Abstract
We have investigated the interaction of quinolones with DNA by a number of methods to establish whether a particular binding mode correlates with quinolone potency. The specificities of the quinolone-mediated DNA cleavage reaction of DNA gyrase were compared for a number of quinolones. Two patterns that depended on the potency of the quinolone were identified. Binding to plasmid DNA was examined by measuring the unwinding of pBR322 by quinolones; no correlation with quinolone potency was observed. Quinolone binding to short DNA oligonucleotides was measured by surface plasmon resonance. The quinolones bound to both single- and double-stranded oligonucleotides in an Mg2+-dependent manner. Quinolones bound to single-stranded DNA with a higher affinity, and the binding exhibited sequence dependence; binding to double-stranded DNA was sequence independent. The variations in binding in the presence of metal ions showed that Mg2+ promoted tighter, more specific binding to single-stranded DNA than softer metal ions (Mn2+ and Cd2+). Single-stranded DNA binding by quinolones correlated with the in vitro quinolone potency, indicating that this mode of interaction may reflect the interaction of the quinolone with DNA in the context of the gyrase-DNA complex.
Quinolones are synthetic antibacterial compounds based on a four-quinolone skeleton (4, 29). Fluoroquinolones have been very successful clinically and are used to treat bacterial infections, including respiratory and urinary tract infections. Quinolones target bacterial type II topoisomerases, generally DNA gyrase in gram-negative bacteria and DNA topoisomerase IV in gram-positive bacteria (9). Type II topoisomerases are enzymes that resolve topological problems in DNA (38). They cleave double-stranded DNA in both strands and then transport another segment of double-stranded DNA through the cleaved DNA segment before religating the DNA. Quinolones inhibit the strand-passage activity of topoisomerases by stabilizing the formation of a cleaved-DNA complex (13, 34).
Quinolone resistance mutations generally cluster around the active-site region of the DNA gyrase A protein, between residues 67 and 106, known as the quinolone resistance-determining region (40). These resistance residues lie within the proposed DNA binding interface in the structure of the DNA cleavage-religation domain of DNA gyrase (23); quinolones are thought to interact with both gyrase and DNA. These quinolone resistance-determining region residues are likely to form a binding pocket for quinolones, which may also include quinolone resistance residues from GyrB (17). Consistent with this, resistance mutations of gyrase show a decrease in drug binding affinity (17, 39).
The interaction between quinolones and DNA is also important within the ternary complex, since DNA is necessary to form a stable gyrase-quinolone complex (7). Quinolones have been shown to bind to single-stranded DNA with a preference for poly(dG) [poly(dG) > poly(dA) > poly(dT) > poly(dC)], and they were also found to bind weakly to double-stranded DNA in a sequence-independent manner (32). The preference for G residues can be explained by a model in which four quinolone molecules bind to a single-stranded region within the gyrase-DNA complex by base pairing (33). Other work also suggests a preferential interaction of quinolones with G residues (6, 15). DNA modeled into the structure of the cleavage-religation domain of DNA gyrase suggests that the DNA is distorted in the active site (23), which may provide a single-stranded region for drug binding. The quinolone-DNA interaction is clearly important for the stability of the ternary complex, but the Shen model is unlikely to be entirely correct since binding studies suggest that two quinolone molecules bind per gyrase-DNA complex (7). This conclusion is supported by the positions of resistance mutations, which appear to form two quinolone-binding pockets, one on either side of the GyrA dimer interface (23). Also, the Shen model does not take into account the fact that Mg2+ is required for the quinolone-DNA interaction (26). Mg2+ has been shown by nuclear magnetic resonance to bind to quinolones via the C-3 and C-4 carboxylate and carbonyl groups (Fig. 1 and Table 1), probably in a 1:1 ratio (20). Quinolones have been proposed to bind to the DNA phophodiester backbone via an Mg2+ bridge coordinated by the C-3 and C-4 groups (27), an interaction that would be stabilized by stacking between the bases and the quinolone.
FIG. 1.
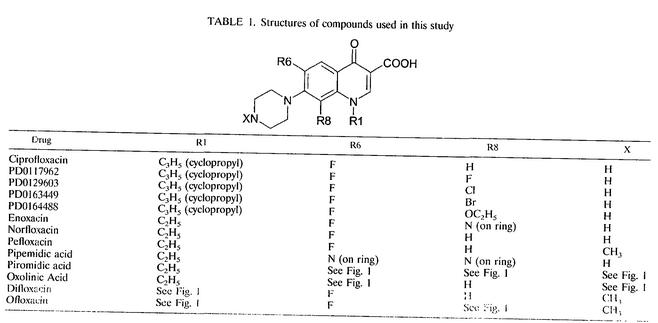
Structures of compounds used in this study.
TABLE 1.
Structures of compounds used in this study
Insight into the mode of interaction of quinolones has also been obtained by examining the sequence of quinolone-induced gyrase cleavage sites. The sites of cleavage of pBR322 DNA by DNA gyrase in the presence of oxolinic acid were used to define a 20-bp consensus sequence (22). An important determinant of the consensus sequence is the 4-base region downstream of the gyrase cleavage site (5′-↓GRYC, where R and Y indicate purine and pyrimidine, respectively, and the arrow indicates the site of cleavage). If the G or C is mutated, DNA cleavage is significantly reduced (11); the major cleavage site of pBR322 is at 990 bp (12).
A model has been described for quinobenzoxazines and Mg2+ binding to the yeast topoisomerase II-DNA complex (10, 19). These compounds appear to require Mg2+ for DNA binding, since they contain the carbonyl and carboxylate groups, but they form a much more stable intercalative complex with DNA than the quinolones do (28). This model proposes that two pairs of quinobenzoxazine molecules bind to DNA via Mg2+ bridges (10). In each pair, one drug molecule intercalates between bases adjacent to the cleavage site and the other binds externally, where it can interact with the protein. On the basis of the observation that the quinolone norfloxacin can form mixed-structure dimers with quinobenzoxazines on DNA, it has been suggested that this mode of binding may be applicable to quinolones (10).
