Abstract
A 23S rRNA gene fragment in domain V was sequenced from 30 clinical isolates of Campylobacter spp., including 22 resistant to macrolides. Two point mutations associated with erythromycin resistance were identified at positions 2074 and 2075 on the 23S rRNA gene (homologous to A2142C and A2143G mutations in Helicobacter pylori) in which an adenine residue is also replaced with a cytosine and a guanine residue, respectively. A combined PCR-restriction fragment length polymorphism technique was developed to detect these mutations by using the BsaI and BceAI enzymes.
Campylobacters are the most common bacterial cause of human gastrointestinal infection worldwide. The species most frequently isolated from humans are Campylobacter jejuni and Campylobacter coli (8). Other species, such as Campylobacter fetus, Campylobacter upsaliensis, and Campylobacter lari, are occasionally found in clinical isolates (7). Macrolides are the drugs of choice for treating Campylobacter infections (15). Campylobacter resistance to macrolides is mainly found in strains of animal origin, especially C. coli from pigs but also from chickens (1, 2, 18). Such strains can be transmitted to humans. Furthermore, macrolide resistance may develop during the course of antibiotic treatment in humans (5).
In a closely related bacterium, Helicobacter pylori, three major point mutations occurring in the peptidyl-encoding region in domain V of the 23S rRNA gene were found to be associated with macrolide resistance (A2142G, A2143G, and less frequently A2142C) (16, 20). The same mechanism was described recently for campylobacters (17). In this study, we sequenced a 23S rRNA gene fragment from 30 clinical isolates received at the French National Reference Center of Campylobacters and Helicobacters and found that two of the mutations described for H. pylori (A2142C and A2143G, equivalent to Escherichia coli coordinates 2058 and 2059) are also associated with macrolide resistance in Campylobacter species. Moreover, to identify these mutations rapidly, we used a combination of PCR and restriction fragment length polymorphism analysis with the BsaI and BceAI (BcefI) restriction enzymes that has already been described for H. pylori (9, 12, 19).
Campylobacter sp. isolates were cultured on Trypticase soy medium containing 5% horse blood under microaerobic conditions at 37°C. Their susceptibility to erythromycin (Sigma, St. Louis, Mo.) was detected by disk diffusion and confirmed by the standard agar dilution method in accordance with the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (3). Strains were considered resistant to erythromycin with a MIC of ≥8 μg/ml. The bacterial cells were harvested and then subjected to DNA extraction with the QIAamp DNA mini kit (Qiagen SA, Courtaboeuf, France).
Since the genetic information for domain V of the 23S rRNA gene of some campylobacters was available in the GenBank database, sequences were aligned by using multiple sequence alignment with hierarchical clustering (4) and a search was carried out to identify conserved regions flanking the sequence that supposedly presented mutations associated with erythromycin resistance. As a result, we selected a set of primers, F1-campy-23S (5′-AAGAGGATGTATAGGGTGTGACG-3′) and R1-campy-23S (5′-AACGATTTCCAACCGTTCTG-3′), designed to amplify a 508-bp sequence corresponding to nucleotides 1825 to 1847 and 2313 to 2332 of the three copies of the 23S rRNA gene of C. jejuni NCTC 11168, respectively (GenBank accession numbers AL139074, AL139075, and AL139076) (13). The amplification reaction was carried out in a final volume of 50 μl containing 1× buffer, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 0.2 μM each of the primers, 1 U of Taq polymerase (Eurobio, Les Ulis, France), and 2 μl of extracted DNA. Amplification was performed in a Perkin-Elmer GeneAmp 9700 thermocycler under the following conditions: 1 cycle of 94°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and 1 cycle of 72°C for 7 min. PCR amplicons were purified by using MicroSpin S-400 HR columns in accordance with the manufacturer's (Amersham Pharmacia Biotech Inc., Uppsala, Sweden) instructions. Sequencing of both strands of the amplified fragments was achieved by using an Applied Biosystems Prism 377 automated sequencer with dRhodamine-labeled terminators (PE Applied Biosystems, Foster City, Calif.).
On the basis of the sequence alignment of the 23S rRNA genes obtained with F1-campy-23S and R1-campy-23S, we designed a new set of primers, F2-campy-23S (5′-AATTGATGGGGTTAGCATTAGC-3′) and R2-campy-23S (5′-CAACAATGGCTCATATACAACTGG-3′), corresponding to nucleotides 1869 to 1890 and 2184 to 2161 of the 23S rRNA gene of C. jejuni NCTC 11168, respectively (13). Amplification with these primers resulted in a 316-bp PCR product. The conditions of amplification were the same as those described above.
