Abstract
Carbapenem antibiotics have been used to counteract resistant strains of bacteria harboring β-lactamases and extended-spectrum β-lactamases. Four enzymes from the class A group of β-lactamases, NMC-A, IMI-1, SME-1, and KPC-1, efficiently hydrolyze carbapenem antibiotics. Sequence comparisons and structural information indicate that cysteines at amino acid residues 69 and 238, which are conserved in all four of these enzymes, form a disulfide bond that is unique to these β-lactamases. To test whether this disulfide bond is required for catalytic activity, the codons for residues Cys69 and Cys238 were randomized individually and simultaneously by PCR-based mutagenesis to create random replacement libraries for these positions. Mutants that were able to confer resistance to ampicillin, imipenem, or cefotaxime were selected from these libraries. The results indicate that positions Cys69 and Cys238 are critical for hydrolysis of all of the antibiotics tested, suggesting that the disulfide bond is generally required for this enzyme to catalyze the hydrolysis of β-lactam antibiotics.
Carbapenems, which include the widely used antibiotics meropenem and imipenem, are used to treat severe infections caused by β-lactamase-harboring bacteria (1, 3). Unfortunately, as with other antibiotics, resistance to carbapenem antibiotics has emerged and several β-lactamases that catalyze the hydrolysis of carbapenems have been identified (9, 11). The production of class B metallo-β-lactamases is the most common mechanism for resistance to carbapenems; however, class D OXA β-lactamases, such as OXA-23, -24, -25, -26, and -27, also provide significant resistance to these antibiotics (6, 11).
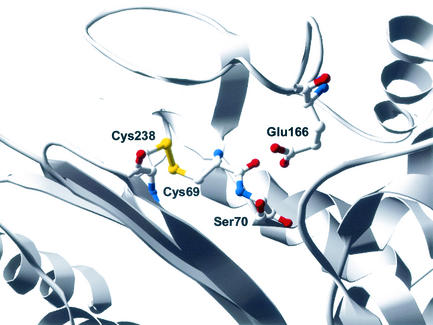
Recent additions to the list of carbapenem-hydrolyzing β-lactamases are four class A β-lactamases, NMC-A, IMI-1, SME-1, and KPC-1 (7, 8, 13, 20). As a group, these enzymes are 45% identical, with nearly 70% amino acid identity between NMC-A and SME-1 and 95% amino acid identity between NMC-A and IMI-1 (8, 20). In addition, these enzymes share approximately 40% amino acid identity with the TEM and SHV β-lactamases; and they possess all the structural features characteristic of class A β-lactamases, such as the omega loop and the SXXK, SDN, and KTG motifs (7, 8, 13, 20). One unique feature of the class A carbapenemases is the presence of a disulfide bond between positions Cys69 and Cys238 (12, 15, 17) (Fig. 1). Many class A β-lactamases, such as the TEM β-lactamase, possess a disulfide bond between residues Cys77 and Cys123, which provides a connection between the all-alpha and the alpha-beta domains of the enzyme, but lack cysteines at positions 69 and 238 (14). Interestingly, the Cys77-Cys123 disulfide bond is not necessary for the TEM-1 enzyme to hydrolyze β-lactam antibiotics (14, 18). The role of the disulfide bond between Cys69 and Cys238 in the unique substrate specificity of class A carbapenemases is yet to be determined. Site-directed mutagenesis of position Cys69 in the SME-1 β-lactamase has been carried out by replacing the cysteine at position 69 with an alanine (15). The Cys69Ala SME-1 mutant was unable to confer resistance to several antibiotics including imipenem, amoxicillin, ticarcillin, cefoxitin, and aztreonam (15). This result suggests that the disulfide bond may be required for the unique substrate specificity of the SME-1 enzyme.
FIG. 1.
Structure of SME-1 β-lactamase (15). Positions 69, 70, 166, and 238 are highlighted. The disulfide bond between positions 69 and 238 is also shown.