In other work a number of quinolones were compared by using quantitative structure-activity relationships (21). Quinolones were suggested to intercalate in double-stranded DNA with the R1 group (Fig. 1 and Table 1) in the major groove and the R7 group in the minor groove, allowing GyrA to interact with the R7 group, possibly by forming hydrogen bonds. Mg2+ binding to the complex was analyzed by molecular dynamics simulation (21). A hydrated Mg2+ ion bound to the complex; the binding was coordinated by the quinolone carboxylate and carbonyl groups, two oxygens from each of two phosphates, and the N-7 and O-6 of a purine and a guanine, respectively.
The diversity of the models discussed above shows that, without a high-resolution structure of the gyrase-quinolone-DNA complex, understanding the quinolone binding interaction is challenging. In order to progress our understanding of the gyrase-quinolone-DNA interaction, we have examined the interaction between quinolones and DNA to see if a particular mode of binding correlates with the in vitro quinolone potency against DNA gyrase. We find that quinolones bind to single-stranded DNA in a sequence-dependent manner that is mediated by Mg2+. This interaction correlates with quinolone potency, indicating that similar binding to the gyrase-DNA complex occurs. Binding to double-stranded DNA was not dependent on the sequence used and did not correlate with the quinolone potency.
MATERIALS AND METHODS
Quinolones, enzymes, and DNA.
All quinolones except enoxacin were dissolved in a 1:5 molar ratio of drug to KOH; enoxacin required equimolar NaOH for dissolution. The PD compounds, ciprofloxacin, norfloxacin, enoxacin, ofloxacin, and difloxacin were gifts from Parke-Davis (PD; Ann Arbor, Mich.), Bayer, Merck, Dainippon, Hoechst, and Abbott Laboratories, respectively. Oxolinic acid, pipemidic acid, and piromidic acid were purchased from Sigma.
GyrB was purified to homogeneity as described previously (24). GyrA and relaxed pBR322 were gifts from A. J. Howells (John Innes Enterprises Ltd.). The oligonucleotides (MWG Biotech) used in this study were based on the preferred quinolone-mediated gyrase DNA-cleavage site of pBR322 (referred to as the 990 site) and were as follows: single-stranded 6-mers (5′→3′) TGG CCT, TAA CCT, TGG TTT, and TAA TTT; single-stranded 24-mer CGA GGC TGG ATG GCC TTC CCC ATT; and hairpin oligonucleotides TGG CCT CCC CAG GCC A and TAA CCT CCC CAG GTT A.
Enzyme assays.
Supercoiling assays for determination of the in vitro potencies of the quinolones were performed as described previously (17, 30). DNA cleavage specificity assays contained 10 μg of relaxed pBR322 DNA per ml, 9 μg of tRNA per ml, 35 mM Tris · HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 1.8 mM spermidine, 1 mM ATP, 0.1 mg of bovine serum albumin per ml, 6.5% (wt/vol) glycerol, and the quinolone at the desired concentration. Samples were preincubated for 5 min at 25°C before gyrase was added at a final concentration of 70 to 100 nM (GyrA monomer). Following a 30-min incubation of the cleavage reaction at 25°C, 0.5 U of EcoRI was added and the incubation was continued at 37°C for 15 min. Sodium dodecyl sulfate and proteinase K were added to final concentrations of 0.2% (wt/vol) and 0.1 mg/ml, respectively, and the incubation was continued for 30 min at 37°C. Samples were prepared for electrophoresis by the addition of 1 volume of 40% sucrose-0.1 M Tris · HCl (pH 7.5)-0.1 M EDTA-bromophenol blue and 2 volumes of chloroform-isoamyl alcohol (24:1). The products were separated on a 1.35% agarose gel.
DNA unwinding assays.
Samples contained 1 U of wheat germ topoisomerase I (Promega), 10 μg of negatively supercoiled pBR322 DNA per ml, 35 mM Tris · HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 6.5% (wt/vol) glycerol, 4 mM dithiothreitol, and the quinolone at the desired concentration in a final volume of 30 μl. The samples were preincubated for 5 min at 37°C before the addition of topoisomerase I to start DNA relaxation. The samples were further incubated at 37°C for 30 min. The reactions were terminated, and the samples were prepared for agarose gel analysis by the addition of 0.5 volume of chloroform-isoamyl alcohol (24:1) and 0.2 volume of 0.2% (wt/vol) sodium dodecyl sulfate-80 mM Tris · acetate-2 mM EDTA-20% (wt/vol) glycerol-bromophenol blue. The products were separated on 1.1% agarose gels containing 0, 0.5, or 1 μg of chloroquine per ml.
SPR.
Surface plasmon resonance (SPR) was performed (at 25°C) by using a Biacore 2000 or a Biacore X instrument (Biacore AB, Uppsala, Sweden) with streptavidin-coated sensor chips. 5′ Biotinylated oligonucleotides were attached in HBS-EP buffer (Biacore) to flow cell 2 by flowing 100 μl of 200 nM oligonucleotide in 300 mM NaCl across the chip at 5 μl/min. Flow cells 1 and 2 were then blocked with 100 μl of 1 mg of biotin per ml. A buffer of 35 mM Tris · HCl (pH 7.5) and 1 mM MgCl2 was flowed across flow cell 1 and then flow cell 2 at 30 μl/min, unless indicated otherwise. The quinolones in the same buffer were injected across flow cell 1 and then flow cell 2; the blank (flow cell 1) signal was subtracted, and the change in amplitude was measured. In order to determine apparent dissociation constants (Kds), the amplitude was measured at a range of drug concentrations, and the data were fitted to a single-site binding equation {[bound drug] = (capacity × [free drug])/(Kd + [free drug])}. To minimize mass transport effects, a fast flow rate was used; experiments carried out at slower flow rates gave comparable results. The resultant binding isotherms (e.g., see Fig. 3) suggested that mass transport correction was unnecessary.