After successful amplification of the 316-bp PCR products, amplicons were precipitated and suspended in 15 μl of H2O and 5 μl of the amplicons was digested with the restriction enzymes BsaI (5 U) and BceAI (1 U) (New England Biolabs, Beverly, Mass.) as previously reported for H. pylori (9). The fragments were incubated overnight at 50°C for BsaI and 37°C for BceAI, separated by electrophoresis on a 10% acrylamide gel, and visualized under UV light after ethidium bromide staining.
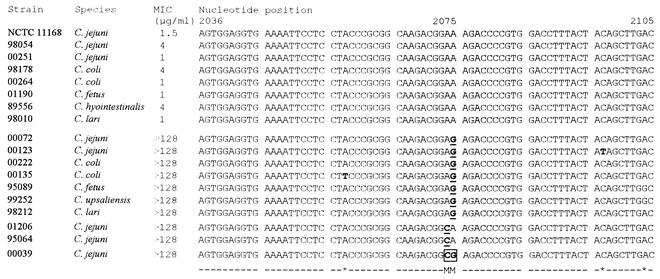
Thirty Campylobacter strains isolated in different areas of France were selected from the collection of the National Reference Center for Campylobacters on the basis of the disk diffusion test results; 22 were erythromycin-resistant (MIC, ≥128 μg/ml) strains of different species, and 8 were susceptible strains. They were chosen randomly from among the different species isolated in humans. The C. jejuni reference strain, NCTC 11168, which is susceptible to erythromycin, was also included. DNA sequences of an internal area of the 23S rRNA gene were determined for the 30 strains. Representative sequences are presented in Fig. 1. At least one mutation was detected in all of the macrolide-resistant strains but in none of the strains susceptible to this group of antibiotics. Mutations were found at positions 2074 and 2075, which are homologous to positions 2142 and 2143 in H. pylori, respectively (16). The predominant mutation was the transition mutation A2075G (20 cases). The transversion mutation A2074C was found in three resistant strains. It was noted that one strain (C. jejuni 00039) carried both the A2075G and A2074C mutations. Finally, none of the strains contained the mutation homologous to the A2142G mutation commonly described in H. pylori.
FIG. 1.
Alignment of the sequences of the 23S rRNA gene associated with erythromycin resistance. The sequences correspond to the partial nucleotide sequences of the 508-bp PCR products from 17 selected Campylobacter sp. strains and C. jejuni reference strain NCTC 11168 (GenBank accession numbers AL139074, AL139075, and AL139076). The strain numbers and the MICs of erythromycin are indicated. M, position of a point mutation associated with resistance to erythromycin; *, spontaneous mutation.
As previously described, each of these two mutations created an additional site for digestion by the restriction enzymes BsaI (for the A2075G mutation) and BceAI (for the A2074C mutation) (9, 20). PCR amplification with the F2-campy-23S and R2-campy-23S primers was performed on the 30 clinical isolates and the reference strain. A 316-bp amplicon was obtained for 26 clinical isolates, whereas no amplification was observed for other Campylobacter clinical isolates, such as C. hyointestinalis (strain 89556), C. fetus (strain 01190), and C. upsaliensis (strain 99252). All of the C. jejuni (n = 10), C. coli (n = 13), and C. lari (n = 3) strains tested, with the exception of C. lari strain 98212, were amplified by this PCR, which seems to be specific for thermophilic campylobacters. To prove the specificity of the primers, we tested various bacterial species, including E. coli (clinical isolates), Salmonella enteritidis (clinical isolates), Salmonella enterica serovar Typhimurium (clinical isolates), Flexispira rappini (CCUG 29176), H. pylori (CCUG 41936 and ATCC 700824), Helicobacter felis (CCUG 28539T), Helicobacter bilis (CCUG 38995BT), Helicobacter hepaticus (CCUG 33637T), Helicobacter muridarum (CCUG 29262T), and Helicobacter pullorum (CCUG 33842 and CCUG 33839), and none of them were amplified (data not shown).
