Abstract
Background
Differential methylation of the two alleles is a hallmark of imprinted genes. Correspondingly, loss of DNA methyltransferase function results in aberrant imprinting and abnormal post-fertilization development. In the mouse, mutations of the oocyte-specific isoform of the DNA methyltransferase Dnmt1 (Dnmt1o) and of the methyltransferase-like Dnmt3L gene result in specific failures of imprint establishment or maintenance, at multiple loci. We have previously shown in humans that an analogous inherited failure to establish imprinting at multiple loci in the female germline underlies a rare phenotype of recurrent hydatidiform mole.
Results
We have identified a human homologue of the murine Dnmt1o and assessed its pattern of expression. Human DNMT1o mRNA is detectable in mature oocytes and early fertilized embryos but not in any somatic tissues analysed. The somatic isoform of DNMT1 mRNA, in contrast, is not detectable in human oocytes. In the previously-described family with multi-locus imprinting failure, mutation of DNMT1o and of the other known members of this gene family has been excluded.
Conclusions
Mutation of the known DNMT genes does not underlie familial hydatidiform mole, at least in the family under study. This suggests that trans-acting factors other than the known methyltransferases are required for imprint establishment in humans, a concept that has indirect support from recent biochemical studies of DNMT3L.
Keywords: autozygosity, hydatidiform mole, imprinting, oogenesis
Background
Genomic imprinting refers to the process whereby in mammals certain genes are differentially expressed from the paternal and maternal alleles. The primary imprint "mark" that distinguishes maternal and paternal alleles has to be placed on a gene during gametogenesis, the only time at which the two alleles are in different cellular environments. This mark could be in the form of methylation or perhaps some other epigenetic modification to the chromatin. The sequelae of the primary imprint, however, are manifested at various developmental stages, and depend not just on the presence of this initial gametic "mark", but also on the postzygotic interpretation of such primary imprints, into the form of stable epigenetic differences between the two alleles.
The expression of a normal pattern of genomic imprinting in the mouse embryo depends on the activity of the DNA methyltransferase Dnmt1 [1]. Consequently, Dnmt1 deficiency is an embryonic lethal [2]. Because Dnmt1 shows preferential activity against hemimethylated DNA, it has been proposed to be involved primarily in maintaining methylation patterns through rounds of DNA replication, rather than establishing them de novo. However, the fact that Dnmt1 predominates at all stages of embryogenesis suggests that it may be involved in de novo methylation as well as maintenance [3,4].
A notable feature of Dnmt1 is the existence of gamete-specific isoforms, a finding that points to unique developmental roles, distinct from those of the ubiquitous somatic isoform. These different isoforms are generated by alternative Dnmt1 promoter usage. The mouse oocyte-specific transcripts initiate in exon 1o, 6 kb upstream of the major somatic exon 1 [5]. Exon 1o lacks an in-frame initiation codon, so that translation of this mRNA species initiates from a downstream ATG in exon 4, yielding the N-truncated Dnmt1o protein. This isoform is enzymatically active and can rescue a Dnmt1 null mutation when expressed in ES cells [6].
During late oogenesis, accumulating Dnmt1o becomes excluded from the nucleus and at fertilization is sequestered in the subcortical cytoplasm. It only re-enters the nucleus transiently at the 8-cell stage, shortly thereafter to be re-excluded. This translocation (dictated by specific nuclear and cytoplasmic localization signals) [7] suggests that Dnmt1o may provide a temporally-critical function early in embryogenesis; this might involve the replication, propagation or interpretation of early methylation patterns at selected loci (in particular imprinted genes) that are then subsequently able to escape the global genomic demethylation that occurs in later preimplantation development [8]. This prediction was strikingly confirmed by deleting the Dnmt1o promoter in the mouse germline [9]. Homozygous mutants show a maternal-effect phenotype in which Dnmt1o fails to accumulate during oogenesis, and their resulting offspring die during late gestation. This is associated with a partial (50%) failure to appropriately maintain the methylation of imprinted alleles post-zygotically. However, there is no defect either of global methylation levels in embryos, or of the establishment of methylation in the oocyte. The latter fact implies that other methylases may be responsible for setting the primary methylation imprint "mark" in the oocyte. In contrast, the accumulated Dnmt1o appears to be required post-zygotically, for the maintenance of imprints during one critical S phase (8–16 cell stage) of preimplantation development.