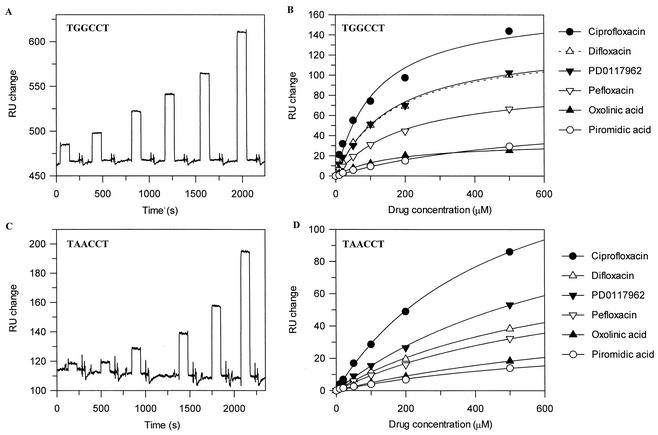
FIG. 3.
SPR sensograms and drug-DNA binding isotherms. (A and C) Examples of the SPR sensograms produced for ciprofloxacin (10, 20, 50, 100, 200, and 500 μM) binding to TGGCCT and TAACCT. Experiments were performed with 1 mM MgCl2. The increase in RUs from the baseline was measured and used to calculate Kdapps. (B and D) Examples of drug-DNA binding isotherms for TGGCCT and TAACCT. The amplitudes from the SPR sensogram experiments were fitted to a single-site binding equation. The stoichiometries (Table 3) were calculated from the ratio of the increase in the numbers of RUs for quinolone binding to the mass (in RUs) of DNA attached to the chip surface.
RESULTS
Quinolone potency in vitro.
To examine the interaction of quinolones with DNA, we selected a range of compounds (Fig. 1 and Table 1), some of which had been used in a previous study (2). The potencies of these compounds were determined in vitro by measuring the 50% inhibitory concentrations (IC50s) for the inhibition of gyrase supercoiling activity (Table 2). The quinolone IC50s for supercoiling activity varied significantly among the quinolones tested. Ciprofloxacin and its derivatives had the lowest IC50s (i.e., were the most potent), and the nonfluorinated compounds oxolinic acid, pipemidic acid, and piromidic acid had the highest (i.e., were the least potent).
TABLE 2.
Properties of drugs
| Drug | IC50 (μM)a | Cleavage specificity | DNA unwinding (ΔLk°/mM)b |
|---|---|---|---|
| PD0117962 | 0.7 | I | 12 |
| PD0129603 | 0.8 | I | 17 |
| PD0163449 | 0.9 | I | 5.4 |
| Ciprofloxacin | 1.1 | I | 33 |
| PD0164488 | 1.7 | I | 4.3 |
| Difloxacin | 2.5 | I | NDc |
| Ofloxacin | 3.5 | I | 9.5 |
| Enoxacin | 4.4 | I | 29 |
| Norfloxacin | 4.8 | I | 13 |
| Pefloxacin | 5.1 | I | 11 |
| Oxolinic acid | 15.1 | II | 6.0 |
| Piromidic acid | 56 | II | ND |
| Pipemidic acid | 70 | II | ND |
Data for all drugs except pefloxacin, difloxacin, pipemidic acid, piromidic acid, and oxolinic acid are from Barnard and Maxwell (2).
ΔLK°/mM, average change in mean linking difference per millimolar drug.
ND, not determined.
Quinolone-dependent DNA cleavage specificity.
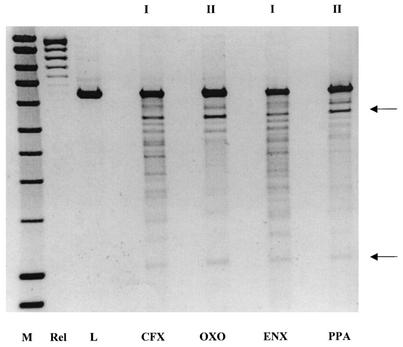
The specificity of DNA cleavage by gyrase in the presence of various different quinolones (Fig. 1 and Table 1) was examined to see if there was a correlation between cleavage specificity and quinolone potency. A relaxed DNA substrate (plasmid pBR322) was used with a high gyrase concentration to produce cleavage at several sites on the DNA. The DNA was then linearized with EcoRI to produce a cleavage pattern that was similar for all the quinolones tested (Fig. 2). All the quinolones showed a preference for cleavage at the 990 site (12). However, three of the quinolones, oxolinic acid, pipemidic acid, and piromidic acid, produced cleavage patterns that were distinctly different. These cleavage patterns are termed set II and were distinct from the other patterns, which are set I (Table 2). Therefore, the nature of the quinolone alters the cleavage site preference of DNA gyrase, indicating that the quinolone-DNA interaction is important within the ternary complex. This suggests that the DNA cleavage pattern is related to drug potency, since piromidic acid, pipemidic acid, and oxolinic acid, the weakest drugs by IC50 (Table 2), were the only compounds that caused cleavage pattern II. These are also the only drugs that lack a fluorine atom at C-6.
FIG. 2.
Cleavage specificity assay. A 1.35% agarose gel shows the cleavage of relaxed pBR322 DNA by gyrase in the presence of quinolone drugs. Lanes: M, hyperladder DNA marker (Bioline); Rel, relaxed pBR322 DNA substrate alone; L, EcoRI-linearized substrate. The remaining lanes show the cleavage patterns for ciprofloxacin (CFX; 5 μM), oxolinic acid (OXO; 50 μM), enoxacin (ENX; 25 μM), and pipemidic acid (PPA; 150 μM), classified as set I or set II. The arrows show the approximate positions of the products generated by cleavage at the 990 site (12).
DNA unwinding.
DNA relaxation by topoisomerase I has previously been used to investigate the DNA unwinding effects of nalidixic acid and norfloxacin (36, 37). In these experiments, closed-circular DNA was incubated with the quinolones and then treated with topoisomerase I, and the extent of DNA unwinding was determined electrophoretically. The unwinding of supercoiled plasmid DNA by norfloxacin was 17-fold higher than that by nalidixic acid. This compares to ∼60- to 120-fold differences in potency, as determined by MICs and IC50s (16, 41). We have examined the DNA unwinding ability of a larger set of quinolones to see if these results correlate with other binding data and the in vitro potency. Each compound was tested at two or three different concentrations, and agarose gels of the topoisomer products were analyzed for the change in linking number (data not shown). A Gaussian function was used to calculate the mean change in linking number of the DNA for each drug at each concentration (3). In control experiments, drugs were titrated into the reaction in order to confirm that the change in linking number was linear with respect to the amount of drug added. The data show that the level of DNA unwinding varies significantly for the quinolones (Table 2). However, there is not an obvious correlation between quinolone potency (as measured by IC50) and DNA unwinding. For example, PD0117962 is more potent than ciprofloxacin (∼1.6-fold) but is a less effective unwinding agent (∼40% compared with ciprofloxacin); oxolinic acid is ∼15-fold less potent than ciprofloxacin but is ∼5-fold less effective as an unwinding agent (Table 2).
