Abstract
A 6-aminoquinolone derivative, WM5, which bears a methyl substituent at the N-1 position and a 4-(2-pyridyl)-1-piperazine moiety at position 7 of the bicyclic quinolone ring system, was previously shown to exhibit potent activity against replication of human immunodeficiency virus type 1 (HIV-1) in de novo-infected human lymphoblastoid cells (V. Cecchetti et al., J. Med. Chem. 43:3799-3802, 2000). In this report, we further investigated WM5's mechanism of antiviral activity. WM5 inhibited HIV-1 replication in acutely infected cells as well as in chronically infected cells. The 50% inhibitory concentrations were 0.60 ± 0.06 and 0.85 ± 0.05 μM, respectively. When the effects of WM5 on different steps of the virus life cycle were analyzed, the reverse transcriptase activity and the integrase and protease activities were not impaired. By using a transient trans-complementation assay to examine the activity of WM5 on the replicative potential of HIV-1 in a single round of infection, a sustained inhibition of Tat-mediated long terminal repeat (LTR)-driven transcription (>80% of controls) was obtained in the presence of 5 μM WM5. Interestingly, the aminoquinolone was found to efficiently complex TAR RNA, with a dissociation constant in the nanomolar range (19 ± 0.6 nM). These data indicate that WM5 is a promising lead compound for the development of a new class of HIV-1 transcription inhibitors characterized by recognition of viral RNA target(s).
Human immunodeficiency virus type 1 (HIV-1) is the etiologic agent of AIDS in humans (6, 20). HIV-1 establishes a persistent infection in human hosts, with the depletion of CD4+ lymphocytes, the major target cells of viral infection in vivo, eventually resulting in defective cellular immunity (16). Combination of antiretroviral drugs (highly active antiretroviral therapy [HAART]) has changed the outcome of HIV-1 infection, leading to a dramatic reduction in AIDS-related morbidity and mortality (38). Treatment with a combination of reverse transcriptase (RT) and protease inhibitors can provide long-term control of viral replication, with reduction of HIV RNA in the plasma of infected individuals to undetectable levels, and can rescue CD4+ cell counts with significant benefit for HIV-seropositive patients and prolonged survival. Unfortunately, complex drug interactions within the HAART schemes, along with complicated dosing schedules, toxicity, and considerable side effects, makes long-term compliance with drug regimens difficult for most patients. In addition, the high rate of genetic variation of the HIV-1 genome, combined with natural selection under therapy, gives rise to the development and outgrowth of virus variants resistant to one or multiple administered agents (25). This has become a major concern that may affect further development of antiretroviral compounds and represents one of the most difficult challenges in the search for effective inhibitors of HIV-specific enzymes (24). One way to circumvent this problem would be the identification of new targets for drug therapy characterized as being essential for viral replication and therefore less prone to mutational changes. Moreover, recent studies showed that infectious HIV-1 persists latently in resting, memory CD4 lymphocytes (10, 18, 19, 56, 59). Viral persistence despite prolonged treatment is due not only to the slow turnover of residually infected memory lymphocytes, but also to the inability of current antiretroviral regimens to completely suppress viral replication, thereby allowing replenishment of this pool of latently infected cells (44). This latent reservoir of HIV-1 represents the major documented hurdle to virus eradication, although other viral sanctuaries may exist (11). Therefore, newer effective drugs and treatment modalities are highly desirable.
Quinolones represent an important class of broad-spectrum antibacterials, the main structural features of which are a 1,4 dihydro-4-oxo-piridinyl moiety bearing an essential carboxyl group at position 3. The quinolones' mechanism of action is due to the inhibition of prokaryotic type II topoisomerases, namely DNA gyrase and, in few cases, topoisomerase IV (21). Quinolones interact directly with the bacterial chromosome, that enzyme inhibition following the interaction with single-stranded DNA (49, 50). Physicochemical and enzymatic studies conducted by our group previously identified the quinolone moieties involved in binding to DNA and DNA gyrase (2, 40, 41). We demonstrated that the interaction between quinolones and the nucleic acid is mediated by magnesium ions and that the complex formation is dependent on ion concentration (39, 54).
Quinolone derivatives have been shown to inhibit HIV-1 replication in de novo- and chronically infected cells (4, 5, 22, 29, 35-37, 55). A new fluoroquinolone, K12, bearing an o-methoxyphenyl-piperazinyl group and a difluoromethoxyl group at positions 7 and 8, respectively, was reported to have strong and selective anti-HIV-1 activity (4). The antiviral activity seemed to be related to an inhibitory effect at the transcriptional level. Two K12 analogues bearing a phenyl-dehydro-piperidinyl moiety at position 7 were effective at inhibiting HIV-1 long terminal repeat (LTR)-driven gene expression, as well as suppressing tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) production in blood mononuclear cells, suggesting a mechanism of action mediated by inhibition of Tat functions (4, 36).
Recently, our group developed a new class of 6-substituted quinolones and tested their antibacterial and anti-HIV-1 activities (9, 51). A 6-aminoquinolone bearing a methyl substituent at the N-1 position and a 4-(2-pyridyl)-1-piperazine moiety at the C-7 position (WM5) was found to be the most active compound in inhibiting HIV-1 replication on de novo-infected cells. In this study, we investigated the mechanism of action of WM5 and examined the effect of this compound on the HIV-1 life cycle at the molecular level.
MATERIALS AND METHODS
Compounds.
The two aminoquinolone derivatives studied, the structure of which is shown in Fig. 1, were synthesized as previously described (9). The compounds were dissolved in dimethyl sulfoxide (DMSO) at a concentration of 25 mM or higher to exclude any antiviral or cytotoxic effect of DMSO after dilution in the appropriate media and kept frozen in aliquots until use. Their concentration was checked by spectroscopy with molar extinction coefficients of 5,800 ± 150 M−1 cm−1 at 355 nm for WM5 and 4,800 ± 350 M−1 cm−1 at 350 nm for WM5E, experimentally determined in a mixture of Tris (10 mM, pH 7.0), NaCl (20 mM), and Mg2+ (1 mM) at 25°C. Tris, EDTA, NaCl, and Mg(ClO4)2 were obtained from Sigma Chemical Co., St. Louis, Mo. Stock solutions of Mg(ClO4)2 were made in Millipore water and filtered, and the correct concentration of magnesium was checked by atomic absorption on a Perkin-Elmer 360 instrument. Each buffer was made in sterile water and filtered through a 5-μm-pore-diameter filter before use to eliminate any particulate material that would interfere with the fluorescence response.