By sequence homology criteria, five members of the mammalian Dnmt family have been recognized [10]. One of these, Dnmt3L, lacks sequence motifs involved in methyltransferase catalytic activity, but has an interesting expression pattern restricted to testis and ovary [11]. Its importance in establishing imprints was again demonstrated by mouse knockouts, in which homozygous mothers produce abnormal conceptuses that lack correct methylation imprints on the maternal allele [12,13].
Several inherited human disorders can result from failure to establish correct genomic imprinting within individual imprinted chromosomal domains. At the Prader-Willi/Angelman syndrome locus (15q11-q13), the exact nature of the responsible mutations has been identified; a small imprinting centre (IC) defined by these mutations controls the setting of imprints across a ~2 Mb genomic region [14]. However, comparable cis-acting mutations at other imprinted loci are implicated in Beckwith syndrome [15], transient neonatal diabetes [16] and pseudohypoparathyroidism type Ib [17]. In contrast to these disorders, we have recently defined a clinical situation in which there is a failure to establish methylation imprints at multiple loci [18]. This failure appears selective for loci that are normally maternally imprinted (the majority of all imprinted genes). The condition manifests as a recurrent form of hydatidiform mole, clinically indistinguishable from the more common, sporadic androgenetic mole, but in which a full haploid chromosome complement from both parents is present. The inheritance of this condition (familial biparental hydatidiform mole, FBHM) suggests that affected women are homozygous for a recessive mutation in a trans-acting factor required for imprint establishment during oogenesis.
The demonstration of the existence of trans-acting imprinting control genes in both humans and mouse raises the issue of the extent to which the components required for correct imprinting are conserved in both species. We have addressed two questions in the present report; (i) we have investigated the existence of a human oocyte-specific DNMT1 isoform and (ii) we have examined the possibility that mutation of one of the human DNMT family members could underlie FBHM.
Results
Female germline-specific DNMT1 isoform
We first investigated the existence of a human equivalent of the mouse oocyte-specific Dnmt1o isoform. EST database entries matching the 5' part of the DNMT1 mRNA were examined for possible alternative 5' ends. These 5' ends were then compared to the DNMT1 locus genomic sequence. This analysis indicated the existence of possible novel first exons at -6 kb (EST BE537788) and -37 kb (ESTs BE902068, AW977685, AI798069) relative to the known somatic exon 1.
To test for oocyte-specific expression, two preparations of cDNA from single mature oocytes were amplified using primers specific for the somatic DNMT1, for EST BE537788, and for EST AI798069. Strong PCR products were obtained from both oocyte cDNAs, only with the BE537788 primers. These oocyte cDNAs were negative for somatic DNMT1 and for the AI798069 EST. The BE537788 primers also yielded a weak product in fetal ovary cDNA (which was also positive for the somatic DNMT1, and very weakly for AI798069). In contrast, no expression of the BE537788 EST could be detected in a range of somatic fetal tissues, although widespread DNMT1 expression was detected in these tissues using primers in exons 4 and 5 or exons 36–39. The BE537788 EST therefore appeared to represent a DNMT1 isoform expressed preferentially in human oocytes, and scarce or absent in other tissues. We therefore refer to transcripts containing the novel exon in BE537788 (Figure 1b) as DNMT1o.