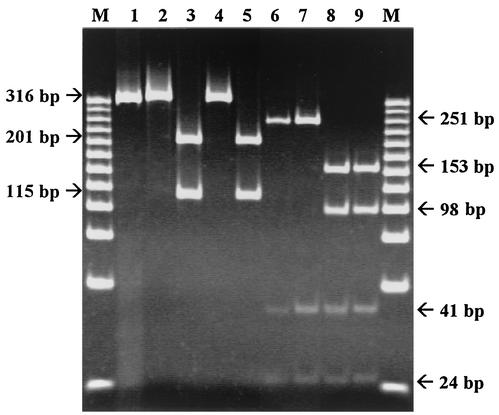
Clinical isolates and the reference strain were subjected to these two sets of restriction enzymes. As shown in Fig. 2, erythromycin-resistant isolates could easily be distinguished from the susceptible ones by a pattern characteristic of the specific substitution. The 316-bp amplicon containing the A2075G mutation was digested by BsaI into two subproducts of 201 and 115 bp (Fig. 2, lanes 3 and 5), in contrast to the uncut wild-type sequence and that with the A2074C mutation (Fig. 2, lanes 2 and 4). This mutation can also be detected by restriction of the 316-bp amplicon with Alw26I (Promega, Madison, Wis.), leading to 145-, 56-, and 115-bp fragments (data not shown). The enzyme BceAI recognized two restriction sites on the wild-type and A2075G mutated sequences, producing three fragments of 41, 24, and 251 bp (Fig. 2, lanes 6 and 7). Mutation A2074C created an additional recognition site, leading to the occurrence of an additional cleavage at position 2086 of the 23S rRNA gene. Consequently, digestion of the PCR product with BceAI resulted in an additional cleavage of the 251-bp fragment into two subproducts of 153 and 98 bp (Fig. 2, lanes 8 and 9). These two subproducts were also detected on a 3% agarose standard gel compared to the 251-bp nonhydrolyzed remaining product identified in the other amplified sequences (data not shown). Campylobacter spp. contain three copies of the rRNA genes (the rrnB operon), and a recent study reported that some C. coli strains of animal origin have different copies of the 23S rRNA gene containing either mutated (A2075G) and nonmutated copies in resistant isolates (6). All of the strains tested in this study were homozygous for the A2074C and A2075G mutations. In the case of heterozygous strains containing the A2075G mutation, restriction with BsaI would lead to an uncut 316-bp PCR product in addition to the 201- and 115-bp bands corresponding to the digested amplicon containing the A2075G mutation (data not shown). When digested with BceAI, a heterozygous strain containing the A2074C mutation would lead to an additional 251-bp product in addition to the 153-, 98-, 41-, and 24-bp bands corresponding to the digested amplicon containing the A2074C mutation (data not shown).
FIG. 2.
PCR-restriction fragment length polymorphism patterns obtained after digestion with BsaI and BceAI. The restriction products were analyzed by electrophoresis on a 10% polyacrylamide gel stained with ethidium bromide. Wild-type C. jejuni reference strain NCTC 11168 and strains 00072, 01206, and 00039, with mutations A2075G, A2074C, and both A2074C and A2075G occurring in domain V of the 23S rRNA gene, were used. Lanes: M, 25-bp DNA Step Ladder molecular size markers (Promega); 1, nondigested PCR products of wild-type C. jejuni strain NCTC 11168; 2 to 5, BsaI-digested PCR products of the wild-type and A2075G, A2074C, and A2074C-A2075G mutant C. jejuni strains, respectively; 6 to 9, BceAI-digested PCR products of the wild-type, A2075G, A2074C, and A2074C-A2075G C. jejuni strains, respectively.
These results confirm that the 23S rRNA mutation that is most frequently found in macrolide-resistant Campylobacter strains is A2075G. It was found in half of the resistant C. jejuni and C. coli strains tested in Japan (11). In this report, we describe for the first time the occurrence of another mutation, the transversion A2074C, as well as the presence of both mutations in one strain. A technique that is easy to carry out in most laboratories is also described. This is an alternative to the line probe assay that has already been described, which detects only half of the mutations occurring in Japanese strains (11). While the rate of macrolide resistance in clinical isolates (≤5%) is not yet alarming (8, 10, 14), a trend toward an increase has been noted both in animal strains that are potentially transmissible to humans via food and in human strains during macrolide treatments. Therefore, the study and monitoring of this resistance have, in turn, become increasingly necessary.
Nucleotide sequence accession numbers.
The partial 23S rRNA gene sequences of strains 98054 (464 bp), 00251 (457 bp), 98178 (464 bp), 00264 (457 bp), 01190 (461 bp), 89556 (451 bp), 98010 (410 bp), 00072 (456 bp), 00123 (464 bp), 00222 (464 bp), 00135 (464 bp), 95089 (344 bp), 99252 (421 bp), 98212 (420 bp), 01206 (394 bp), 95064 (384 bp), and 00039 (423 bp) have been submitted to the GenBank database and assigned accession numbers AY190985 to AY191001, respectively.