To determine if the cysteine residues are the only combination of amino acids that can occur at positions 69 and 238, a codon randomization and functional selection approach was used. In this study, the codons for positions Cys69 and Cys238 in the blaSME gene were randomized by PCR-based mutagenesis (4). The codons were randomized individually as well as simultaneously in order to identify other amino acid interactions (i.e., hydrophobic interactions, salt bridges, etc.) that may functionally replace the disulfide bond between positions Cys69 and Cys238 in the SME-1 β-lactamase. The results presented here indicate that the disulfide bond is critical not only for carbapenem hydrolysis but also for the hydrolysis of penicillins and cephalosporins.
MATERIALS AND METHODS
Site-directed randomization and random library construction.
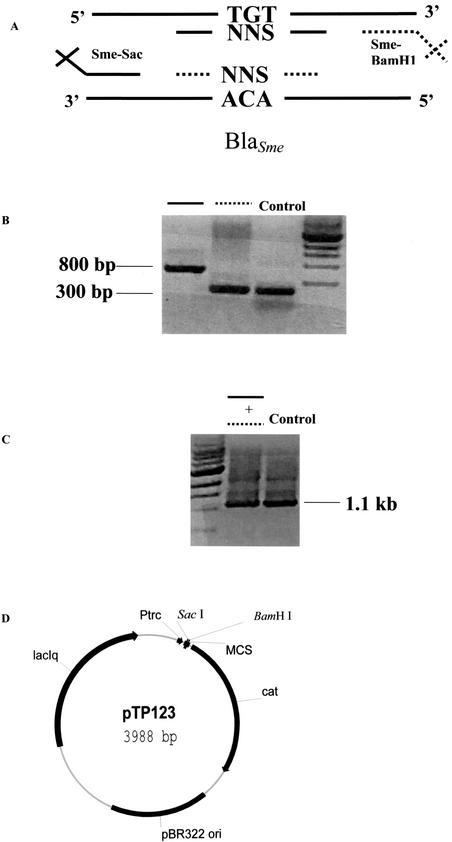
Site-directed mutagenesis was carried out through a two-step PCR with Pfu polymerase (Stratagene, La Jolla, Calif.). Primers were designed such that positions 69 and 238 were randomized as NNS codons, where N is any nucleotide and S is either a cytosine or a guanine. The primer sequences are listed in Table 1; and the expected sizes of the PCR products are as follows: for the SME-Sac plus SME-Cys69-Bot primer pair, 298 bp; for the SME-BamHI plus SME-Cys69-Top primer pair, 836 bp; for the SME-Sac plus SME-Cys238-Bot primer pair, 813 bp; for the SME-BamHI plus SME-Cys238-Top primer pair, 318 bp; and for the SME-Sac plus SME-BamHI primer pair, 1,100 bp.
TABLE 1.
Oligonucleotides used for randomization and cloning of blaSME gene
| Primer name | Sequence | Use |
|---|---|---|
| SME-Sac | 5′-GGGGCGGAGCTCAACTCATTCAACACTCGG-3′a | Cloning |
| SME-BamHI | 5′-GGGGCGGGATCCGCGTCAAGGCCACAGTCAGCTCTAACGC-3′a | Cloning |
| SME-Cys69-Bot | 5′-CCTTTAAATGAACTSNNTAAAGGGAACCGCTCATC-3′b | Randomizing |
| SME-Cys69-Top | 5′-GATGAGCGGTTCCCTTTANNSAGTTCATTTAAAGG-3′b | Randomizing |
| SME-Cys238-Bot | 5′-CGCAGTACCTATAGCCCCSNNGCTCCCAGTTTTGT-3′b | Randomizing |
| SME-Cys238-Top | 5′-ACAAAACTGGGAGCNNSGGGGCTATAGGTACTGCG-3′b | Randomizing |
The SacI and BamHI restriction sites (underlined) were used to clone the PCR products into pTP123.
