Abstract
This study investigated macrolide resistance mechanisms in clinical Haemophilus influenzae strains with different levels of susceptibility to macrolides. A total of 6,382 isolates were collected during the Alexander Project from 1997 to 2000. For 96.9% of these isolates, the azithromycin MICs were 0.25 to 4 μg/ml, and these were defined as baseline strains. For 1.8% of the isolates, the azithromycin MICs were lower (<0.25 μg/ml), and for 1.3% of the isolates, the MICs were higher (>4 μg/ml). These isolates were defined as hypersusceptible and high-level macrolide-resistant strains, respectively. To identify the mechanisms associated with these three susceptibility patterns, representative strains were studied for the presence of macrolide efflux pumps and for ribosomal alterations. Macrolide efflux was studied by measuring the accumulation of radioactive azithromycin and clarithromycin in the presence or absence of carbonyl cyanide m-chlorophenylhydrazone (CCCP), a protonophore. Treatment with CCCP increased the accumulation of macrolides in baseline as well as high-level resistant strains, demonstrating the presence of an efflux mechanism, but not in the 20 hypersusceptible strains tested. Among the 31 strains studied that showed high-level resistance to both azithromycin and clarithromycin, 28 had ribosomal alterations, 7 had mutations in ribosomal protein L4, 11 had mutations in L22, 2 had mutations in 23S rRNA, 8 had multiple mutations, and 3 had no mutations. From these results, we conclude that the vast majority (>98%) of H. influenzae strains have a macrolide efflux mechanism, with a few of these being hyperresistant (1.3%) due to one or several ribosomal mutations. Occasional hypersusceptible strains (1.8%) were found and had no macrolide resistance mechanisms and appeared to be the only truly macrolide-susceptible variants of H. influenzae.
Haemophilus influenzae strains are often associated with community-acquired respiratory tract infections. These bacteria are the main cause of acute exacerbations of chronic bronchitis (17) and are the second most common cause of community-acquired pneumonia (8), sinusitis (19), and otitis media (5). Nontypeable H. influenzae strains are associated with these respiratory tract infections, while the prevalence of bacteremia and meningitis due to H. influenzae has decreased in countries such as the United States that use vaccines against type b H. influenzae (2). Although macrolides have gained wide clinical acceptance for treatment of H. influenzae infections, the results of pharmacokinetic-pharmacodynamic, experimental, and double-tap otitis media studies cast doubt on their efficacy against these strains (10).
Macrolide resistance is mainly due to target modification or active efflux and rarely to antibiotic inactivation. Target site modification is achieved by methylation of specific residues in 23S rRNA (11), by specific methylases encoded by the erm class of genes, or by different mutations in 23S rRNA and ribosomal proteins L4 and L22 (3, 21) in certain bacterial species.
Macrolides are inhibitors of protein synthesis, but they are not active against many species of gram-negative bacteria. The ribosomes from these strains are susceptible to macrolides, but decreased cell membrane permeability and/or multidrug efflux pumps make macrolides inactive against these gram-negative bacteria (24). H. influenzae, a gram-negative rod, is more susceptible to macrolides than are other gram-negative bacteria; however, the level of susceptibility is less than those of macrolide-susceptible, gram-positive bacteria. High-level resistance to macrolides in H. influenzae, with macrolide MICs higher than current NCCLS breakpoints, is currently rare (10). The prevalences of azithromycin and clarithromycin resistance among clinical isolates have been reported to be 0.5 and 1.9%, respectively (7).
Macrolide resistance due to acquired efflux pumps among clinically significant pathogens such as Streptococcus pneumoniae, Streptococcus pyogenes, and Staphylococcus aureus has been reported (12). Among gram-negative bacteria, intrinsic macrolide resistance is due to impermeability or the active efflux of these antibiotics. In Escherichia coli, the acrAB gene cluster has been found to be responsible for a macrolide efflux pump associated with resistance. The acrAB gene clusters have recently been found in H. influenzae, and inactivation of either one of these genes has been reported to cause hypersusceptibility to some drugs such as macrolides, as well as to dyes such as ethidium bromide (18).
The aim of this study was to correlate the macrolide susceptibility of clinical isolates of H. influenzae with mechanisms of macrolide resistance.
MATERIALS AND METHODS
Bacterial strains and chemicals.