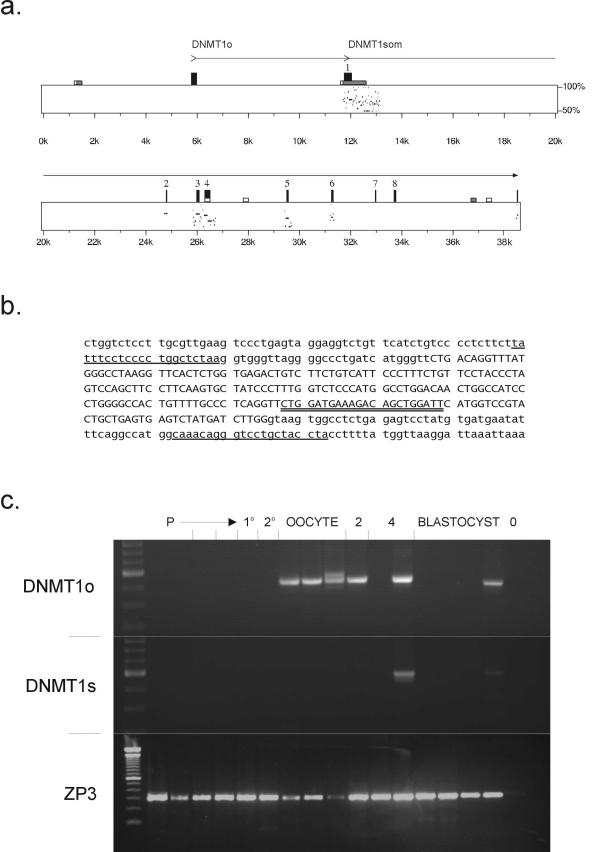
Figure 1.
a. PIP (percentage identity plot) diagram for human and murine DNMT1. The horizontal axis represents the human genomic sequence and has been annotated to show the position of the novel exon 1o, which unlike the previously recognized DNMT1 exons, shows no sequence conservation relative to mouse. Tall black rectangles indicate the positions of the known human DNMT1 exons. Low grey and white rectangles indicate regions with CpG-island like features (CpG/GpC respectively ≥ 0.75 and ≥ 0.60). b. Alternative first exon (1o) of human DNMT1. The exon is in upper case. Underlining indicates the PCR primers used for genomic sequence analysis (flanking) and double underlining for RT-PCR analysis of oocyte material. c. Detection of DNMT1 expression in cDNA from human oocytes and embryos. The panel labelled DNMT1o shows PCR reactions using the upstream primer indicated in panel b, unique to the novel isoform of human DNMT1. In the DNMT1s panel, an upstream primer in the previously defined exon 1 was used. The ZP3 panel shows control reactions that detect the mRNA for zona pellucida protein ZP3, which is expressed at all stages of oogenesis. Lanes 1–2, primordial follicles (P); 3, mix of primordial and early primary; 4, mix of early primary and primary (1°); 5, primary; 6, secondary (2°); 7–9, ovulated oocytes; 10, 2-cell embryos (2); 11–12, 4-cell embryos (4); 13–16, blastocysts; 17, negative control (0).
As noted above, this novel exon (nt 145549–145765 of AC011511) lies, similarly to the murine Dnmt1o exon, 6 kb upstream of the somatic exon 1, and splices onto exon 2. Despite this, there is little or no sequence similarity between the human and murine sequences in the region of the oocyte-specific exon (Figure 1a). This may not be entirely unexpected, given that in both species the predicted translation initiation site for the oocyte-specific mRNA is farther downstream in exon 4. This presumably results in a lack of evolutionary pressure for maintaining the sequence of the oocyte-specific exon.
We next attempted to assess the time course of DNMT1o in human oocyte development. A panel of cDNAs representing stages from primordial oocyte through to early blastocyst was examined (Figure 1c). DNMT1o was consistently detectable in mature oocytes, and also in some early embryo samples (2- and 4-cell stage). Somatic DNMT1 was absent from the oocyte cDNAs, though detected in one 4-cell and one blastocyst stage embryo.
DNMTs as candidate genes in inherited hydatidiform mole
As described in the Introduction, DNMT1o and DNMT3L are good candidates for involvement in FBHM. In one family with this rare disorder, we have demonstrated a genome-wide failure to establish methylation imprints in the female germline [18]. We therefore performed mutation analysis of the index case in this family, who has had six successive molar pregnancies and is the product of a consanguineous mating.
A DNMT1o PCR product of normal sequence was obtained from this patient, who was also heterozygous for the nearby microsatellite markers D19S916 and D19S413. Since the causative mutation in this family is expected to be homozygous by descent, this makes involvement of DNMT1 unlikely.