Single-stranded DNA binding specificity.
In order to assess the binding of quinolones to single-stranded DNA, we synthesized a series of 6-mer oligonucleotides, based on the preferred gyrase cleavage site of pBR322, at nucleotide 990 (5′→3′): TGGCCT, TAACCT, TGGTTT , and TAATTT. Binding was measured by SPR. In preliminary experiments with norfloxacin and PD0163449 (Table 1), we found that TGGCCT, the canonical 990 site sequence, bound most tightly and that TAACCT showed the weakest binding (data not shown). In subsequent experiments with a range of other drugs (Fig. 1 and Table 1), only these two oligonucleotides were used. (It was assumed that the oligonucleotides were single stranded on the chip surface since the melting temperature for each is ∼20°C, and experiments were performed at 25°C. Also, there was no loss of mass from the surface of the chip when the temperature was increased to 30°C.)
The SPR sensograms did not produce classic on-off binding curves but showed a rapid increase in response units (RUs) when a quinolone was present, and this returned to the baseline when the injection was stopped (Fig. 3). The results for quinolone titrations show that the interactions are noncompetitive and saturable, indicating that this is a genuine interaction with a fast on-and-off rate. Other interactions with a rapid on-and-off rate have been shown to produce similar results by SPR (5, 25); by using the steady-state level of the RU change, it is possible to derive binding data from experiments carried out with a range of drug concentrations.
DNA binding was measured with a range of drug concentrations (10 to 500 μM) for both oligonucleotides; the data for ciprofloxacin are shown in Fig. 3. The data were fitted to a single-site binding equation, since although there may be more than one drug binding site, the sites are likely to be independent (Fig. 3). Kdapps were calculated from the fitted data (Table 3). It is apparent from the shape of the curves in Fig. 3B and D that binding to TAACCT is weaker than that to TGGCCT; the Kdapps for most compounds were more than fourfold higher for the substituted sequence (Table 3).
TABLE 3.
Drug binding to 6-mer single-stranded DNA oligonucleotides
| Drug | Apparent Kd (μM)a
|
Calculated stoichiometry
|
||
|---|---|---|---|---|
| TGGCCT | TAACCT | TGGCCT | TAACCT | |
| PD0117962 | 163.7 ± 21.3 | 789.5 ± 102.7 | 1.8 ± 0.10 | 1.8 ± 0.16 |
| PD0129603 | 87.6 ± 18.6 | 684.9 ± 92.0 | 1.8 ± 0.14 | 2.2 ± 0.19 |
| PD0163449 | 88.6 ± 22.6 | 837.5 ± 125.3 | 1.8 ± 0.16 | 2.4 ± 0.25 |
| Ciprofloxacin | 115.4 ± 24.0 | 480.1 ± 23.0 | 2.4 ± 0.19 | 2.3 ± 0.07 |
| PD0114488 | 48.4 ± 15.3 | 750.4 ± 116.6 | 1.6 ± 0.15 | 2.0 ± 0.21 |
| Difloxacin | 164.7 ± 12.5 | 693.0 ± 98.9 | 1.5 ± 0.05 | 1.0 ± 0.10 |
| Ofloxacin | 234.2 ± 55.9 | 550.2 ± 51.4 | 1.6 ± 0.19 | 1.0 ± 0.06 |
| Enoxacin | 240.4 ± 36.9 | 1180.1 ± 203.2 | 2.2 ± 0.16 | 2.9 ± 0.38 |
| Norfloxacin | 336.4 ± 87.1 | 1599.0 ± 352.1 | 3.5 ± 0.48 | 5.9 ± 1.04 |
| Pefloxacin | 196.0 ± 15.4 | 752.3 ± 136.0 | 1.3 ± 0.04 | 1.1 ± 0.14 |
| Oxolinic acid | 164.4 ± 21.2 | 1057.0 ± 256.5 | 0.6 ± 0.03 | 1.0 ± 0.18 |
| Piromidic acid | 604.3 ± 112.3 | 778.2 ± 242.6 | 1.0 ± 0.12 | 0.6 ± 0.12 |
Kdapp were determined from quinolone-binding titrations by SPR (Fig. 3).
The fitted data also gave a value (in RUs) for the quinolone-DNA binding capacity (the maximum amount of quinolone bound to the DNA). This RU increase is proportional to the mass bound to the DNA. Therefore, if the mass of DNA bound to the chip is also known in terms of the numbers of RUs, then the mass of a quinolone that binds to the DNA can be calculated. For the compounds with the set II cleavage pattern, this corresponds to a stoichiometry of ∼1 per oligonucleotide, whereas for most of the compounds with the set I cleavage pattern, this is ∼2 per oligonucleotide (Table 3). The exception is norfloxacin, which has the highest binding capacity (a stoichiometry of 3 to 4 to TGGCCT), suggesting that norfloxacin may exhibit a different mode of binding and may be less sequence selective.
Double-stranded DNA binding.