The macrolide susceptibilities of 6,382 clinical H. influenzae isolates tested within the Alexander Project from 1997 to 2000 were analyzed, and isolates with various phenotypic features were selected for further investigation (for details, see Results). The strains selected were recovered from storage at −70°C and subcultured three times before being studied. CCCP (carbonyl cyanide m-chlorophenylhydrazone) was purchased from Sigma (St. Louis, Mo.). CCCP was chosen to represent a protonophore whose addition to the cell results in instantaneous dissipation of the electrochemical gradient of protons across the cytoplasmic membrane. Radioactive clarithromycin ([6-O-methyl-3H]clarithromycin; 350 mCi/mmol; 2.14 mg/ml) was obtained from Moravek Biochemicals, Inc. (Brea, Calif.), and radioactive azithromycin ([N-methyl-3H]azithromycin; 3.5Ci/mmol) was obtained from Perkin-Elmer Life Sciences, Inc. (Boston, Mass.). [N-methyl-3H] azithromycin was diluted 10-fold with unlabeled azithromycin to reach a specific activity of 350 mCi/mmol, 2.22 mg/ml, before use.
Susceptibility determinations.
Azithromycin and clarithromycin MICs for the strains studied were retested by the NCCLS microdilution method using freshly prepared Haemophilus test medium (16). Inoculum was prepared from chocolate agar plates incubated for a full 24 h. The standard quality control strains H. influenzae ATCC 49766 and H. influenzae ATCC 49247 were used as controls on each day of testing. Inoculum checks were done, and only suspensions yielding 3 × 105 to 7 × 105 CFU/ml were used. Microdilution trays were incubated at 35°C for 20 to 24 h in ambient air.
DNA amplification and sequencing.
Strains were studied for the presence of genes conferring resistance to macrolides [erm(A), erm(B), mef(A), and ere(A)], as described previously by Sutcliffe et al. (20), and for mutations in the genes coding for ribosomal proteins L4 and L22 and domain V of 23S rRNA. Nucleotide sequences of 23S rRNA, L4, and L22 ribosomal genes were obtained from the website of The Institute for Genomic Research (http://www.tigr.org) by using the total genome sequence of H. influenzae Rd, and specific primers were designed for this purpose (Table 1). The PCR conditions were as follows: 94°C for 5 min for 1 cycle, 94°C for 30 s, 53°C for 30 s, 72°C for 45 s for 35 cycles, and 72°C for 7 min for 1 cycle. The PCR products were purified by using a QIAquick PCR purification kit (QIAGEN, Valencia, Calif.) and were sequenced by using an Applied Biosystems model 373 DNA sequencer.
TABLE 1.
Specific primers used for PCR and their positions
| Gene product | Primer name | 5′ primer positiona | Sequence | Product size (bp) |
|---|---|---|---|---|
| 23S rRNA | 23S-3 | 1902 | 5′-CGGCGGCCGTAACTATAACG | 1,001 |
| 23S-4 | 2902 | 5′-TTGGATAAGTCCTCGAGCTATT | ||
| HF2330 | 2331 | 5′-GTATAAGCAAGCTTAACTG | 441 | |
| HF2771 | 2770 | 5′-CAAGTTTCGTGCTTAGATG | ||
| L4 | L4-1 | −38 | 5′-TTAAGCCGGCAGTTAAAGC | 662 |
| L4-2 | +21 | 5′-CACTTAGCAAACGTTCTTG | ||
| L22 | L22-1 | −45 | 5′-CGGCAGATAAGAAAGCTAAG | 296 |
| L22-2 | +38 | 5′-TGGATGTACTTTTTGACCC |
23S rRNA primer positions are expressed in terms of the E. coli 23S rRNA numbering system, and those for ribosomal protein L4 and L22 bases are expressed relative to start and stop codons.
Efflux assays.
Efflux of macrolide antibiotics was determined indirectly by measuring the accumulation of radioactive [6-O-methyl-3H] clarithromycin and [N-methyl-3H]azithromycin. The accumulation was done as described by Wondrack et al., with slight modifications as described below (23). Forty milliliters of freshly made HTM Haemophilus test medium was inoculated with 2 ml of an overnight culture of the selected strain. After the optical density at 600 nm reached 0.2 to 0.4, half of the culture was exposed to CCCP (25 μg/ml) for 10 min, and 0.2 μg of radioactive antibiotic was then added to both samples. Four milliliters of culture was removed from each sample at 5, 10, 20, and 30 min and filtered through a Whatman GF/C glass microfiber filter previously wetted with saline containing 1 μg of appropriate unlabeled macrolide antibiotic/ml. Filters were washed twice with saline macrolide mixture (total of 10 ml per filter) and were dried at room temperature. Radioactivity was then determined by liquid scintillation counting.