We next deduced the human DNMT3L gene structure [11] from the genomic sequence AP001753 and designed primers for PCR-amplification of each exon. No mutation in the index FBHM patient was found on sequencing all 11 coding exons. One heterozygous A to G substitution in exon 9 was found in the patient, predicting the amino-acid change R278G. R278 is conserved in all other DNMT3 family members [11], but despite this, we found that R278G was a common polymorphic variant; a diagnostic SmaI digest demonstrated the presence of this change on 8 out of 42 alleles. Lack of identity by descent in the FBHM family was confirmed for both affected individuals for whom DNA was available, using the closely flanking markers D21S1411 and D21S1912. Together with the heterozygous intragenic polymorphism, this effectively rules out the possibility of an undetected DNMT3L mutation (assuming homozygosity by descent).
It has been suggested that Dnmt3L is involved in imprint establishment by interacting with Dnmt3b and/or Dnmt3a [13]. The cognate human DNMT3A and DNMT3B genes therefore required assessment. Markers adjacent to DNMT3A (D2S171, D2S174) were heterozygous in the index case in our FBHM family, making the involvement of this gene highly unlikely. Around DNMT3B, however, there appeared to be a region of probable homozygosity by descent in both affected individuals. We therefore sequenced this gene in the index case. No pathogenic mutation was identified in exons 1b-23. (We were unable to amplify the very GC-rich exon 1a.) Several previously reported polymorphic variants from the reference genomic sequence (AL035071) were noted. Three new variants were also noted; homozygous changes C→T at nt55630 (IVS12 +30) and G→A at nt65452 (IVS21 +139), and more importantly a heterozygous silent change in exon 18 (nt60758). The latter suggested that the DNMT3B genomic region was not in fact homozygous by descent in the index case. We confirmed this by identifying and typing a new polymorphic GT repeat at nt21057-21106. The index case in the FBHM family was heterozygous, which since this marker lies only 1.3 kb upstream of exon 1a, effectively eliminates the possibility of homozygosity for an exon 1a mutation.
The remaining DNMT family member, DNMT2 [19] is of unknown function. Six markers spanning the 6 cM interval D10S1864-D10S203, around DNMT2, were all homozygous in the FBHM index case, suggesting homozygosity by descent for this region. The human DNMT2 structure was again deduced (from genomic sequence of AL133415) and all eleven exons were sequenced. Four polymorphic substitutions were identified (Table 1), only one of which predicts an amino-acid change (H101Y). No changes specific to the FBHM patient were identified.
Table 1.
Coding region polymorphisms, defined during DNMT mutation screening. Nucleotide numbering refers to cDNA sequence AF012128 (DNMT2) or AF194032 (DNMT3L).
| Gene | nt change (cDNA) | Exon | Effect | Frequency |
| DNMT2 | G175→A | 2 | silent | A = 6, G = 4 |
| DNMT2 | T217→C | 3 | silent | T = 8, C = 2 |
| DNMT2 | C308→T | 4 | His101→Tyr | T = 6, C = 4 |
| DNMT2 | G556→T | 8 | silent | G = 8, T = 2 |
| DNMT3L | A1316→G | 9 | Arg278→Gly | A = 34, G = 8 |
Frequencies were those observed among 10 alleles sequenced (DNMT2) or assessed by diagnostic restriction digest (DNMT3L).
Discussion
In the mouse, paternally imprinted genes are required for correct development of extraembryonic membranes and trophoblast [20]. In contrast, the lack of maternally imprinted genes, in androgenetic conceptions, result in poor embryo development, while trophoblast develops comparatively well [21]. In humans, the phenotype resulting from androgenesis is distinctive, comprising a complete hydatidiform mole (CHM) in which embryo development is poor or absent while abnormal trophoblast tissue proliferates [22]. We have, further, recently shown that rare cases of CHM that are biparental may arise through failure of the maternal genome to acquire imprints during oogenesis. This failure affects multiple imprinted loci, and therefore presumably reflects deficiency of an oocyte-specific trans-acting factor required for imprint establishment [18]. Based on recent studies of mouse knockouts, the DNMT3L, DNMT3A, DNMT3B and DNMT1 (DNMT1o isoform) loci all represent candidates for involvement in human trans-acting imprinting defects [9,12,13]. Recessive mutations of the human DNMT3B gene, however, are known to cause ICF syndrome (MIM 242860) [23-25]. This disorder, in which juxta-centromeric repeat methylation is abnormal, bears no phenotypic resemblance to FBHM, and DNMT3B might therefore be dismissed as a FBHM candidate. However, all ICF syndrome patients have mutations of specific residues in the C-terminal catalytic domain, with no patients homozygous either for clearly null mutations (this being presumed lethal; [26] or for missense mutations in the non-catalytic regions of the protein. Thus it still remained possible that different mutations altering DNMT3B interaction with chromatin or other proteins might underlie FBHM.