The randomizing primers were used to replace the target codon by NNS randomization, where N represents any nucleotide and S represents guanine or cytosine.
This approach eliminates two of three stop codons and reduces the number of clones needed to form a complete randomized library. Outside primers were designed to allow cloning into the pTP123 cloning vector by restriction digestion (10). In the first step, the outside primers and the randomizing primers for positions Cys69 and Cys238 were used to amplify each half of the blaSME gene. The PCR products generated from the first step were then mixed together and amplified by using only the outside primers to regain the full blaSME genes randomized at position Cys69 or Cys238 (4). To randomize both positions in the same gene, full-length PCR products from each individual randomization reaction were mixed together with outside and randomizing primers from the opposing position. The resulting PCR products were then amplified with the outside primers to regain the full-length blaSME gene. The outside primers contained SacI and BamHI restriction sites for cloning into the pTP123 vector. This experiment is diagramed in Fig. 2. All primers were obtained from Integrated DNA Technologies, Inc. (Coralville, Iowa).
FIG. 2.
Construction of random libraries. Random libraries for positions 69 and 238 and the combination of positions 69 and 238 in SME-1 were created by a PCR-based approach. (A) Primers were designed to randomize the targeted codon (NNS) and to introduce restriction sites SacI and BamHI at the ends of the PCR products for cloning into the pTP123 vector. (B) The first PCR consisted of the use of one internal mutagenic primer and one outside cloning primer (SME-Sac or SME-BamHI). (C) The PCR products from the first set of reactions were mixed together and amplified by using only the outside cloning primers to regain the full-length blaSME gene with the target position randomized. (D) This PCR product was digested with SacI and BamHI and cloned into the pTP123 vector. MCS, multicloning site.
Bacterial strains and cloning of random libraries.
All blaSME genes that were randomized at either codon Cys69 or Cys238, or both, were cloned as a SacI-BamHI fragment into the multicloning site of pTP123. pTP123 is a derivative of the pTrc 99A vector (Amersham Pharmacia Biotech, Piscataway, N.J.), which possesses the cat gene for chloramphenicol (CMP) resistance (10) (Fig. 2D). The three randomized libraries were electroporated into Escherichia coli XL-1 Blue {recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′::Tn10(Tetr) proAB ΔlacIq (laqZ)M15]} (2). After electroporation, cells were spread on Luria-Bertani (LB) agar plates supplemented with 12.5 μg of CMP per ml (LB-CMP). The colonies were pooled in 1 ml of LB medium. Plasmid DNA was harvested from these bacteria by using a Qiaprep Spin Miniprep kit (Qiagen, Valencia, Calif.). Both the SacI and the BamHI enzymes were obtained from New England Biolabs (Beverley, Mass.).
Antibiotic selection.
Three antibiotics, ampicillin, imipenem, and cefotaxime, were used to select functional clones from the Cys69X, Cys238X, and Cys69X-Cys238X random libraries. E. coli XL-1 Blue cells were electroporated with plasmid DNA from the random library, and transformed cells were spread on LB-CMP plates. To carry out selection of the randomized libraries, a sterile blank paper disk (Becton Dickinson, Cockeysville, Md.) soaked in either 10 mg of ampicillin (Sigma Chemical Co., St. Louis, Mo.) per ml, 4 mg of imipenem (Merck Research Laboratories, Rahway, N.J.) per ml, or 2 mg of cefotaxime (Sigma) per ml was placed in the center of the library culture spread on the LB agar plate; and the bacteria were allowed to grow overnight at 37°C. Colonies were picked from the zone of clearing around the disk and replated on LB-CMP plates. The blaSME gene was amplified from each colony by PCR for DNA sequencing. Taq polymerase (Promega, Madison, Wis.) was used for the colony PCR. An alternative approach was taken for selection on cefotaxime with the SME Cys69-Cys238X double library because no functional clones were obtained by the disk diffusion method. In this approach, cefotaxime was directly added to the LB-CMP plates. Bacterial growth occurred on the plate with CMP concentrations up to 0.1 μg/ml; and colonies of various sizes were picked, and the blaSME gene was sequenced to identify substitutions that may have occurred at positions Cys69 and Cys238.
