXY in strain K1525 significantly affected susceptibility to the glycylcyclines as a group, increasing it to a level approaching that of the mexAB-oprM strain (K1119), while having much less impact on susceptibility to the nonglycyl derivatives. Loss of mexXY and mexB together (K1542) increased susceptibility to the nonglycyl derivatives to a level similar to that of the mexAB-oprM strain (K1119) (Table 4). Therefore, minocycline and doxycycline were exported efficienty by MexAB-OprM alone, with a modest contribution from MexXY-OprM. This is in contrast to tigecycline, DMG-MINO, DMG-DMDOT, and PAM-MINO, which were exported primarily by MexXY-OprM (Table 4), with seemingly less of a contribution from MexAB-OprM. Consistent with a reduction in MexAB-OprM-mediated efflux of glycyl-substituted tetracyclines, the MIC of minocycline and doxycycline for a MexAB-OprM overproducer lacking MexXY (XY3) rose to 64 μg/ml while those of the glycylcyclines did not exceed 8 μg/ml. Similarly, for strains BXY1 and M1, where resistance to tigecycline is mediated by MexCD-OprJ in the absence of both MexAB-OprM and MexXY-OprM, the MIC of minocycline and doxycycline was very high at 128 μg/ml while the glycyl derivatives all had MICs not exceeding 8 to 16 μg/ml (Table 4). Therefore, efflux of glycyl-substituted tetracyclines by MexCD-OprJ also appears to be less efficient.
FIG. 4.
Structures of tetracycline and the derivatives used in this study.
TABLE 4.
Effects of glycylcycline substitutions on efflux
| Strain | Deletion(s) | MIC (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|---|
| MIN | DOX | TET | TGC | DMG-MINO | DMG-DMDOT | PAM-MINO | ||
| K767 | None (wild type) | 16 | 16 | 16 | 8 | 8 | 8 | 4 |
| K1525 | mexXY | 4 | 4 | 4 | 0.5 | 0.5 | 0.5 | 0.5 |
| XY1 | 128 | 128 | 32 | 4 | 8 | 16 | 8 | |
| XY3 | 64 | 64 | 64 | 4 | 8 | 8 | 4 | |
| K1542 | mexB/mexXY | 0.5 | 0.25 | 0.5 | 0.5 | 0.25 | 0.125 | 0.25 |
| BXY1 | 128 | 128 | 64 | 8 | 16 | 16 | 8 | |
| BXY4 | 128 | 64 | 32 | 4 | 16 | 16 | 8 | |
| K1119 | mexAB-oprM | 0.5 | 0.25 | 0.25 | 0.5 | 0.25 | 0.125 | 0.25 |
| M1 | 128 | 128 | 64 | 8 | 16 | 16 | 8 | |
| M9 | 128 | 128 | 64 | 8 | 16 | 16 | 8 | |
Abbreviations: DOX, doxycycline; MIN, minocycline; TET, tetracycline, TGC, tigecycline. See Fig. 4 for structures of DMG-MINO, DMG-DMDOT, and PAM-MINO.
In conclusion, efflux by MexXY-OprM is responsible for reducing the susceptibility of P. aeruginosa PAO1 to tigecycline (and other glycylcyclines). Inhibition of MexXY-OprM activity would readily select for compensatory efflux by overexpressed MexAB-OprM or MexCD-OprJ and possibly other efflux pumps. Therefore, in contrast to previous observations for tetracycline-specific efflux pumps, glycyl substitution does not overcome efflux by RND family pumps. However, it did result in a shift in the relative contribution of particular pumps mediating intrinsic resistance, from MexAB-OprM for semisynthetic tetracyclines to MexXY-OprM for glycyl derivatives. Moreover, the efficiency of efflux by individual pumps appears to be reduced for the glycyl derivatives. This suggests the possibility that RND efflux pump-mediated drug resistance can be overcome through chemical modification of existing, otherwise effective drugs.