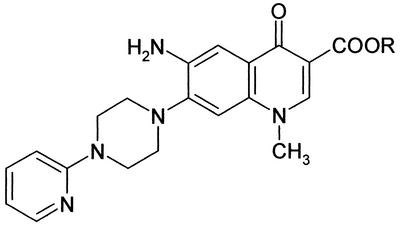
FIG. 1.
Chemical structures of 6-aminoquinolones WM5 and WM5E. R = H for WM5 and C2H5 for WM5E. CC50 = 56.24 ± 5.24 μM for WM5 and 58.36 ± 8.84 μM for WM5E (as measured by the MTT method [see Materials and Methods]).
The catechol derivative RDS 1028 was synthesized as previously described (3). The HIV-1 protease inhibitor saquinavir (Ro 31-8959) was kindly supplied by I. Duncan (Roche Products, Welwyn Garden City, Hertsfordshire, United Kingdom) (13).
RNA.
TAR RNA was obtained by in vitro transcription as described previously (9). Before every titration, the RNA was denatured at 95°C and cooled down in the presence of magnesium. The concentration was checked by spectrophotometric measurement at 260 nm with a molar extinction coefficient (per average residue) of 8,000 M−1 cm−1, while the concentration (molecular mass) of TAR was assumed to be 34,200 Da.
Cell cultures and virus.
The human T-lymphoid Jurkat cell line was maintained in RPMI 1640 medium (Life Biotechnologies, Gaithersburg, Md.) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Life Biotechnologies), 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Life Biotechnologies). 293T cells (adenovirus 5-transformed human embryonic kidney 293 cells constitutively expressing the simian virus 40 large T antigen) were maintained in Dulbecco's modified Eagle's medium (Life Biotechnologies) containing 10% FBS and antibiotics. HIV-1IIIB chronically infected H9 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and antibiotics. Cell activation was achieved through an initial 3-day stimulation with 1% (vol/vol) purified phytohemagglutinin (PHA) and subsequent culture with 10 U of recombinant human IL-2 per ml (Collaborative Research, Becton Dickinson, Bedford, Mass.).
HIV-1 virus stock was produced by transient transfection of Jurkat cells with the pSVC21 plasmid containing the infectious HXB2 molecular clone of HIV-1 (45) by the DEAE-dextran method as described previously (14) and stored at −80°C until use. Viral titer was measured as 50% tissue culture infective doses (TCID50) per milliliter on C8166 cells by the end point dilution method of Reed and Muench (46).
Cytotoxicity assay.
The cytotoxicities of the compounds for the Jurkat cell line were based on the cell viability measured by the colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (34). Briefly, Jurkat cells (104 per well) were cultured in triplicate in a 96-well plate in the absence or presence of various concentrations of the test compounds. After 72 h of incubation at 37°C, MTT was added to each well, and the mixture was incubated for 4 h before addition of the solubilization solution for another 12 h. The plates were then read at 550 nm with an enzyme-linked immunosorbent assay plate reader (LP200 Diagnostics Pasteur).
Antiviral assay.
The compound's effect on acute HIV-1 infection was based on the inhibition of RT activity in Jurkat cell culture supernatants. Jurkat cells (106 cells) were incubated with virus stock at multiplicities of infection (MOI) of 0.1 or 0.01 TCID50 per cell. After 2 h of incubation at 37°C, the cultures were washed twice and maintained in the absence or presence of various concentrations of the test compounds, with a 50% medium change every 3 days. Virus replication after infection was monitored by RT activity in cell-free culture supernatants (47). Where specified, virus replication was measured, evaluating the viral load in cell-free culture supernatants, by a viral load assay performed according to the manufacturer's specifications (AMPLICOR HIV-1 MONITOR, version 1.5; Roche Diagnostics, Branchburg, N.J.). The effect on chronic HIV-1 infection was based on the inhibition of RT production in HIV-1IIIB H9 cells. HIV-1IIIB chronically infected H9 cells (activated with PHA and IL-2) were washed twice to remove cell-free virus and resuspended at a density of 5 × 104 cells per well in 48-well plates in fresh medium containing the test compound at the appropriate concentration. After 24 h of incubation at 37°C, cell-free supernatants were collected to determine viral growth by measuring RT activity. The 50% inhibitory concentration (IC50) was determined as the concentration of compound required to inhibit virus replication by 50% and was calculated by nonlinear regression analysis with Sigma Plot (Jandel Scientific, Corte Madera, Calif.).
env complementation assay.
A single round of infection was assayed in a previously described env complementation assay (23). Briefly, 293T cells were cotransfected by the calcium phosphate method with 20 μg of the pHXBH10ΔenvCAT plasmid and 5 μg of pSVIIIenv plasmids expressing the HIV-1 HXBc2 or the 89.6 envelope glycoproteins and Rev to produce recombinant virions. The pHXBH10ΔenvCAT plasmid contains an HIV-1 provirus carrying a deletion in the envelope (env) gene and with the chloramphenicol acetyltransferase (CAT) gene replacing the nef gene. At 12 h following transfection, cells were washed and cultured in RPMI 1640 medium supplemented with 10% FBS and antibiotics. Conditioned medium containing recombinant viruses was harvested and filtered (0.45-μm-pore-size filter) 24 h later. Jurkat cells were incubated with 30,000 3H cpm RT units of recombinant CAT reporter viruses at 37°C and then maintained in the absence or presence of the compounds. Cells were lysed 4 days after infection, and CAT activity was determined, indicating the efficiency of infection.
Inhibition of viral enzymes in vitro. (i) Inhibition of RT activity.
Supernatants from HIV-1 chronically infected H9 cell lines were pelleted, lysed, and incubated in the presence or in the absence of the compound at 37°C for 15 min, and subsequently, the RT inhibition assay was performed as described previously (47).