The binding of quinolones to double-stranded DNA was examined by SPR. A self-complementary hairpin oligonucleotide was constructed: TGGCCTCCCCAGGCCA (5′→3′). This oligonucleotide formed a hairpin with a tetracytosine loop; the double-stranded region is the same as the 990 site of pBR322. Such hairpin oligonucleotides have been used previously in SPR experiments (1, 35); we found that there was no increase in the numbers of RUs when a complementary oligonucleotide was flowed across the chip, implying that the hairpin structure was maintained on the chip surface. Cytosines were used in the hairpin loop since quinolones show the weakest affinity for poly(dC) (32). Binding was compared to a second hairpin, TAACCTCCCCAGGTTA (5′→3′), containing the sequence of the single-stranded oligonucleotide that had previously shown the weakest quinolone binding affinity. The Kdapps for most of the quinolones are similar for both hairpin oligonucleotides (Table 4), indicating that the interaction with double-stranded DNA is less sequence dependent. Furthermore, there is less variation between the Kdapps for different quinolones than those determined with single-stranded DNA (Table 3), suggesting that quinolone potency is not correlated with binding to double-stranded DNA. In general, the Kdapps for binding to the hairpins (Table 4) are increased compared to those for binding to the single-stranded oligonucleotides (Table 3), suggesting that the quinolones, particularly those with lower IC50s, bind preferentially to single-stranded DNA.
TABLE 4.
Drug binding affinity to double-stranded DNA hairpin oligonucleotides
| Drug | Apparent Kd (μM)
|
|
|---|---|---|
| 5′-TGGCCTCC 3′-ACCGGACC | 5′-TAACCTCC 3′-ATTGGACC | |
| PD0117962 | 329.5 ± 38.7 | 369.3 ± 55.0 |
| PD0129603 | 342.3 ± 47.2 | 336.4 ± 40.3 |
| PD0163449 | 427.9 ± 73.1 | 438.3 ± 61.6 |
| Ciprofloxacin | 320.2 ± 64.6 | 348.0 ± 68.0 |
| PD0164488 | 287.0 ± 43.0 | 362.3 ± 58.0 |
| Difloxacin | 346.4 ± 34.2 | 534.6 ± 58.6 |
| Ofloxacin | 362.0 ± 49.1 | 465.4 ± 30.2 |
| Enoxacin | 264.0 ± 38.3 | 335.6 ± 51.0 |
| Norfloxacin | 309.0 ± 53.6 | 333.9 ± 45.9 |
| Pefloxacin | 291.8 ± 30.1 | 322.9 ± 67.6 |
| Oxolinic acid | 504.4 ± 145.3 | 335.9 ± 68.4 |
| Piromidic acid | 497.3 ± 106.3 | 681.1 ± 183.2 |
Correlations between in vitro potency and DNA binding.
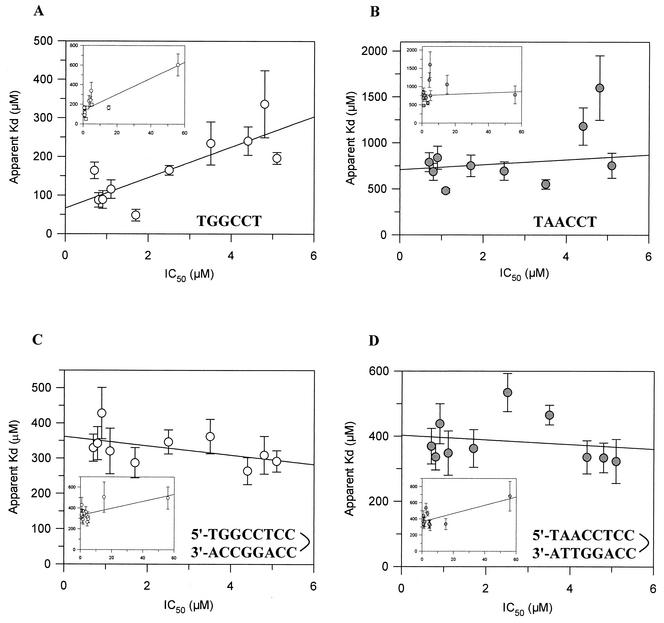
The IC50s for in vitro quinolone potency were plotted against the Kdapps for binding to single- and double-stranded DNA to determine whether there is a systematic correlation. The Kdapps for binding to TGGCCT (the 990 site sequence) and TAACCT were plotted (Fig. 4A and B insets). The data for TGGCCT appear to show a correlation, but this is heavily influenced by the data for the low-potency drugs; little or no correlation exists for TAACCT. Therefore, the data were replotted by excluding those for oxolinic acid and piromidic acid, which had the highest IC50s (Fig. 4A and B). Again, the data for TGGCCT appear to show a correlation: as the potency increases, the DNA binding affinity also increases. Therefore, the mode of binding to single-stranded DNA is apparently related to quinolone potency, suggesting that quinolones bind to single-stranded DNA in the gyrase-DNA complex. The results for norfloxacin are outliers, particularly for binding to TAACCT, which may be because it has a different mode of binding to single-stranded DNA since it has a higher binding stoichiometry (Table 3).
FIG. 4.
Correlations between IC50s and Kdapps for single-stranded and double-stranded DNAs. Correlations were plotted for the single-stranded 990 site sequence of pBR322 (TGGCCT) (A), the single-stranded substituted sequence (TAACCT) (B), the hairpin that formed the 990 site sequence (C), and the hairpin that formed the substituted sequence (D). The plots include all the datum points (insets) or the datum points for just the compounds with cleavage pattern I (main plots). The correlation coefficients for the main plots were as follows: 0.80 (A), 0.54 (B), −0.50 (C), and −0.18 (D).
The IC50s were also plotted against the Kdapp values for binding to the hairpin oligonucleotides, which are equivalent to double-stranded DNA (Fig. 4C and D). However, there is no obvious correlation between quinolone potency and double-stranded DNA affinity. This indicates that the binding to double-stranded DNA is not related to the quinolone potency against DNA gyrase. Therefore, it is unlikely that binding to double-stranded DNA reflects the mode of quinolone interaction with the gyrase-DNA complex.
Metal ion dependence of DNA binding.