REFERENCES
- 1.Aarestrup, F. M., E. M. Nielsen, M. Madsen, and J. Engberg. 1997. Antimicrobial susceptibility patterns of thermophilic Campylobacter spp. from humans, pigs, cattle, and broilers in Denmark. Antimicrob. Agents Chemother. 41:2244-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chuma, T., T. Ikeda, T. Maeda, H. Niwa, and K. Okamoto. 2001. Antimicrobial susceptibilities of Campylobacter strains isolated from broilers in the southern part of Japan from 1995 to 1999. J. Vet. Med. Sci. 63:1027-1029. [DOI] [PubMed] [Google Scholar]
- 3.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2002. Concentrations, diamètres critiques et règles de lecture interprétative pour Campylobacter spp., p 44. Comité de l'Antibiogramme de la Société Française de Microbiologie, Paris, France.
- 4.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funke, G., R. Baumann, J. L. Penner, and M. Altwegg. 1994. Development of resistance to macrolide antibiotics in an AIDS patient treated with clarithromycin for Campylobacter jejuni diarrhea. Eur. J. Clin. Microbiol. Infect. Dis. 13:612-615. [DOI] [PubMed] [Google Scholar]
- 6.Jensen, L. B., and F. M. Aarestrup. 2001. Macrolide resistance in Campylobacter coli of animal origin in Denmark. Antimicrob. Agents Chemother. 45:371-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lastovica, A. J., and M. B. Skirrow. 2000. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli, p. 89-120. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 8.Mégraud, F. 1999. Les infections à Campylobacter en France (1986-1997). Bull. Epidemiol. Ann. 2:83-84. [Google Scholar]
- 9.Ménard, A., M. Oleastro, A. Santos, and F. Mégraud. 2002. PCR-restriction fragment length polymorphism can also detect point mutation A2142C in the 23S rRNA gene, associated with Helicobacter pylori resistance to clarithromycin. Antimicrob. Agents Chemother. 46:1156-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore, J. E., M. Crowe, N. Heaney, and E. Crothers. 2001. Antibiotic resistance in Campylobacter spp. isolated from human faeces (1980-2000) and foods (1997-2000) in Northern Ireland: an update. J. Antimicrob. Chemother. 48:455-457. [DOI] [PubMed] [Google Scholar]
- 11.Niwa, H., T. Chuma, K. Okamoto, and K. Itoh. 2001. Rapid detection of mutations associated with resistance to erythromycin in Campylobacter jejuni/coli by PCR and line probe assay. Int. J. Antimicrob. Agents 18:359-364. [DOI] [PubMed] [Google Scholar]
- 12.Occhialini, A., M. Urdaci, F. Doucet-Populaire, C. M. Bébéar, H. Lamouliatte, and F. Mégraud. 1997. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob. Agents Chemother. 41:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. Quail, M. A. Rajandream, K. M. Rutherford, A. VanVliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez, R., V. Fernandez-Baca, M. D. Diaz, P. Muñoz, M. Rodriguez-Creixems, and E. Bouza. 1994. Evolution of susceptibilities of Campylobacter spp. to quinolones and macrolides. Antimicrob. Agents Chemother. 38:1879-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 16.Taylor, D. E., Z. Ge, D. Purych, T. Lo, and K. Hiratsuka. 1997. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob. Agents Chemother. 41:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trieber, C. A., and D. E. Taylor. 2001. 23S rRNA mutations and macrolide resistance in Campylobacter. Int. J. Med. Microbiol. 291(Suppl. 31):5. [Google Scholar]
- 18.Van Looveren, M., G. Daube, L. De Zutter, J. M. Dumont, C. Lammens, M. Wijdooghe, P. Vandamme, M. Jouret, M. Cornelis, and H. Goossens. 2001. Antimicrobial susceptibilities of Campylobacter strains isolated from food animals in Belgium. J. Antimicrob. Chemother. 48:235-240. [DOI] [PubMed] [Google Scholar]
- 19.Versalovic, J., M. S. Osato, K. Spakovsky, M. P. Dore, R. Reddy, G. G. Stone, D. Shortridge, R. K. Flamm, S. K. Tanaka, and D. Y. Graham. 1997. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J. Antimicrob. Chemother. 40:283-286. [DOI] [PubMed] [Google Scholar]
- 20.Versalovic, J., D. Shortridge, K. Kibler, M. V. Griffy, J. Beyer, R. K. Flamm, S. Ken Tanaka, D. Y. Graham, and M. F. Go. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40:477-480. [DOI] [PMC free article] [PubMed] [Google Scholar]