The MICs of ampicillin, imipenem, and cefotaxime were determined by the twofold dilution method. Intermediate imipenem concentrations of 25, 50, 75, and 100 μg/ml were included to increase the sensitivity of the assay. Similarly, ampicillin MICs were determined at 100-μg/ml intervals to provide increased sensitivity.
Sequencing.
The sequences of the blaSME genes from several selected clones were determined by automated DNA sequencing with a model 3100 automated sequencer (Applied Biosystems Instruments, Foster City, Calif.). All protocols were followed according to the specifications of the manufacturer.
RESULTS AND DISCUSSION
A codon randomization and functional selection approach was used in this study to determine the role of the unique disulfide bond found between positions Cys69 and Cys238 in the class A carbapenemase SME-1. Random libraries for positions Cys69, Cys238, and Cys69 together with Cys238 were created by PCR-based randomization (19). Prior to the selection of functional mutants, the clones from the naïve SME Cys69X, Cys238X, and Cys69X-Cys238X libraries were sequenced to ensure that there was no obvious bias for any codon in the starting libraries. As seen in Table 2, a diverse set of sequences is present in the naïve libraries. Because the naïve libraries are very diverse, any bias in the sequences of the clones obtained from antibiotic selection must be a result of the selection process.
TABLE 2.
Sequencing results for naïve SME Cys69X, SME Cys238X, and SME Cys69X-Cys238X libraries
| SME library | Amino acid (codon)
|
|
|---|---|---|
| Cys69 | Cys238 | |
| Cys69X | Cys69 (TGC)a | |
| Ala4b (GCG) | ||
| Leu (CTC) | ||
| Pro (CCG) | ||
| Cys2 (TGC) | ||
| Thr (ACG) | ||
| Arg2 (AGG) | ||
| Arg (CGC) | ||
| Lys (AAG) | ||
| Trp (TGG) | ||
| Tyr (TAC) | ||
| Cys238X | Cys238 (TGT)a | |
| Ala2 (GCC) | ||
| Gly (GGG) | ||
| Ile (ATC) | ||
| Leu (TTC) | ||
| Pro (CCG) | ||
| Val (GTG) | ||
| Ser (TCG) | ||
| Glu2 (GAG) | ||
| Tyr (TAC) | ||
| Stop (TAG) | ||
| Cys69X-Cys238Xc | Cys69 (TGC)a | Cys238 (TGT)a |
| Trp (TGG) | Arg (AGG) | |
| Asp (GAC) | Lys (AAG) | |
| His (CAC) | Lys (AAG) | |
| Ala (GCG) | Stop (TAG) | |
| Cys (TGC) | Trp (TGG) | |
| Pro (CCC) | Asn (AAC) | |
| Val (CTC) | Asn (AAC) | |
| Cys (TGC) | Glu (GAG) | |
| Gly (GGC) | Lys (AAG) | |
| His (CAC) | Ser (AGC) | |
| Gly (GGC) | Ala (GCC) | |
| Asp (GAC) | Ala (GCC) | |
| Leu (CTC) | Leu (TTG) | |
The amino acid and codon found in the wild-type SME-1 gene.
The superscript numbers represent the numbers of clones sequenced.
Cys69X and Cys238X were connected by —CH2-S—S-CH2—.
The SME-1 β-lactamase efficiently hydrolyzes many β-lactam antibiotics including ampicillin and imipenem (12). The MICs of ampicillin and imipenem for E. coli cells harboring the blaSME gene on the pTP123 vector were ≥2,000 and 32 μg/ml, respectively. In contrast, the SME-1 enzyme confers only a low level of resistance to cefotaxime, with an MIC of 0.2 μg/ml (Table 3). The concentration of antibiotic used for each selection was based on the MICs for E. coli cells harboring the SME-1 β-lactamase gene listed in Table 3.