(ii) Integrase assay.
The following oligonucleotides representing the terminal 21 nucleotides of the HIV-1 U5 LTR were used: B (5′-ACTGCTAGAGATTTTCCACAC-3′ [minus strand]) and C (5′-GTGTGGAAAATCTCTAGCA-3′ [plus strand]). Oligonucleotide C was annealed with oligonucleotide B in 0.1 M NaCl by being heated at 80°C and then slowly cooled to room temperature overnight. This double-stranded substrate was labeled by introducing at the 3′ end of C the two missing nucleotides with [α-32P]dGTP, cold dTTP, and Klenow polymerase. Unincorporated [α-32P]dGTP was separated from the duplex substrate by two consecutive runs through G-25 Sephadex quick spin columns. The reaction mixtures contained 40 mM NaCl, 10 mM MnCl2, 25 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 2% glycerol, 1 nM duplex B:C labeled at the 3′ end, and 5 nM integrase (IN) (considered as monomer, purified as previously described) (53). Reaction mixtures were incubated at 37°C for 1 h in a volume of 15 μl and stopped by adding 3 μl of sample buffer (96% formamide, 20 mM EDTA, 0.08% bromophenol blue, 0.25% xylene cyanol). Samples were heated at 100°C for 3 min, and 10 μl of each of them was layered onto a denaturing 15% polyacrylamide gel (7 M urea, 0.09 M Tris borate [pH 8.3], 2 mM EDTA, 15% acrylamide) and run for 1 h at 80 W. Reaction products were visualized and quantified by a Bio-Rad FX Phosphoimager.
(iii) Protease inhibition by fluorometric assay.
The ability of the compounds to inhibit HIV-1 protease was assessed by using the fluorescent peptide substrate aminobenzoyl-Thr-Ile-Nle-Phe(NO2)-Gln-Arg-NH2 (the scissile bond is underlined). Recombinant HIV-1 protease was expressed in Escherichia coli, purified, and refolded (26, 33). Assays were carried out as previously described (32, 33, 52). Briefly, the assay mixture contained 100 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH 5.5), 400 mM NaCl, 0.5% dimethyl sulfoxide, 1.5 to 20 nM protease, 0 to 100 nM protease inhibitor saquinavir (Ro 31-8959), and 10 μM fluorogenic substrate. The IC50 of the test compounds for HIV-1 protease activity was determined by nonlinear regression analysis with Sigma Plot (Jandel Scientific, Corte Madera, Calif.). The protease inhibitor saquinavir, used as reference in this assay, had a measured IC50 of 2 nM.
Binding to nucleic acids.
To evaluate the formation of complexes between the quinolones and RNA (DNA) in the presence of magnesium ions, we performed fluorometric titrations, exploiting the drugs' high fluorescence yield upon excitation at 350 nm for WM5 and 355 nm for WM5E. Fluorescence spectra were recorded at 25°C on a Luminescence Spectrophotometer LS50B (Perkin-Elmer). Briefly, weighted amounts of nucleic acid solutions were added to quinolone solution in Tris-HCl (10 mM [pH 7.0]), NaCl (20 mM), and Mg(ClO4)2 (1 mM) directly in the cuvette, and the emission spectra were recorded after allowing 5 min for equilibration. Hence, the fraction of bound drug (ν) can be directly obtained from the spectra according to the equation (31) ν = [I(f) − I]/[I(f) − I(b)], where I(f) and I(b) are the fluorescence intensities of free and RNA-bound drug, respectively, and I is the fluorescence response of the mixture of free and bound drug being examined.
RESULTS
Effect of WM5 on HIV-1 replication in acutely and chronically infected cells.
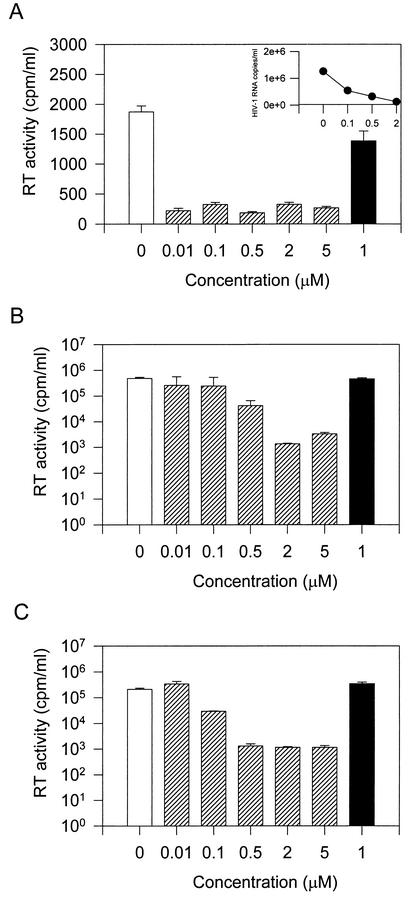
In a previous study we showed that a 6-aminoquinolone, WM5 (Fig. 1), was able to inhibit HIV-1 replication on the de novo-infected C8166 human lymphoblastoid T-cell line (9). Among the members of the quinolone structural class of compound, WM5 appears to be one of the most effective anti-HIV-1 agents so far described. This property prompted us to further extend our studies. To investigate the mechanism of action of WM5 at the molecular level, among a variety of human lymphoblastoid cell lines tested, we selected the human CD4+ T-cell line Jurkat, which is highly permissive for HIV-1 replication. Jurkat cells were exposed to HIV-1 at MOI of 0.1 and 0.01 TCID50 per cell, cultured in the presence of WM5, and monitored for virus replication by measuring RT activity in the culture supernatants. As shown in Fig. 2, WM5 significantly inhibits viral replication in Jurkat cells at both MOI without affecting cell viability (concentration of compound required to reduce Jurkat cell viability by 50% [CC50] = 56.24 μM, as reported in Fig. 1). At the higher MOI, the inhibitory effect was more striking when virus replication was monitored by viral load in the culture supernatants. At an MOI of 0.1, the IC50 was 0.60 ± 0.06 μM after 12 days of infection. When viral infection was maintained in the presence of WM5E, the 3-ethyl-esterified form of WM5, no effect on virus replication was observed. This finding is consistent with our previous observations obtained with the C8166 cell line (9), indicating the critical contribution of the carboxylic acid at the C-3 position of the quinolone moiety to the antiviral activity. To test the effect of WM5 on chronic infection, HIV-1IIIB chronically infected H9 cells, activated with PHA and IL-2, were incubated in the presence or absence of various concentrations of WM5 for 24 h, and HIV-1 growth was monitored by assessing RT activity in the culture supernatants. WM5 inhibited HIV-1 replication in these cells with an IC50 of 0.85 ± 0.05 μM. Cell viability in H9-treated cultures was essentially similar to that of control cells, as determined by MTT assay (data not shown).