To further examine the quinolone-DNA interaction, binding of 100 μM ciprofloxacin was measured by SPR in the presence of different divalent metal ions. Binding was measured by using a single-stranded 24-mer oligonucleotide based on the 990 site of pBR322 to increase the DNA binding signal-to-noise ratio. The experiments were performed by altering the metal ion concentration in both the buffer used to prime the chip surface and the quinolone solution that was injected across it. For ciprofloxacin the optimal metal ion concentration was about 0.5 to 1 mM, and the level of binding was greater for all the softer Lewis acids tested than for Mg2+ (data not shown). This was surprising because quinolones require Mg2+ for DNA binding (26), possibly by forming an Mg2+ bridge to phosphates in the DNA backbone (27). Softer Lewis acids, such as Cd2+, tend to bind to the nitrogens of bases in the minor groove. This suggests that the manner of binding may differ in the presence of softer metal ions. In support of this idea, the shapes of the sensograms produced with the softer metal ions were different from those produced with Mg2+ (data not shown). A significant proportion of the binding has a slower on-and-off rate, but the sensograms still do not resemble a typical binding curve, since there is a large amount of very rapid binding. Therefore, there may be more than one mode of binding of quinolones to DNA in the presence of softer metal ions: rapid, weak binding and slower, tighter binding.
The Mg2+ dependence of double-stranded DNA binding was also measured. Binding of 100 μM ciprofloxacin to the 990 site hairpin sequence was measured in the presence of increasing MgCl2 concentrations (data not shown). Significant binding required the presence of 0.5 to 1 mM MgCl2, similar to binding to a single-stranded oligonucleotide. Therefore, quinolone binding to double-stranded DNA is also Mg2+ dependent.
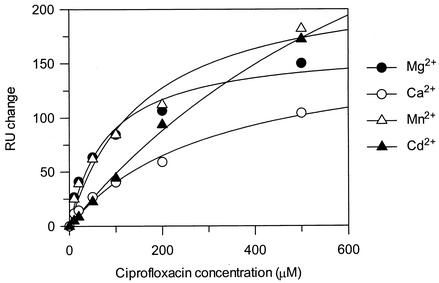
The fact that an increased level of binding to single-stranded DNA is seen for ciprofloxacin with softer metal ions suggests that the stoichiometry of binding is increased. However, it does not give any indication of the affinity of the interaction. Therefore, ciprofloxacin binding to the 990 site 6-mer (TGGCCT) was measured in the presence of Mg2+, Ca2+, Mn2+, and Cd2+. The manner of ciprofloxacin binding in the presence of Ca2+ appears to be similar to that in the presence of Mg2+, with a similar binding stoichiometry but a weaker Kdapp (Table 5; Fig. 5). The binding interaction with the softer metal ions appears to be different, with a higher binding stoichiometry and a weaker Kdapp, indicating that binding is less specific. Mn2+, which is softer than Mg2+ and Ca2+, increases the binding stoichiometry slightly to ∼3, whereas Cd2+, a soft divalent metal ion (14), causes a dramatic increase in the binding stoichiometry. The binding stoichiometry of oxolinic acid to the 6-mer with the various metal ions was similar to that of ciprofloxacin (data not shown).
TABLE 5.
Binding affinities of ciprofloxacin for single-stranded TGGCCT by using different divalent metal ions
| Metal ion | Kdapp (μM) | Calculated stoichiometry |
|---|---|---|
| Mg2+ | 82.5 ± 18.5 | 2.1 ± 0.16 |
| Ca2+ | 309.5 ± 74.6 | 2.1 ± 0.26 |
| Mn2+ | 157.5 ± 44.0 | 2.9 ± 0.34 |
| Cd2+ | 904.0 ± 171.5 | 6.1 ± 0.81 |
FIG. 5.
Ciprofloxacin binding to the 6-mer oligonucleotide (TGGCCT). Experiments were performed with 1 mM MgCl2, CaCl2, MnCl2, or CdCl2; and the data were fitted to a single-site binding equation.
DISCUSSION
The aim of this work was to explore the interaction of quinolones with DNA to attempt to find a mode of binding that correlated with the potencies of the drugs for their target, DNA gyrase. Initially, we compared the effects of a number of quinolones on the DNA cleavage specificity of DNA gyrase. The cleavage specificities were classified into set I or set II. Pattern II compounds included oxolinic acid, pipemidic acid, and piromidic acid, which have relatively high IC50s for the inhibition of gyrase supercoiling. Therefore, it is likely that the cleavage specificity is dependent on the quinolone potency; more potent drugs have less specificity at DNA cleavage sites, while less potent drugs cleave mainly at the 990 site. The obvious difference between the two sets is a fluorine atom at C-6, which is known to increase quinolone potency (8, 18). The 990 site of pBR322 is the major cleavage site for all drugs, suggesting that this particular sequence is less dependent on the quinolone structure than other sites.
The DNA unwinding abilities of the quinolones were measured. We found that there was no obvious correlation between the unwinding ability and the IC50. The amount of unwinding presumably indicates the degree of intercalation. Therefore, it is likely that compounds such as PD0163449 and PD0164488, which have bulky groups at C-8, do not intercalate as well as ciprofloxacin, which has H at C-8. This suggests that this mode of interaction does not reflect the manner of the quinolone-DNA interaction within the gyrase-DNA complex.
The affinities of the various quinolones for single-stranded DNA were measured by SPR. Binding to a 6-mer oligonucleotide, TGGCCT, the sequence of the preferred gyrase cleavage site of pBR322, and to the substituted sequence, TAACCT, was measured. All of the quinolones tested had a much greater affinity for the 990 site sequence, with most having 4- to 10-fold lower Kdapps. This is consistent with previous findings suggesting a preference for a G residue 5′ to the quinolone-induced cleavage site (6, 15).
The affinities of the quinolones for double-stranded DNA were also measured by SPR. Two double-stranded hairpin DNA sequences were constructed, both of which formed 6 bases of double-stranded DNA. TGGCCTCCCCAGGCCA formed the sequence of the 990 site of pBR322, while TAACCTCCCCAGGTTA contained a substitution of the first two guanines. The quinolones bound to either sequence with similar affinities, suggesting that binding to double-stranded DNA is not sequence dependent. The quinolones with lower IC50s, such as ciprofloxacin and the PD compounds, showed higher Kdapps for binding to double-stranded DNA than for the preferred single-stranded oligonucleotide, with at least two- to fivefold stronger binding to the single-stranded oligonucleotide. Other compounds such as piromidic acid showed a similar affinity for single- or double-stranded DNA. This indicates that the more potent compounds have a greater preference for single-stranded DNA than for double-stranded DNA than the less potent compounds. This difference may explain in part the differences in quinolone potencies against DNA gyrase. The different quinolones tested bound to a double-stranded DNA hairpin with similar affinities, suggesting that the mode of interaction with double-stranded DNA is less dependent on quinolone structure and potency than the mode of interaction with single-stranded DNA. Taken together, these data suggest that quinolones bind to a single-stranded DNA within the gyrase-DNA-quinolone complex.