TABLE 3.
MICs for E. coli XL-1 Blue cells harboring SME-1, SME variants, TEM-1, and pTP123 vector
| β-Lactamase | MIC (μg/ml)
|
||
|---|---|---|---|
| Ampicillin | Imipenem | Cefotaxime | |
| SME-1 | 2,000 | 50 | 0.2 |
| SME-1 (Cys238-TGC) | 2,000 | 50 | 0.2 |
| TEM-1 | 2,000 | 1 | 0.05 |
| pTP123 | 2 | 0.0625 | 0.05 |
Selection of the SME Cys69X library on agar plates containing ampicillin or imipenem resulted in the possession of the wild-type TGC codon by all clones selected (Table 4). Similarly, when the SME Cys69X library was selected on agar plates containing cefotaxime, all clones picked for sequencing contained the cysteine TGC codon (Table 4). Initially, it was predicted that disruption of the disulfide bond may allow larger substrates, such as cefotaxime, to fit into the active site of SME-1, but this does not seem to be the case, as alternate substitutions that would disrupt the disulfide bond were not found in any of the functional clones.
TABLE 4.
Sequencing results from selection of SME Cys69X, SME Cys238X, and SME Cys69X-Cys238X libraries on plates with ampicillin, imipenem, and cefotaxime
| SME library and antibiotic used for selection | Amino acid (codon)
|
|
|---|---|---|
| Cys69 | Cys238 | |
| Cys69X | Cys69 (TGC)a | |
| Ampicillin | Cys10b (TGC) | |
| Imipenem | Cys20 (TGC) | |
| Cefotaxime | Cys10 (TGC) | |
| Cys238X | Cys238 (TGT)a | |
| Ampicillin | Cys15 (TGC) | |
| Imipenem | Cys12 (TGC) | |
| Cefotaxime | Cys12 (TGC) | |
| Cys69X-Cys238Xc | ||
| Ampicillin | Cys16 (TGC) | Cys16 (TGC) |
| Imipenem | Cys15 (TGC) | Cys15 (TGC) |
| Cefotaxime | Cys28 (TGC) | Cys28 (TGC) |
The amino acid and codon found in the wild-type SME-1 gene.
The superscript numbers represent the numbers of clones sequenced.
Cys69X and Cys238X were connected by —CH2-S—S-CH2—.
Selection of the SME Cys238X library was also carried out on plates with ampicillin, imipenem, and cefotaxime. The codon sequence for the SME-1 β-lactamase gene at position Cys238 is TGT. On the basis of the sequences of the primers designed for randomization (see Materials and Methods), only the non-wild-type TGC codon would appear if cysteine were required for function at this position. Similar to position Cys69, the cysteine residue is critical for hydrolysis of all three antibiotics, in that only the TGC codon was identified at position 238 among clones selected on agar plates containing any of these substrates (Table 4).
The fact that a wide variety of codons were identified among clones from the naïve libraries, while only the TGC cysteine codon was identified among clones selected for antibiotic resistance, strongly suggests that the intact disulfide bond between positions Cys69 and Cys238 is essential for function. Interestingly, the requirement for the disulfide bond is not unique to imipenem hydrolysis but, rather, is required for all antibiotics tested. It has been shown that replacement of the cysteine at position 69 results in an enzyme unable to confer resistance to several antibiotics (15). The effect of substitutions at position 238 or whether other pairs of substitutions may compensate for the loss of the disulfide bond is not known, however. It has been shown in the Arc repressor that salt bridges within the enzyme can be replaced by hydrophobic interactions to form a more stable enzyme, provided that the residues participating in the salt bridge are substituted simultaneously (19). Although the randomization experiments with single residues suggest that the disulfide bond is critical for function, it is possible that other amino acid combinations resulting in different types of interactions, such as hydrophobic interactions or the formation of a salt bridge, may compensate for the loss of the disulfide bridge by maintaining the overall structural orientations of amino acids around the active site and thereby confer resistance to β-lactam antibiotics. To test this possibility, the amino acids at positions Cys69 and Cys238 were randomized simultaneously, and functional clones were selected on plates with ampicillin, imipenem, and cefotaxime, as described above.