FIG. 2.
Effect of WM5 on replication-competent HIV-1 in Jurkat lymphocytes. Cultures were infected with the HXBc2 isolate at MOI of 0.1 (A and B) and 0.01 (C) TCID50 per cell for 2 h, washed, and maintained in the absence (open bars) or presence (hatched bars) of WM5 at the concentration shown over time. Virus replication was monitored by RT production in cell-free supernatants 5 (A) and 12 (B and C) days after infection. As a control, WM5E, the 3-ethyl-esterified form of WM5, was included (solid bars). Where indicated (A [inset]) in the same culture, virus production was also determined by measuring the viral load.
Effect of WM5 on a single round of HIV-1 replication.
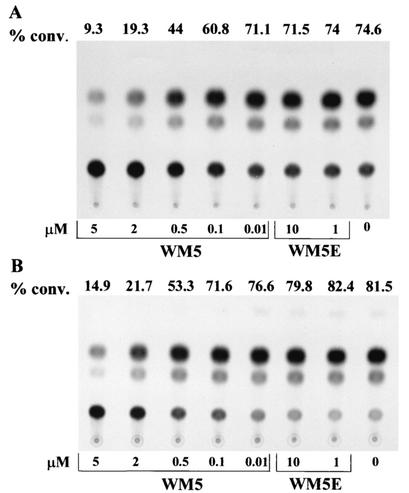
To study the replicative properties of HIV-1, we used an env complementation assay, which measures the efficiency of the early events in a single round of infection (23). HIV-1 infection of target cells requires the sequential binding of envelope glycoprotein to CD4 and one of the chemokine receptors. In this assay, an env-defective provirus encoding the bacterial CAT gene was complemented by the envelope glycoprotein. The level of CAT expression in the target cells reflects the efficiency of a single round of the retroviral infection cycle. The use of the env complementation system has several advantages for these studies. First, precise measurements of early-phase replicative ability can be made. Second, since all measurements are performed with a single cycle of infection initiated by clonal viral genes, the possibility of changes in viral phenotype during virus propagation is eliminated. Because the T-cell line we used in this assay expresses the CXCR-4 chemokine receptor, the envelope glycoprotein was derived from the laboratory-adapted T-cell-tropic strain HXBc2, which uses CXCR-4 as a coreceptor (17). Recombinant HIV-1 virions expressing CAT and containing the HXBc2 envelope glycoprotein were harvested 36 h after transfection of 293T cells, normalized to 30,000 cpm of RT activity, and used to infect Jurkat cells. Cell cultures were then maintained in the absence or presence of the compounds. CAT transfer by recombinant viruses was significantly inhibited by WM5 at concentrations of 5 and 2 μM (IC50 =0.64 ± 0.05 μM) (Fig. 3). In contrast, WM5E did not exert any effect on HIV-1 replication. These results indicate that WM5 specifically inhibits early events in HIV-1 infection.
FIG. 3.
CAT activities in Jurkat cells infected with HIV-1 CAT reporter viruses. Cultures were infected with HIV-1 recombinant CAT viruses expressing envelope glycoproteins from a laboratory-adapted T-tropic virus, HXBc2 (A), and a primary dualtropic virus, 89.6 (B), and then maintained in the absence or presence of WM5 or WM5E at the concentrations shown. Four days later, CAT activity in cell lysates was measured. The results are reported as percent conversion (% conv.) of [14C]chloramphenicol to acetylated forms above the spots. A representative experiment of two performed is shown.
It is possible that the observed inhibition of HIV-1 replication by WM5 might be strain specific. To address this issue, we examined the ability of WM5 to inhibit other HIV-1 strains. We generated recombinant HIV-1 viruses containing the envelope glycoprotein of the primary HIV-1 isolate 89.6, a dualtropic strain that can use either CXCR-4 or CCR5 as a coreceptor (12, 15). Figure 3 shows that WM5, but not WM5E, significantly abrogated replication of recombinant virions expressing the 89.6 envelope glycoprotein to levels comparable to those of recombinant virions containing the HXBc2 envelope glycoprotein. The IC50 was similar to that obtained with the HXBc2 envelope glycoprotein (0.60 ± 0.04 μM).
Inhibition of RT activity in vitro.
To examine the effects of WM5 at different steps of the virus life cycle, we first tested the effect of the compound on the activity of the RNA-dependent DNA polymerase enzyme. To this end, we used an in vitro RT inhibition assay performed in the presence of various concentrations of WM5. As shown in Table 1, no significant inhibitory effect on RT activity was observed in the presence of WM5.
TABLE 1.
Anti-HIV-1 activity of WM5 on virus-encoded enzymes
| Compound | IC50 (μM)a
|
|||
|---|---|---|---|---|
| RT | IN
|
PR | ||
| 3′ processing | Strand transfer | |||
| WM5 | 800 ± 150 | 60 ± 20 | 75 ± 15 | 71 ± 21 |
| WM5E | >100 | >100 | ||
| RDS 1028 | 0.4 ± 0.2 | 0.3 ± 0.2 | ||
| Saquinavir | 0.002 ± 0.00035 | |||
IC50, concentration of compound required to reduce the HIV-1 enzyme-catalyzed reactions by 50%. PR, protease. All data represent means ± standard deviations obtained in three separate experiments.
Effect of WM5 on HIV-1 IN.