Our present results, though, rule out known members of the DNA methyltransferase-like group as the gene mutated in our FBHM family. Although based only on one family, this result is significant, because it implies that other oocyte-specific components participate in establishing the epigenetic modifications needed for correct imprinting in the human female germline. Recent studies of DNMT3L are interesting in this regard. These studies show that in vitro, DNMT3L can mediate transcriptional repression by recruiting histone deacetylase via the conserved PHD (plant homeodomain) motif of DNMT3L [27,28]. It remains to be seen, though, whether such interactions are relevant to the in vivo mechanism through which Dnmt3L mediates maternal imprinting in the germline. Indeed, other recent work indicates the possibility of a wider role for DNMT3L as a stimulatory cofactor for de novo methylation by DNMT3A [29].
Finally, FBHM may, additionally, be a genetically heterogeneous condition. Two previously described families appear to map to a locus on chromosome 19q [30], whereas linkage to this region has been previously excluded in our family [18]. It is currently unknown whether a defect of methylation of imprinted genes is present in molar tissue of 19q-linked families. It therefore also remains possible that there is further genetic heterogeneity, and that DNMT mutations could result in a FBHM phenotype in other families.
Conclusions
A human oocyte-specific form of the DNMT1 mRNA has been identified, and may correspond to murine Dnmt1o. Mutation of known DNA methyltransferase genes does not underlie a trans-acting imprinting defect that manifests as familial biparental hydatidiform mole. This suggests the existence of other factors required for specifying imprinting in the female germline.
Methods
RT-PCR analysis of DNMT1 expression in oocytes
Detailed descriptions of the harvesting of human oocyte material and cDNA preparation will be published elsewhere (Huntriss et al., in preparation). Briefly, follicle samples were obtained from an ovarian cortex biopsy donated for research (22 year-old woman), and were measured and staged microscopically. Metaphase II oocytes and preimplantation embryos were surplus to clinical requirements under IVF or ICSI procedures. All samples were obtained with informed consent under ethically approved protocols including a licence from the Human Fertilisation and Embryo Authority (HFEA). RNA was prepared by binding of lysates to oligo(dT)25 magnetic beads (Dynal Biotech, Oslo, Norway), according to the manufacturer's protocols. cDNA was synthesized using a SMART™ protocol (Clontech, Palo Alto, U.S.A.) and pre-amplified using PCR primers matching the SMART IV and CDS III/3' sequences in the cDNA (SMART cDNA library construction kit).
Individual PCR reactions were then performed on this cDNA, using the following cycling conditions: 5 min at 94°C followed by 9 cycles of 94°C for 30 s, 70°C (reduced by 1°C each cycle) for 30 s, 72°C for 60 s and then 32 cycles of 94°C for 30 s, 61°C for 30 s, 72°C for 60 s. The DNMT1s-specific upstream primer (exon 1) was dTCTGCTGAAGCCTCCGAGATG and the DNMT1o-specific primer (Figure 1b) was dCTGGATGAAAGACAGCTGGATTC.
The common downstream primer (exon 5) was dCCTGGTGCTTTTCCTTGTAATC.
Mutation analysis
Mutation analysis was performed by direct cycle-sequencing of PCR products, using either 33P-ddNTP labelling (Amersham Biosciences, Uppsala, Sweden) or BigDye fluorescent terminators and an ABI377 sequencer (Applied Biosystems, Foster City, U.S.A.).