As was discovered for the libraries with single codons, all of the functional clones selected from the Cys69X-Cys238X library possessed cysteines at positions Cys69 and Cys238. Again, the wild-type codon at position Cys238 was replaced with the non-wild-type TGC codon. Therefore, it is critical for the SME-1 β-lactamase to possess a disulfide bond just outside the active-site pocket in order to function as a penicillinase, carbapenemase, and cephalosporinase.
From the collective data for the single and combinatorial libraries, it is evident that there is an absolute requirement for the presence of a disulfide bond between positions Cys69 and Cys238 in the SME-1 β-lactamase. Structural studies have shown that the nucleophilic Ser70 base and the Glu166 general base are in close proximity, resulting in the absence of a structural water molecule that bridges these residues in most other class A β-lactamases (15). Alteration of this proximity by removal of the disulfide bond may distance Ser70 and Glu166 too far from each other and prevent the enzyme from functioning. Alternatively, the overall stability of the enzyme may be reduced through the loss of the disulfide bond between Cys69 and Cys238.
Ampicillin and imipenem are efficiently hydrolyzed by SME-1 and the other carbapenem-hydrolyzing class A β-lactamases, making them effective penicillinases and carbapenemases (12). The carbapenemase activities of these enzymes distinguish them from other class A β-lactamases. A unique feature of all these enzymes is the disulfide bond between residues Cys69 and Cys238. Interestingly, this structural characteristic does not uniquely influence the carbapenemase activity of the SME-1 β-lactamase but is required in general for the enzyme to function as a β-lactamase. Three other class A β-lactamases, GES-1, GES-2, and IBC-1, also possess cysteines at positions 69 and 238 but do not have significant carbapenemase activities (9). These enzymes are characterized by their activities toward extended-spectrum cephalosporins (9). It is not yet known if the disulfide bond is required for the catalytic activities of these enzymes. We have demonstrated that the disulfide bond is necessary for all β-lactamase activities for SME-1, not simply for carbapenemase activity. Taken together, the results indicate that the disulfide bond does not uniquely provide for the hydrolysis of carbapenem antibiotics. Rather, the results suggest that a subtle reorganization of the SME-1 active-site pocket is responsible for the carbapenemase activity of the enzyme. It is known that very small movements of the active-site groups can drastically alter the catalytic properties of an enzyme (5). In addition, other residues may contribute more specifically to carbapenem hydrolysis. For example, residue Ser237 contributes specifically to the carbapenemase activity of SME-1, in that replacement of serine at position 237 with an alanine results in an enzyme that is able to hydrolyze penicillin, but not imipenem (16).
Infectious bacteria harboring the SME-1 β-lactamase or any of the class A carbapenemases are a threat to the efficacies of β-lactam antibiotics, which are critical to the treatment of infections. A molecular understanding of the enzymes involved in such resistance will facilitate the design of new antibiotics and inhibitors.
Acknowledgments
We thank Patrice Nordmann for providing us with Serratia marcescens S6, which contains the SME-1 β-lactamase gene, and Merck Research Laboratories for providing imipenem. We also thank Hariharan Jayaram for assistance with Fig. 1 and Matthew McKevitt for assistance with Fig. 2D.
T.G.P. is supported by National Institutes of Health grant AI32956.