RESULTS
Clinical strain analysis and macrolide resistance phenotypes.
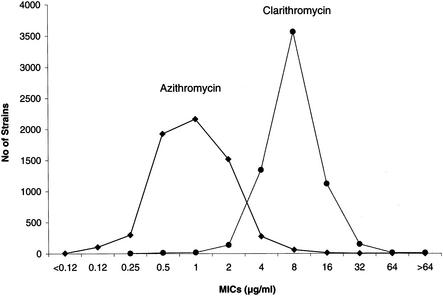
A total of 6,382 H. influenzae clinical isolates were collected during a 4-year period. Analysis of their susceptibilities to macrolide showed unimodal distributions of MICs of azithromycin and clarithromycin (Fig. 1). The mode was 1 μg/ml for azithromycin and 8 μg/ml for clarithromycin. When NCCLS breakpoints for azithromycin (≤4 μg/ml, susceptible; >4 μg/ml, nonsusceptible) and clarithromycin (≤8 μg/ml, susceptible; 16 μg/ml, intermediate; >16 μg/ml, resistant) were applied, 1.3 and 2.5% of isolates, respectively, were found to be resistant (Table 2). For 6,187 (96.9%) of the 6,382 isolates tested, the azithromycin MICs were 0.25 to 4 μg/ml, and these isolates were defined as baseline strains. For 113 (1.8%) isolates, the MICs were lower (<0.25 μg/ml), and these were defined as hypersusceptible strains; for 82 (1.3%) strains, the MICs were higher (>4 μg/ml), and these were defined as high-level macrolide-resistant strains. Twenty baseline, 20 hypersusceptible, and 49 high-level resistant strains were selected for further investigation. Of these 49 strains, 31 were resistant to both azithromycin and clarithromycin and 18 were resistant to clarithromycin but susceptible to azithromycin.
FIG. 1.
Distribution of macrolide MICs for 6,382 H. influenzae isolates from the Alexander Project (1997 to 2000).
TABLE 2.
Macrolide susceptibility among 6,382 H. influenzae isolates
| Drug | No. of strains
|
||
|---|---|---|---|
| Hypersusceptiblea | Baselineb | High-level resistantc | |
| Azithromycin | 113 (1.8%) | 6,187 (96.9%) | 82 (1.3%) |
| Clarithromycin | 37 (0.6%) | 6,185 (96.9%) | 160 (2.5%) |
Azithromycin MICs of <0.25 μg/ml; clarithromycin MICs of <2 μg/ml.
Azithromycin MICs of 0.25 to 4 μg/ml; clarithromycin MICs of 2 to 16 μg/ml.
Azithromycin MICs of >4 μg/ml; clarithromycin MICs of >16 μg/ml.
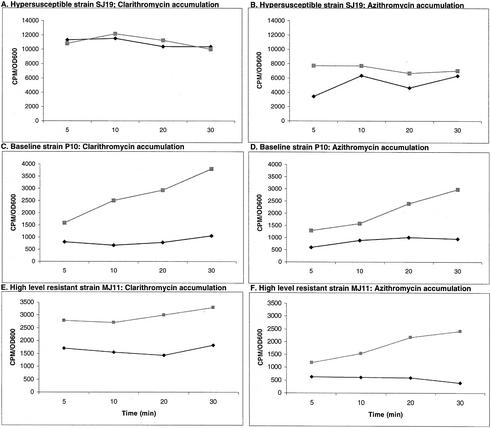
Macrolide accumulation in 20 hypersusceptible, 20 baseline, and 20 of 31 high-level azithromycin- and clarithromycin-resistant strains was determined. By contrast, all 49 high-level resistant strains were tested for ribosomal alterations. Figure 2 shows the levels of accumulation of radioactive azithromycin and clarithromycin in representative strains from each susceptibility group. In hypersusceptible strains, macrolide accumulation levels were high without CCCP treatment, which indicates the absence of a macrolide efflux mechanism. In baseline or high-level macrolide-resistant strains, initially low levels of macrolide accumulation became higher after CCCP treatment. This indicates the presence of a macrolide efflux mechanism in these strains.
FIG. 2.
Accumulation of radioactive clarithromycin (A, C, and E) and azithromycin (B, D, and F) in representative strains in the presence (▪) or absence (⧫) of CCCP. CCCP treatment increases radioactive antibiotic accumulation in baseline and high-level macrolide-resistant strains, while it had no effect in the hypersusceptible strain because the accumulation level was already high. OD600, optical density at 600 nm.