Primers for amplification and sequencing of DNMT3L:
Exon 1: dGCTGACGGCTTTCTCACATCT dTGGGATCAGCATCCTAAGTGA
Exon 2: dTTTGGAAAACGAATTGTGATGC dTTGTCTGGTGTGGGCCAGTAT
Exons 3–4: dTTTGACAGCACTGGGAAAACA dGAATGAGTAGCTGAAAGGATGG
Exon 5: dCCAAAACAAGGCTCAATCAGG dGTAAGTCAGGGCCTGCTGCT
Exon 6: dGGCACAATGTCCTGTTTTGGT dTCCATGTGTGTCTTTGGGACA
Exon 7: dACAATCCACGCTCACCCTCT dGTCTGAGAGTTCACGGCGGTA
Exon 8: dACTGCTGCTGCCTCTCAGTG dGACAGGCTGCCAGAACTGAA
Exon 9: dCTCTCAAGCCCCAGTTTCCTT dACTGAGCTCCACCAGAAAAGC
Exon 10: dTTACCGTCCAGGTCCCCTGA dATGTTCCCCTTGACCCTGAAC
Exon 11: dCTGCGCTTGCTCACAACCTT dACGTGAGGAAAGTTGCCTTTG
Exon 4 sequencing primer: dGGATGACACAAGCCAGCCT
Primers for amplification and sequencing of DNMT3B:
For exon 1b, PCR primers were dGACAGGCCAGCAGACCAATG and dCAGGATGCATTCTTAGGGTGAT. Other PCR primers were as previously described [25]http://humgen.med.uu.nl/research/ICF/default.html. To amplify a polymorphic (GT)n sequence upstream of exon 1a, dTCACCTGGTAAGAGATAC and dTGCCCAGATCTTTGCAGG were used.
Primers for amplification and sequencing of DNMT2 (for each exon, the first two primers given were used for PCR, others for sequencing):
Exon 1: dGAGACGGGGGAACTGAAACG dGAGAAGGGAGTCAGCGTCTG dGCAGGCCTAGCTCCGGGGCT
Exon 2: dCCACTGACAGTCTGAGATGAGG dCAATCACATTTCCCTTGCTACA dTCTATAATGAACTTAAGGAC
Exon 3: dACCAAAGTTGATTTGACAAGCA dGCATGAAAATGGACCAATACAA dTCCCCATTCTATCCATGTTT
Exon 4: dTTTTTGTGAGGACCAACTTATTTTT dTTCCAGACCTGGAGTTTCAAAT dAGTGATTTGACATCTTTTTATC
Exon 5: dTCTCCAGCCTTATAAAATTCCA dTGTCAGAATGTATGCACTGGTTT dCTTAGTAATGAGATATTTAGAAT
Exon 6: dCCTGGGCAACAGAGTGAGACT dAAGCCATTCTGCTTATCGAATC dAATAAGAACTGCCTTTCCAAG
Exon 7: dAAGCTAAATGTAGGCTCGTTTGA dAACAGCCTTGAAGATGGACCTA dTGCTGTTTAACTTGGTGCTAA
Exon 8: dTTTAATGGCAACCCCATAAAG dGGCAATTAAGGGAAAATACCTCA dCGTAATGTTGACTGGACATTG dTGGATACAGGATCAATAGATC
Exon 9: dTTCAGAAGGATGTAACTGTGTAGATTG dCATGGCCAGTTGAATGCATAA dACACATACATCAGGTGCTGCA
Exon 10: dCTTTGCCACTGTGTGTTACAGC dGCAAGTCCTAGACACACAAGTCC dAATCTAGTTGCACATTACTT
Exon 11: dTGGCTCCAAATTCTCTTTTGTT dTGCCTGTTATGACACTTCACAAG dAAGGATACATCCTTTCATCTC dTACTCATGTAGACACTAAAGC
Authors' contributions
B.E.H. designed and performed the bioinformatics and experimental identification of human DNMT1o, as well as the RT-PCR analysis. M. De V. performed DNMT3B mutation analysis and some of the microsatellite marker analysis. H.J. performed DNMT2 mutation analysis and some of the microsatellite marker analysis. D.H. participated in the microsatellite and sequence analysis. J.H. performed the oocyte RNA preparation and SMART PCR cDNA synthesis. H.M.P. performed the staging of human oocytes. E.S. obtained and interpreted clinical and pedigree data and clinical samples. D.T.B. coordinated the different aspects of the study and wrote the manuscript. The final version was read and approved by all authors.