The quinolone affinity for single- or double-stranded DNA and the in vitro quinolone potency were compared to see if there was a correlation. There did not appear to be a relationship between double-stranded DNA affinity and quinolone potency. However, when the single-stranded DNA affinity and the IC50s were compared, there appeared to be a meaningful correlation; as the quinolone IC50 increased, the Kdapp also increased. This suggests that drug binding to single-stranded DNA within the gyrase-DNA complex correlates with the inhibition of gyrase supercoiling activity, i.e., the greater the affinity for single-stranded DNA, the greater the potency.
The interaction with both single- and double-stranded DNA is Mg2+ dependent, with an optimal MgCl2 concentration of 0.5 to 1 mM. Surprisingly, when Mg2+ was replaced by softer Lewis acids for quinolone binding to single-stranded DNA, the level of binding increased. The Kdapps for ciprofloxacin and oxolinic acid binding in the presence of Mg2+, Ca2+, Mn2+, and Cd2+ were calculated (Table 5). Mg2+ is necessary for the higher-affinity sequence-specific interaction, since Ca2+ increased the Kdapp similarly to the increase after alteration of the GpG sequence. The manner of binding with Mg2+ and Ca2+ is likely to be similar since the binding stoichiometries were very similar. Binding in the presence of softer metal ions was less specific. Mn2+, a softer Lewis acid than Mg2+ and Ca2+, showed an increased binding stoichiometry, while Cd2+, the softest Lewis acid tested, had a significantly higher binding stoichiometry, indicating nonspecific binding. This suggests that Mg2+ is required for the higher-affinity sequence-specific quinolone-single-stranded DNA interaction. Therefore, it may mediate guanine-specific contacts by forming a bridge between the quinolone and the DNA.
Substitution of the GpG step of the 990 site oligonucleotide significantly affected the affinities of the quinolones for single-stranded DNA, since all of the quinolones showed a significantly reduced affinity for the altered sequence (TAACCT). This is slightly surprising since GpG has been replaced by ApA and indicates that the presence of guanines is important for the higher-affinity interaction. We suggest that Mg2+ may mediate the sequence-dependent quinolone binding to the GpG step. It is interesting that a recent double-stranded A-form DNA dodecamer crystal structure shows a hydrated Mg2+ bound to two GpG sequences within ACCGGCCGGT (31). Three water molecules of the hydrated Mg2+ form hydrogen bonds to the N-7 and O-6 groups of the two guanines. This mode of interaction is reminiscent of the quinolone-DNA model suggested by Llorente et al. (21), in which Mg2+ binding is coordinated by the C-3 and C-4 groups of the drugs and the O-6 and N-7 groups of guanines.
We should point out that in the present study we have only looked at a single gyrase cleavage site (the major gyrase cleavage site of plasmid pBR322) and made only limited changes, and it is not certain that the conclusions with regard to sequence specificity would be generally applicable. Also, it is important that drug potency be determined by the efficiency of formation of cleavage complexes and subsequent replication fork arrest, issues that we have not specifically addressed in this work. Additionally, quinolones target topoisomerase IV in some organisms, and it is not known whether quinolone-DNA binding affects drug potency in the case of this enzyme. It is also important to point out that the SPR experiments and the enzyme assays described in this work were not carried out under identical conditions (e.g., they were carried out at different temperatures) and that this might affect the correlations that we have made.
In summary, we have found that there are various modes of quinolone binding to DNA but that interaction with single-stranded DNA, mediated by Mg2+, best correlates with drug potency. We suggest that single-stranded DNA, revealed by distortion of the bound DNA duplex in the gyrase active site, forms part of the drug binding pocket.
Acknowledgments
We thank Parke-Davis, GlaxoSmithKline (Harlow, United Kingdom), and BBSRC (Swindon, United Kingdom) for financial support.
REFERENCES
- 1.Bailly, C., C. Tardy, L. Wang, B. Armitage, K. Hopkins, A. Kumar, G. B. Schuster, D. W. Boykin, and W. D. Wilson. 2001. Recognition of ATGA sequences by the unfused aromatic dication DB293 forming stacked dimers in the DNA minor groove. Biochemistry 40:9770-9779. [DOI] [PubMed] [Google Scholar]
- 2.Barnard, F. M., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob. Agents Chemother. 45:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, A. D., and A. Maxwell. 1993. DNA topology. IRL Press, Oxford, United Kingdom.