REFERENCES
- 1.Bradley, J. S., J. Garau, H. Lode, K. V. Rolston, S. E. Wilson, and J. P. Quinn. 1999. Carbapenems in clinical practice: a guide to their use in serious infection. Int. J. Antimicrob. Agents 11:93-100. [DOI] [PubMed] [Google Scholar]
- 2.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL-1 Blue: a high efficiency plasmid transforming recA Eschericia coli strain with β-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 3.Hellinger, W. C., and N. S. Brewer. 1999. Carbapenems and monobactams: imipenem, meropenem, and aztreonam. Mayo Clin. Proc. 74:420-434. [DOI] [PubMed] [Google Scholar]
- 4.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 5.Koshland, D. E., Jr. 1998. Conformational changes: how small is big enough? Nat. Med. 4:1112-1114. [DOI] [PubMed] [Google Scholar]
- 6.Livermore, D. M., and N. Woodford. 2000. Carbepenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 7.Naas, T., and P. Nordmann. 1994. Analysis of a carbapenem-hydrolyzing class A beta-lactamase from Enterobacter cloacae and of its LysR-type regulatory protein. Proc. Natl. Acad. Sci. USA 91:7693-7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naas, T., L. Vandel, W. Sougakoff, D. M. Livermore, and P. Nordmann. 1994. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, SME-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 38:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 10.Petrosino, J., G. W. Rudgers, H. Gilbert, and T. Palzkill. 1999. Contribution of aspartate 49 and phenylalanine 142 residues of a tight binding inhibitory protein of β-lactamases. J. Biol. Chem. 274:2394-2400. [DOI] [PubMed] [Google Scholar]
- 11.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing beta-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 12.Raquet, X., J. Lamotte-Brasseur, F. Bouillenne, and J.-M. Frere. 1997. A disulfide bridge near the active site of carbapenem hydrolyzing class A β-lactamases might explain their unusual substrate profile. Proteins 27:47-58. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen, B. A., K. Bush, D. Keeney, Y. Yang, R. Hare, C. O'Gara, and A. A. Medeiros. 1996. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 40:2080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz, S. C., G. Dalbadie-McFarland, J. J. Neitzel, and J. H. Richards. 1987. Stability of wild-type and mutant RTEM-1 beta-lactamases: effect of the disulfide bond. Proteins 2:290-297. [DOI] [PubMed] [Google Scholar]
- 15.Sougakoff, W., G. L'Hermite, L. Pernot, T. Naas, V. Guillet, P. Nordmann, V. Jarlier, and J. Delettre. 2002. Structure of the imipenem-hydrolyzing class A beta-lactamase SME-1 from Serratia marcescens. Acta Crystallogr. Sect. D Biol. Crystallogr. 58(Pt 2):267-274. [DOI] [PubMed] [Google Scholar]
- 16.Sougakoff, W., T. Naas, P. Nordmann, E. Collatz, and V. Jarlier. 1999. Role of Ser-237 in the substrate specificity of the carbapenem-hydrolyzing class A beta-lactamase SME-1. Biochim. Biophys. Acta 1433:153-158. [DOI] [PubMed] [Google Scholar]
- 17.Swaren, P., L. Maveyraud, X. Raquet, S. Cabantous, C. Duez, J.-D. Pedelacq, S. Mariotte-Boyer, L. Mourey, R. Labia, M.-H. Nicolas-Chanoine, P. Nordmann, J.-M. Frere, and J.-P. Samama. 1998. X-ray analysis of the NMC-A β-lactamase at 1.64-Å resolution, a class A carbapenemase with broad spectrum specificity. J. Biol. Chem. 273:26714-26721. [DOI] [PubMed] [Google Scholar]
- 18.Vanhove, M., G. Guillaume, P. Ledent, J. H. Richards, R. H. Pain, and J. M. Frere. 1997. Kinetic and thermodynamic consequences of the removal of the Cys-77—Cys-123 disulphide bond for the folding of TEM-1 beta-lactamase. Biochem. J. 321(Pt 2):413-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldburger, C. D., J. F. Schildbach, and R. T. Sauer. 1995. Are buried salt bridges important for protein stability and conformational specificity? Nat. Struct. Biol. 2:122-128. [DOI] [PubMed] [Google Scholar]
- 20.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]