Acknowledgments
Acknowledgements
M. De Vos is a Medical Research Council Clinical Research Training Fellow.
Contributor Information
B E Hayward, Email: b.hayward@leeds.ac.uk.
M De Vos, Email: m.devos@leeds.ac.uk.
H Judson, Email: hannahjudson@hotmail.com.
D Hodge, Email: d.hodge@leeds.ac.uk.
J Huntriss, Email: j.huntriss@leeds.ac.uk.
H M Picton, Email: h.m.picton@leeds.ac.uk.
E Sheridan, Email: eamonn.sheridan@leedsth.nhs.uk.
DT Bonthron, Email: d.t.bonthron@leeds.ac.uk.
References
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–5. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270:385–95. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002–10. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–97. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Talbot D, Leonhardt H, Jaenisch R. A short DNA methyltransferase isoform restores methylation in vivo. J Biol Chem. 1998;273:32725–9. doi: 10.1074/jbc.273.49.32725. [DOI] [PubMed] [Google Scholar]
- Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–82. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–38. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Minoshima S, Krohn K, Antonarakis SE, Shimizu N, Kudoh J, Peterson P. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–8. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–9. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–93. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- Reik W, Brown KW, Schneid H, Le Bouc Y, Bickmore W, Maher ER. Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2-H19 domain. Hum Mol Genet. 1995;4:2379–85. doi: 10.1093/hmg/4.12.2379. [DOI] [PubMed] [Google Scholar]
- Gardner RJ, Mackay DJ, Mungall AJ, Polychronakos C, Siebert R, Shield JP, Temple IK, Robinson DO. An imprinted locus associated with transient neonatal diabetes mellitus. Hum Mol Genet. 2000;9:589–96. doi: 10.1093/hmg/9.4.589. [DOI] [PubMed] [Google Scholar]
- Liu J, Litman D, Rosenberg MJ, Yu S, Biesecker LG, Weinstein LS. A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest. 2000;106:1167–74. doi: 10.1172/JCI10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson H, Hayward BE, Sheridan E, Bonthron DT. A global disorder of imprinting in the human female germ line. Nature. 2002;416:539–42. doi: 10.1038/416539a. [DOI] [PubMed] [Google Scholar]
- Hung MS, Karthikeyan N, Huang B, Koo HC, Kiger J, Shen CJ. Drosophila proteins related to vertebrate DNA (5-cytosine) methyltransferases. Proc Natl Acad Sci U S A. 1999;96:11940–5. doi: 10.1073/pnas.96.21.11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–50. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature. 1984;311:374–6. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- Kajii T, Ohama K. Androgenetic origin of hydatidiform mole. Nature. 1977;268:633–4. doi: 10.1038/268633a0. [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–91. doi: 10.1038/46214. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–7. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Hansen RS, Gimelli G, Bjorck EJ, Davies EG, Valentine D, Belohradsky BH, van Dongen JJ, Smeets DF, van den Heuvel LP, Luyten JA, Strengman E, Weemaes C, Pearson PL. Genetic variation in ICF syndrome: evidence for genetic heterogeneity. Hum Mutat. 2000;16:509–17. doi: 10.1002/1098-1004(200012)16:6<509::AID-HUMU8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J Mol Biol. 2001;309:1201–8. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- Aapola U, Liiv I, Peterson P. Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity. Nucleic Acids Res. 2002;30:3602–8. doi: 10.1093/nar/gkf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Brenner C, Burgers WA, Putmans P, Kouzarides T, de Launoit Y, Fuks F. Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res. 2002;30:3831–8. doi: 10.1093/nar/gkf509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chédin F, Lieber MR, Hsieh C-L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99:16916–21. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglabey YB, Kircheisen R, Seoud M, El Mogharbel N, Van den Veyver I, Slim R. Genetic mapping of a maternal locus responsible for familial hydatidiform moles. Hum Mol Genet. 1999;8:667–71. doi: 10.1093/hmg/8.4.667. [DOI] [PubMed] [Google Scholar]