- 4.Bhanot, S. K., M. Singh, and N. R. Chatterjee. 2001. The chemical and biological aspects of fluoroquinolones: reality and dreams. Curr. Pharm. Des. 7:311-335. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco, C., H. Vezin, W. D. Wilson, J. Ren, J. B. Chaires, and C. Bailly. 2001. DNA binding properties of the indolocarbazole antitumor drug NB-506. Anticancer Drug Des. 16:99-107. [PubMed] [Google Scholar]
- 6.Cove, M. E., A. P. Tingey, and A. Maxwell. 1997. DNA gyrase can cleave short DNA fragments in the presence of quinolone drugs. Nucleic Acids Res. 25:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Critchlow, S. E., and A. Maxwell. 1996. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry 35:7387-7393. [DOI] [PubMed] [Google Scholar]
- 8.Domagala, J. M. 1994. Structure-activity and structure-side-effect relationships for quinolone antibacterials. J. Antimicrob. Chemother. 33:685-706. [DOI] [PubMed] [Google Scholar]
- 9.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, J.-Y., D. Sun, H. Yu, S. M. Kerwin, and L. H. Hurley. 1995. Self-assembly of a quinobenzoxazine-Mg2+ complex on DNA: a new paradigm for the structure of a drug-DNA complex and implications for the structure of the quinolone bacterial gyrase-DNA complex. J. Med. Chem. 38:408-424. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, L. M., H. A. Barot, and M. E. Cullen. 1986. DNA gyrase complex with DNA: determinants for site-specific DNA breakage. EMBO J. 5:1411-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, L. M., K. Mizuuchi, M. H. O'Dea, H. Ohmori, and M. Gellert. 1981. Site-specific interaction of DNA gyrase with DNA. Proc. Natl. Acad. Sci. USA 78:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glusker, J. P. 1991. Structural aspects of metal liganding to functional groups in proteins. Adv. Protein Chem. 42:1-76. [DOI] [PubMed] [Google Scholar]
- 15.Gmünder, H., K. Kuratli, and W. Keck. 1997. In the presence of subunit A inhibitors DNA gyrase cleaves DNA fragments as short as 20 bp at specific sites. Nucleic Acids Res. 25:604-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallett, P., and A. Maxwell. 1991. Novel quinolone resistance mutations of the Escherichia coli DNA gyrase A protein: enzymatic analysis of the mutant proteins. Antimicrob. Agents Chemother. 35:335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heddle, J., and A. Maxwell. 2002. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob. Agents Chemother. 46:1805-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhlmann, J., A. Dalhoff, H.-J. Zeiler, et al. 1998. Quinolone antibacterials, vol. 127. Springer-Verlag, Berlin, Germany.
- 19.Kwok, Y., Q. P. Zeng, and L. H. Hurley. 1999. Structural insight into a quinolone-topoisomerase II-DNA complex. Further evidence for a 2:2 quinobenzoxazine-Mg2+ self-assembly model formed in the presence of topoisomerase II. J. Biol. Chem. 274:17226-17235. [DOI] [PubMed] [Google Scholar]
- 20.Lecomte, S., M. H. Baron, M. T. Chenon, C. Coupry, and N. J. Moreau. 1994. Effect of magnesium complexation by fluoroquinolones and their antibacterial properties. Antimicrob. Agents Chemother. 38:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llorente, B., F. Leclerc, and R. Cedergreen. 1996. Using SAR and QSAR analysis to model the activity and structure of the quinolone-DNA complex. Bioorg. Med. Chem. 4:61-71. [DOI] [PubMed] [Google Scholar]
- 22.Lockshon, D., and D. R. Morris. 1985. Sites of reaction of Escherichia coli DNA gyrase on pBR322 in vivo as revealed by oxolinic acid-induced plasmid linearization. J. Mol. Biol. 131:63-74. [DOI] [PubMed] [Google Scholar]
- 23.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Structure of the DNA breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 24.Noble, C. G., and A. Maxwell. 2002. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two-metal-ion mechanism. J. Mol. Biol. 318:361-371. [DOI] [PubMed] [Google Scholar]
- 25.Ohlson, S., M. Strandh, and H. Nilshans. 1997. Detection and characterization of weak affinity antibody antigen recognition with biomolecular interaction analysis. J. Mol. Recognit. 10:135-138. [DOI] [PubMed] [Google Scholar]
- 26.Palu', G., S. Valisena, G. Ciarrocchi, B. Gatto, and M. Palumbo. 1992. Quinolone binding to DNA is mediated by magnesium ions. Proc. Natl. Acad. Sci. USA 89:9671-9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palumbo, M., B. Gatto, Z. Zagotto, and G. Palu'. 1993. On the mechanism of action of quinolone drugs. Trends Microbiol. 1:232-235. [DOI] [PubMed] [Google Scholar]
- 28.Permana, P. A., R. M. Snapka, L. L. Shen, D. T. W. Chu, J. J. Clement, and J. J. Plattner. 1994. Quinobenzoxazines: a class of novel antitumor quinolones and potent mammalian DNA topoisomerase II catalytic inhibitors. Biochemistry 33:11333-11339. [DOI] [PubMed] [Google Scholar]
- 29.Peterson, L. R. 2001. Quinolone molecular structure-activity relationships: what we have learned about improving antimicrobial activity. Clin. Infect. Dis. 33(Suppl. 3):S180-S186. [DOI] [PubMed] [Google Scholar]
- 30.Reece, R. J., and A. Maxwell. 1989. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264:19648-19653. [PubMed] [Google Scholar]
- 31.Robinson, H., Y. G. Gao, R. Sanishvili, A. Joachimiak, and A. H. Wang. 2000. Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes. Nucleic Acids Res. 28:1760-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen, L. L., J. Baranowski, and A. G. Pernet. 1989. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: specificity and cooperativity of drug binding to DNA. Biochemistry 28:3879-3885. [DOI] [PubMed] [Google Scholar]
- 33.Shen, L. L., L. A. Mitscher, P. D. Sharma, T. J. O'Donnell, D. T. W. Chu, C. S. Cooper, T. Rosen, and A. G. Pernet. 1989. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: a cooperative drug-DNA binding model. Biochemistry 28:3886-3894. [DOI] [PubMed] [Google Scholar]
- 34.Sugino, A., C. L. Peebles, K. N. Kruezer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanious, F. A., W. D. Wilson, D. A. Patrick, R. R. Tidwell, P. Colson, C. Houssier, C. Tardy, and C. Bailly. 2001. Sequence-dependent binding of bis-amidine carbazole dications to DNA. Eur. J. Biochem. 268:3455-3464. [DOI] [PubMed] [Google Scholar]
- 36.Tornaletti, S., and A. M. Pedrini. 1988. DNA unwinding induced by nalidixic acid binding to DNA. Biochem. Pharmacol. 37:1881-1882. [DOI] [PubMed] [Google Scholar]
- 37.Tornaletti, S., and A. M. Pedrini. 1988. Studies on the interaction of 4-quinolones with DNA by DNA unwinding experiments. Biochim. Biophys. Acta 949:279-287. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J. C. 1998. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31:107-144. [DOI] [PubMed] [Google Scholar]
- 39.Willmott, C. J. R., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces the binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida, H., T. Kojima, J. Yamagishi, and S. Nakamura. 1988. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol. Gen. Genet. 211:1-7. [DOI] [PubMed] [Google Scholar]