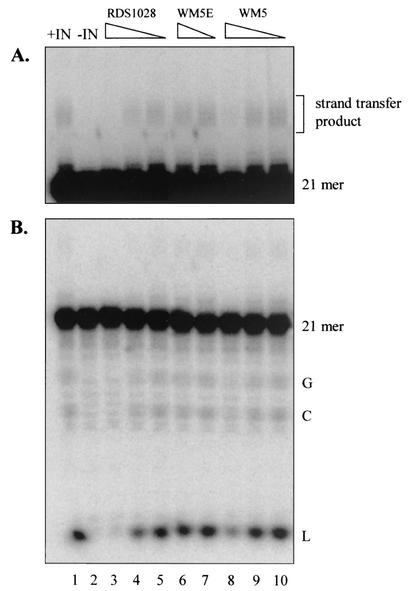
We then investigated the effect of WM5 on the virus integration process. HIV-1 IN catalyzes the integration of the viral DNA into the host chromosome through a two-step mechanism. In the cytoplasm, IN recognizes the LTR termini and removes the last two nucleotides (GT), leaving two recessed 3′-OH ends (3′-processing reaction). In the nucleus, IN catalyzes the joining of the 3′-processed viral 3′ ends into the host DNA through a transesterification reaction (strand-transfer reaction). In vitro, the IN sequence-specific removal of the labeled terminal dinucleotide of the 21-mer double-stranded oligonucleotide substrate by intervention of nucleophiles, such as glycerol, 3′-OH groups of the substrate DNA, and water, generates 5′-glycerol-phosphate-GTOH (G), cyclic GT (C), and 5′-phosphate-GTOH (L), respectively, as reaction products. Subsequently, IN catalyzes the insertion of the 19-mer 3′ processing product into a labeled 21-mer unprocessed substrate, leading to strand transfer products of different lengths. When assayed for anti-IN activity, WM5 showed an ability to inhibit both 3′ processing and strand transfer activities at high micromolar concentrations (Fig. 4 and Table 1), whereas WM5E was completely inactive. A catechol derivative (RDS 1028) was used as a reference control (3).
FIG. 4.
Inhibition of HIV-1 IN-catalyzed 3′-processing and strand transfer reactions by WM5 and WM5E. The strand transfer products migrate more slowly than the 21-mer substrate (A, darker exposure) and the 3′-processing products G, C, and L (B, lighter exposure). Lanes: 1, DNA and IN without drugs; 2, DNA alone; 3 to 10, DNA, IN, and a titration of RDS 1028 (1, 0.33, and 0.1 μM in lanes 3 to 5, respectively), WM5E (100 and 33 μM in lanes 6 and 7, respectively), and WM5 (100, 33, and 11 μM in lanes 8 to 10, respectively).
Effect of WM5 on protease activity.
The late phases of the virus life cycle were analyzed by using an in vitro protease inhibition assay previously reported by our group (32). The wild-type HIV-1 protease, expressed and purified in E. coli, was employed to test its activity on hydrolysis of a fluorogenic substrate. As reported in Table 1, WM5 showed the ability to inhibit protease activity at high micromolar concentrations.
Binding to nucleic acids.
To evaluate the binding of quinolones to nucleic acids, we performed fluorometric titrations. Upon addition of TAR to WM5 solutions in the presence of constant (1 mM) magnesium ions, substantial variations in the compound's fluorescence quantum yield are observed, indicative of a complex formation at low RNA/ligand ratios (up to 0.8), whereas no variations in the emission spectra of WM5E were observed at the same or higher ratios of TAR to quinolone.
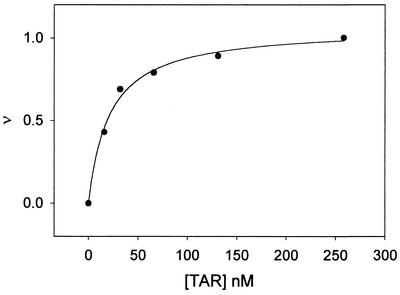
The complex curve formation (fraction of bound quinolone, ν, versus TAR concentration) is shown in Fig. 5. The apparent affinity of WM5 to this nucleic acid, expressed as the concentration of TAR needed to complex 50% of WM5, normalized for quinolone concentration, corresponded to 19 ± 0.6 nM. Lack of interaction with unrelated nucleic acid targets was confirmed by the very poor affinity of WM5 (and WM5E) for single- or double-stranded DNA structures as well as for a tRNA preparation. In particular, binding of the quinolone to DNA yields an appreciable variation in the fluorescence spectra, variation that was not observed upon addition of tRNA. Accordingly, the latter should not exhibit appreciable affinity for the quinolone. The apparent binding constant of WM5 to calf thymus DNA was found to be 0.72 mM (per average base residue), similar to that obtained by the titration with single-stranded DNA (0.71 mM). These data are indeed indicative of a low affinity of aminoquinolones for single- and double-stranded DNA, in accordance with what was previously obtained for other aminoquinolones (51), which bind the nucleic acid five- to sixfold less than the clinically used bacterial fluoroquinolones (39).
FIG. 5.
Binding of WM5 to TAR RNA. Shown is the fraction of bound quinolone (ν) versus the TAR concentration for WM5 as inferred by fluorometric titrations (see text). Experiments were performed with a mixture of Tris-HCl (10 mM, pH 7.0), NaCl (20 mM), and Mg(ClO4)2 (1 mM) at 25°C.
DISCUSSION
Despite the recent success of HAART, the search for new anti-HIV-1 compounds continues to be a major challenge in the development of antiviral molecules. Among a series of 6-aminoquinolone derivatives, we recently identified WM5 as a potent inhibitor of HIV-1 replication (9). Here, we examine, at the molecular level, the basis of this antiviral activity. The studies presented here provide evidence that WM5 exerts its effect at the transcriptional level.