Acknowledgments
We thank Keith Poole, Queen's University, Kingston, Ontario, Canada, for strains and anti-OprM antibody; Paul Dunman, Daphne Macapagal, and Alexey Bulychev for helpful discussions; and David Fruhling for nucleotide sequencing.
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beinlich, K. L., R. Chuanchuen, and H. P. Schweizer. 2001. Contribution of multidrug efflux pumps to multiple antibiotic resistance in veterinary clinical isolates of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 198:129-134. [DOI] [PubMed] [Google Scholar]
- 3.Chuanchuen, R., K. Beinlich, T. T. Hoang, A. Becher, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 45:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean, C. R., C. V. Franklund, J. D. Retief, M. J. Coyne, Jr., K. Hatano, D. J. Evans, G. B. Pier, and J. B. Goldberg. 1999. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 181:4275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean, C. R., and K. Poole. 1993. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J. Bacteriol. 175:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuda, H., M. Hosaka, S. Iyobe, N. Gotoh, T. Nishino, and K. Hirai. 1995. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:790-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg, J. B., and G. B. Pier. 2000. The role of the CFTR in susceptibility to Pseudomonas aeruginosa infections in cystic fibrosis. Trends Microbiol. 8:514-520. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg, E. P. 2000. Bacterial genomics. Pump up the versatility. Nature 406:947-948. [DOI] [PubMed] [Google Scholar]
- 10.Hirikata, Y., R. Srikumar, K. Poole, N. Gotoh, T., and S. K. Suematsu, S. Kamihira, R. E. W. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa infections. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoiby, N., S. S. Pedersen, G. H. Shand, G. Doering, and I. A. Holder (ed.). 1989. Pseudomonas aeruginosa infection. Antibiot. Chemother. 42:1-300. [DOI] [PubMed] [Google Scholar]
- 12.Jakics, E. B., S. Iyobe, K. Hirai, H. Fukuda, and H. Hashimoto. 1992. Occurrence of the nfxB type mutation in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:2562-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J. C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 15.Li, X.-Z., N. Barre, and K. Poole. 2000. Influence of the MexA-MexB-oprM multidrug efflux system on expression of the MexC-MexD-oprJ and MexE-MexF-oprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 46:885-893. [DOI] [PubMed] [Google Scholar]
- 16.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, X.-Z., L. Zhang, and K. Poole. 2000. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:433-436. [DOI] [PubMed] [Google Scholar]
- 18.Li, X.-Z., L. Zhang, R. Srikumar, and K. Poole. 1998. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomovskaya, O., and W. Watkins. 2001. Inhibition of efflux pumps as a novel approach to combat drug resistance in bacteria. J. Mol. Microbiol. Biotechnol. 3:225-236. [PubMed] [Google Scholar]
- 21.Mao, W., M. S. Warren, A. Lee, A. Mistry, and O. Lomovskaya. 2001. MexXY-OprM efflux pump is required for antagonism of aminoglycosides by divalent cations in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda, N., and S. Ohya. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NCCLS. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. NCCLS, Wayne, Pa.
- 27.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1):S32-S41. [DOI] [PubMed] [Google Scholar]
- 28.Okazaki, T., and K. Hirai. 1992. Cloning and nucleotide sequence of the Pseudomonas aeruginosa nfxB gene, conferring resistance to new quinolones. FEMS Microbiol. Lett. 76:197-202. [DOI] [PubMed] [Google Scholar]
- 29.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen, P. J., N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa. 1999. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. Yamagishi, X. Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 32.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pumbwe, L., and L. J. Piddock. 2000. Two efflux systems expressed simultaneously in multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2861-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 35.Schumacher, M. A., and R. G. Brennan. 2002. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45:885-893. [DOI] [PubMed] [Google Scholar]
- 36.Shiba, T., K. Ishiguro, N. Takemoto, H. Koibuchi, and K. Sugimoto. 1995. Purification and characterization of the Pseudomonas aeruginosa NfxB protein, the negative regulator of the nfxB gene. J. Bacteriol. 177:5872-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srikumar, R., X. Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 40.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziha-Zarifi, I., C. Llanes, T. Kohler, J. C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]