In this study, in addition to replication-competent virus, we used a transient transcomplementation assay to assess the replicative potential of HIV-1 in a single round of virus replication (23). The use of recombinant viruses pseudotyped by the envelope glycoprotein allows us to examine early events in the infection process. Because the viral proteins are expressed in a context similar to that occurring in the authentic provirus, the levels of gene expression achieved are expected to resemble those in HIV-1-infected cells. Consistent with our previous observation (9), WM5 inhibited HIV-1 replication in acutely infected cell cultures. Moreover, inhibition of HIV-1 was also found in chronically infected cells. The observation that WM5, but not WM5E, affects virus replication implies that the carboxylic acid group at the C-3 position of the quinolone molecule plays a crucial role in the antiviral effect. These results indicate that this portion of the molecule, along with the presence of small substituents at the N-1 position, a small polar group at the C-6 position, and bulky substituents at the C-7 position, contributes to the antiviral activity, as we previously reported (9). Our results indicate that between 82 and 88% inhibition of infection by recombinant viruses pseudotyped with the HXBc2 and the 89.6 envelope glycoproteins, in a single round of infection of Jurkat cells, was obtained in the presence of 5 μM WM5 compared to the level in control cell cultures. In dissecting specific steps of the early phase of the viral life cycle that might be targeted by WM5, we have evidence that RNA-dependent DNA polymerase and IN activities in vitro, as well as proviral DNA synthesis (data not shown), were not affected by the compound. On the other hand, we found that late phases of the replication life cycle, like those dependent on protease activity, were not impaired by WM5. Altogether, these results suggest that the most likely process affected by WM5 is HIV-1 transcription. This finding correlates with our previous observation that WM5 binds to TAR RNA stem-bulge region of the viral RNA transcript (9). This conclusion is further supported by our results indicating that WM5 exhibits a great affinity to TAR RNA, as suggested by the dissociation constant of the WM5-TAR complex that was found to be in the range of 19 nM. Our results therefore suggest that WM5 may effectively sequester TAR RNA affecting HIV-1 transcription. Fluoroquinolone derivatives have recently been shown to be potent and selective inhibitors of HIV-1 replication (5, 22, 29, 36, 55). A representative compound of the series, K12, has been reported to inhibit HIV-1 transcription, reducing the synthesis of HIV-1 mRNA in chronically infected cells without significantly affecting Tat activity (5). Since Tat-mediated HIV-1 activation involves complex interaction with known and unknown cellular factors (60), it has been suggested that K12 might target cellular factors that play a key role in HIV-1 transcription either by influencing their expression or by interfering with their function in HIV-1 transactivation (55). Recently, a K12 derivative, K37 has been reported to be more active than K12 in inhibiting HIV-1 replication by affecting Tat-mediated gene expression in HIV-1 LTR-driven reporter gene assay (36). Our TAR-quinolone binding experiments instead suggest a newer nucleic acid-targeted mechanism of action. Although we cannot exclude the possibility that WM5 might target cellular factors, the drug's ability to bind TAR with prominent affinity may have an advantage over fluoroquinolone derivatives in terms of possible cytotoxic side effects.
Disruption of Tat-TAR complex has been the goal of rational anti-HIV-1 strategies, based on either a peptidomimetic approach (30) or the screening of combinatorial peptide libraries (27). Recent nuclear magnetic resonance studies have demonstrated that critical groups recognized by Tat are presented to the protein in a unique spatial arrangement created by conformational rearrangement in the TAR RNA that occurs during binding (1, 43). Metal ions play an important role in stabilizing a variety of RNA structural motifs by interacting with RNA functional groups. Mg2+ ions have been shown to be required for folding and function of hammerhead ribozyme (7, 8, 42, 48). TAR RNA can also bind divalent ions (such as Ca2+), as was found in the X-ray structure of model TAR (28). It has been proposed that the bulge region of TAR is a metal binding site, as supported by hydrodynamic studies of the influence of metal ion (Mg2+) binding on TAR RNA bulge region conformation (57, 58). We previously demonstrated how quinolones target single-stranded nucleic acids and the essential contribution of magnesium ions to the mechanism of antibacterial activity (39, 41, 51, 54). Given the ability of quinolones to interact with nucleic acids isolated or complexed to proteins, a similar mechanism of action on nucleic acid-protein complexes may be relevant for antiviral activity. Indeed, here we show the ability of WM5 to bind TAR RNA with high affinity, possibly interfering with Tat-mediated transcriptional activation. In conclusion, WM5 represents a promising lead for the development of a new group of anti-HIV-1 compounds, the synthetic and pharmacological features of which might make them candidates for combinations with clinically available antiretroviral agents.
Acknowledgments
This work was supported by AIDS grants from the Istituto Superiore di Sanità (Rome-AIDS Projects no. 40B.72 and 30.57), the Fondazione Cassa di Risparmio di Padova e Rovigo, Regione Veneto, MURST, CNR Target Project on Biotechnology, and AIRC.
We thank F. Salandin for artwork.
REFERENCES
- 1.Aboul-ela, F., J. Karn, and G. Varani. 1995. The structure of the human immunodeficiency virus type 1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 253:313-332. [DOI] [PubMed] [Google Scholar]
- 2.Antonello, C., E. Uriarte, M. Palumbo, S. Valisena, C. Parolin, and G. Palù. 1993. Synthesis and biological activity of new quinolone derivatives. Eur. J. Med. Chem. 28:291-296. [Google Scholar]
- 3.Artico, M., R. Di Santo, R. Costi, E. Novellino, G. Greco, S. Massa, E. Tramontano, M. E. Marongiu, A. De Montis, and P. La Colla. 1998. Geometrically and conformationally restrained cinnamoyl compounds as inhibitors of HIV-1 integrase: synthesis, biological evaluation, and molecular modeling. J. Med. Chem. 41:3948-3960. [DOI] [PubMed] [Google Scholar]
- 4.Baba, M., M. Okamoto, M. Kawamura, M. Makino, M. Higashida, T. Takashi, Y. Kimura, T. Ikeuchi, T. Tetsuka, and T. Okamoto. 1998. Inhibition of human immunodeficiency virus type 1 replication and cytokine production by fluoroquinoline derivatives. Mol. Pharmacol. 53:1097-1103. [PubMed] [Google Scholar]
- 5.Baba, M., M. Okamoto, M. Makino, Y. Kimura, T. Ikeuchi, T. Sakaguchi, and T. Okamoto. 1997. Potent and selective inhibition of human immunodeficiency virus type 1 transcription by piperazinyloxoquinoline derivatives. Antimicrob. Agents Chemother. 41:1250-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barre-Sinoussi, F., J. C. Chermann, F. Rey, M. T. Nugeyre, S. Chamaret, J. Gruest, C. Dauget, C. Axler-Bin, F. Vezinet-Brun, C. Rouzioux, W. Rozenbaum, and L. Montagnier. 1983. Isolation of a T-lymphocyte retrovirus from a patient at risk for acquired immunodeficiency syndrome (AIDS). Science 220:868-871. [DOI] [PubMed] [Google Scholar]
- 7.Bassi, G. S., N. E. Mollegaard, A. I. Murchie, and D. M. Lilley. 1999. RNA folding and misfolding of the hammerhead ribozyme. Biochemistry 38:3345-3354. [DOI] [PubMed] [Google Scholar]
- 8.Bassi, G. S., N. E. Mollegaard, A. I. Murchie, E. von Kitzing, and D. M. Lilley. 1995. Ionic interactions and the global conformations of the hammerhead ribozyme. Nat. Struct. Biol. 2:45-55. [DOI] [PubMed] [Google Scholar]
- 9.Cecchetti, V., C. Parolin, S. Moro, T. Pecere, E. Filipponi, A. Calistri, O. Tabarrini, B. Gatto, M. Palumbo, A. Fravolini, and G. Palù. 2000. 6-Amino quinolones as new potential anti-HIV agents. J. Med. Chem. 43:3799-3802. [DOI] [PubMed] [Google Scholar]
- 10.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, J. 1998. Exploring how to get at—and eradicate—hidden HIV. Science 279:1854-1855. [DOI] [PubMed] [Google Scholar]
- 12.Collman, R., J. W. Balliet, S. A. Gregory, H. Friedman, D. L. Kolson, N. Nathanson, and A. Srinivasan. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig, J. C., I. B. Duncan, D. Hockley, C. Grief, N. A. Roberts, and J. S. Mills. 1991. Antiviral properties of Ro 31-8959, an inhibitor of human immunodeficiency virus (HIV) proteinase. Antivir. Res. 16:295-305. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, B. R. 1987. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 152:684-703. [DOI] [PubMed] [Google Scholar]
- 15.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 16.Fauci, A., A. Macher, D. Longo, H. C. Lane, A. Rook, H. Masur, and E. Gelmann. 1984. Acquired immunodeficiency syndrome: epidemiologic, clinical, immunologic, and therapeutic considerations. Ann. Intern. Med. 100:92-106. [DOI] [PubMed] [Google Scholar]
- 17.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:809-810. [DOI] [PubMed] [Google Scholar]
- 18.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosemberg, B. Walker, S. Cange, J. Gallant, and R. F. Siciliano. 1999. Latent infection of CD4+ T-cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 19.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siciliano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 20.Gallo, R. C., S. Z. Salahuddin, M. Popovic, G. M. Shearer, M. Kaplan, B. F. Haynes, T. J. Palker, R. Redfield, J. Oleske, B. Safai, G. White, P. Foster, and P. D. Markham. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500-503. [DOI] [PubMed] [Google Scholar]
- 21.Gatto, B., G. Capranico, and M. Palumbo. 1999. Drugs acting on DNA topoisomerases: recent advances and future perspectives. Curr. Pharm. Des. 5:195-215. [PubMed] [Google Scholar]
- 22.Hagihara, M., H. Kashiwase, T. Katsube, T. Kimura, T. Komai, K. Momota, T. Ohmine, T. Nishigaki, S. Kimura, and K. Shimada. 1999. Synthesis and anti-HIV activity of arylpiperazinyl fluoroquinolones: a new class of anti-HIV agents. Bioorg. Med. Chem. Lett. 9:3063-3068. [DOI] [PubMed] [Google Scholar]
- 23.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 64:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-infection: recommendations for an International AIDS Society-USA Panel. JAMA 282:2417-2426. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch, M. S., B. Conway, R. T. D'Aquila, V. A. Johnson, F. Brun-Vezinet, B. Clotet, L. M. Demeter, S. M. Hammer, D. M. Jacobsen, D. R. Kuritzkes, C. Loveday, J. W. Mellors, S. Vella, and D. D. Richman. 1998. Antiretroviral drug resistance testing in adults with HIV infection. JAMA 279:1984-1991. [DOI] [PubMed] [Google Scholar]
- 26.Hui, J. O., A. G. Tomasselli, I. M. Reardon, J. M. Lull, D. P. Brunner, C. S. Tomich, and R. L. Heinrikson. 1993. Large scale purification of HIV-1 protease from Escherichia coli inclusion bodies. J. Protein Chem. 12:323-327. [DOI] [PubMed] [Google Scholar]
- 27.Hwang, S., N. Tamilarasu, K. Ryan, I. Huq, S. Richter, W. C. Still, and T. M. Rana. 1999. Inhibition of gene expression in human cells through small molecule-RNA interactions. Proc. Natl. Acad. Sci. USA 96:12997-13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ippolito, J. A., and T. A. Steitz. 1998. A 1.3-A resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl. Acad. Sci. USA 95:9819-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashiwase, H., K. Momota, T. Ohmine, T. Komai, T. Kimura, T. Katsube, T. Nishigaki, S. Kimura, K. Shimada, and H. Furukawa. 1999. A new fluoroquinolone derivative exhibits inhibitory activity against human immunodeficiency virus type 1 replication. Chemotherapy 45:48-55. [DOI] [PubMed] [Google Scholar]
- 30.Litovchick, A., A. G. Evdokimov, and A. Lapidot. 2000. Aminoglycoside-arginine conjugates that bind TAR RNA: synthesis, characterization and antiviral activity. Biochemistry 39:2838-2852. [DOI] [PubMed] [Google Scholar]
- 31.Marshall, A. G. 1978. Biophysical chemistry. Principles, techniques and applications, p. 70-72. John Wiley & Sons, New York, N.Y.
- 32.Maschera, B., G. Darby, G. Palù, L. L. Wright, M. Tisdale, R. Myers, E. Blaire, and E. S. Furfine. 1996. Human immunodeficiency virus. Mutations in the viral protease that confer resistance to saquinavir increase the dissociation rate constant of the protease-saquinavir complex. J. Biol. Chem. 271:33231-33235. [DOI] [PubMed] [Google Scholar]
- 33.Maschera, B., E. Furfine, and E. D. Blair. 1995. Analysis of resistance to human immunodeficiency virus type 1 protease inhibitors by using matched bacterial expression and proviral infection vectors. J. Virol. 69:5431-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mossmann, T. 1993. Rapid colorimetric assay for cell growth and survival. Application to proliferation and cytotoxicity. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 35.Ohmine, T., T. Katsube, Y. Tsuzaki, M. Kazui, N. Kobayashi, T. Komai, M. Hagihara, T. Nishigaki, A. Iwamoto, T. Kimura, H. Kashiwase, and M. Yamashita. 2002. Anti-HIV-1 activities and pharmacokinetics of new arylpiperazinyl fluoroquinolones. Bioorg. Med. Chem. Lett. 12:739-742. [DOI] [PubMed] [Google Scholar]
- 36.Okamoto, H., T. P. Cujec, M. Okamoto, B. M. Peterlin, M. Baba, and T. Okamoto. 2000. Inhibition of the RNA-dependent transactivation and replication of human immunodeficiency virus type 1 by a fluoroquinoline derivative K-37. Virology 272:402-408. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto, M., T. Okamoto, and M. Baba. 1999. Inhibition of human immunodeficiency virus type 1 replication by combination of transcription inhibitor K-12 and other antiretroviral agents in acutely and chronically infected cells. Antimicrob. Agents Chemother. 43:492-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 39.Palù, G., S. Valisena, G. Ciarrocchi, B. Gatto, and M. Palumbo. 1992. Quinolone binding to DNA is mediated by magnesium ions. Proc. Natl. Acad. Sci. USA 89:9671-9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palù, G., S. Valisena, C. Parolin, G. Ciarrocchi, B. Gatto, and M. Palumbo. 1991. Further insight into the mechanism of action of quinolones. Eur. J. Clin. Microbiol. Infect. Dis. XX(Suppl. I):350-351.
- 41.Palumbo, M., B. Gatto, G. Zagotto, and G. Palù. 1993. On the mechanism of action of quinolone drugs. Trends Microbiol. 1:232-235. [DOI] [PubMed] [Google Scholar]
- 42.Pley, H. W., K. M. Flaherty, and D. B. McKay. 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372:68-74. [DOI] [PubMed] [Google Scholar]
- 43.Puglisi, J. D., R. Tan, B. J. Calnan, A. D. Frankel, and J. R. Williamson. 1992. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 257:76-80. [DOI] [PubMed] [Google Scholar]
- 44.Ramratnam, B., J. E. Mittler, L. Zhang, D. Boden, A. Hurley, F. Fang, C. A. Macken, A. S. Perelson, M. Markowitz, and D. D. Ho. 2000. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med. 6:82-85. [DOI] [PubMed] [Google Scholar]
- 45.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. J. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, L. Ivanoff, S. R. Petteway, Jr., M. L. Pearson, J. A. Lauteenberger, T. S. Papas, J. Ghrayeb, N. T. Chang, R. C. Gallo, and F. Wong-Staal. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-283. [DOI] [PubMed] [Google Scholar]
- 46.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 47.Rho, H. M., B. Poiesz, F. Ruscetti, and R. C. Gallo. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355-360. [DOI] [PubMed] [Google Scholar]
- 48.Scott, W. G., J. B. Murray, J. R. Arnold, B. L. Stoddard, and A. Klug. 1996. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science 274:2065-2069. [DOI] [PubMed] [Google Scholar]
- 49.Shen, L., J. Baranowski, and A. Pernet. 1989. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: specificity and cooperativity of drug binding to DNA. Biochemistry 28:3879-3885. [DOI] [PubMed] [Google Scholar]
- 50.Shen, L., L. Mitscher, P. Sharma, T. O'Donnel, D. Chu, C. Cooper, T. Rosen, and A. Pernet. 1989. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: a cooperative drug-DNA binding model. Biochemistry 28:3886-3894. [DOI] [PubMed] [Google Scholar]
- 51.Sissi, C., M. Andreolli, V. Cecchetti, A. Fravolini, B. Gatto, and M. Palumbo. 1998. Mg2+-mediated binding of 6-substituted quinolones to DNA: relevance to biological activity. Bioorg. Med. Chem. 6:1551-1561. [DOI] [PubMed] [Google Scholar]
- 52.Toth, M. V., and G. R. Marshall. 1990. A simple, continuous fluorometric assay for HIV protease. Int. J. Pept. Protein Res. 36:544-550. [DOI] [PubMed] [Google Scholar]
- 53.Tramontano, E., P. La Colla, and Y.-C. Cheng. 1998. Biochemical characterization of the HIV-1 integrase 3′-processing activity and its inhibition by phosphorothioate oligonucleotides. Biochemistry 37:7237-7242. [DOI] [PubMed] [Google Scholar]
- 54.Valisena, S., M. Palumbo, C. Parolin, G. Palù, and G. A. Meloni. 1990. Relevance of ionic effects on norfloxacin uptake by Escherichia coli. Biochem. Pharmacol. 40:431-436. [DOI] [PubMed] [Google Scholar]
- 55.Witvrouw, M., D. Daelemans, C. Pannecouque, J. Neyts, G. Andrei, R. Snoeck, A. M. Vandamme, J. Balzarini, J. Desmyter, M. Baba, and E. De Clercq. 1998. Broad-spectrum antiviral activity and mechanism of antiviral action of the fluoroquinolone derivative K-12. Antivir. Chem. Chemother. 9:403-411. [DOI] [PubMed] [Google Scholar]
- 56.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 57.Zacharias, M., and P. J. Hagerman. 1995. Bulge-induced bends in RNA: quantification by transient electric birefringence. J. Mol. Biol. 247:486-500. [DOI] [PubMed] [Google Scholar]
- 58.Zacharias, M., and P. J. Hagerman. 1995. The bend in RNA created by the trans-activation response element bulge of human immunodeficiency virus is straightened by arginine and by Tat-derived peptide. Proc. Natl. Acad. Sci. USA 92:6052-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesamen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, Q., and P. A. Sharp. 1995. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 14:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]