The mean values for accumulation of radioactive azithromycin and clarithromycin for all 60 strains tested are shown in Table 3. Accumulation was expressed as the ratio between the radioactive counts with CCCP and those without CCCP after 30 min of exposure to radioactive antibiotic, and 1 was subtracted from the ratio to normalize the no-change value to 0. The means of this ratio for hypersusceptible isolates were 0.04 and 0.13 for clarithromycin and azithromycin, respectively; for baseline strains, the mean values were 0.88 and 1.43, respectively, and for high-level resistant strains, they were 0.88 and 2.48, respectively. Treatment with CCCP therefore significantly increased macrolide accumulation in baseline and high-level macrolide-resistant strains but not in hypersusceptible strains. These differences were statistically significant for baseline versus hypersusceptible strains, as well as for high-level resistant versus hypersusceptible strains (P < 0.001 for azithromycin and clarithromycin; Student's t test). There was no difference between the clarithromycin accumulation values for baseline and high-level resistant strains (P = 0.97). A difference was observed in the azithromycin accumulation values between baseline and high-level resistant strains (P = 0.01). This observation may indicate the probable importance of an efflux pump(s) in strains with high-level resistance to azithromycin. The possibility of overexpression of proteins involved in macrolide efflux in these strains cannot be excluded and is under investigation.
TABLE 3.
Accumulation of radioactive clarithromycin and azithromycin among H. influenzae isolates with different susceptibility patterns
| Drug and susceptibility | Accumulation [(cpm with CCCP/cpm without CCCP) −1]
|
||
|---|---|---|---|
| Mean | Minimum | Maximum | |
| Clarithromycin | |||
| Hypersusceptible | 0.04 | −0.26 | 0.39 |
| Baseline | 0.88 | 0.39 | 3.12 |
| High-level resistant | 0.88 | 0.36 | 1.64 |
| Azithromycin | |||
| Hypersusceptible | 0.13 | −0.44 | 0.39 |
| Baseline | 1.43 | 0.38 | 2.94 |
| High-level resistant | 2.48 | 0.90 | 6.18 |
Macrolide resistance mechanisms in high-level resistant isolates.
The 31 strains with high-level resistance to both azithromycin and clarithromycin as well as the 18 strains which were resistant to clarithromycin but susceptible to azithromycin based on NCCLS breakpoints were studied for ribosomal mutations and the presence of macrolide resistance genes. Amplifications by PCR with specific primers for the erm(A), erm(B), mef(A), or ere(A) gene were negative for all of these strains. Among the 18 strains with high-level resistance to clarithromycin only, no mutations were detected in studied portions of 23S rRNA or ribosomal proteins L4 and L22. Twenty-eight of the 31 strains for which the azithromycin and clarithromycin MICs were high had modifications in 23S rRNA and/or ribosomal proteins L4 and L22. Two strains had a replacement of adenine by guanine at position 2058 (A2058G) of 23S rRNA. One of these mutants had an additional G2160U change. The MICs of azithromycin and clarithromycin for these strains were >64 μg/ml (Table 4). Seven isolates had mutations in ribosomal protein L4 only. Six of these strains had point mutations: one strain had a K61Q change, one had a T64K change, one had an A69S change, one had a T82I change, and two had G → D changes at position 65. An insertion after position 63, involving duplication of two amino acids (GT), was observed in the sixth strain. For the L4 mutant strains, the range of azithromycin MICs was 8 to 64 μg/ml and the range of clarithromycin MICs was 32 to >64 μg/ml (Table 4).
TABLE 4.
MICs for and detected mutations in 31 high-level macrolide-resistant H. influenzae isolates
| Strain | MIC (μg/ml) of:
|
Mutation(s)a
|
|||
|---|---|---|---|---|---|
| Azithromycin | Clarithromycin | L4 | L22 | 23S rRNA | |
| S30 | >64 | >64 | A2058G | ||
| S43 | >64 | >64 | A2058G, G2160U | ||
| S4 | 8 | 64 | INS 63GT | ||
| S27 | 16 | >64 | G65D | ||
| S37 | 16 | 64 | K61Q | ||
| S39 | 16 | 64 | T64K | ||
| S47 | 8 | 64 | G65D | ||
| S50 | 32 | 32 | A69S | ||
| S54 | 64 | 64 | T82I | ||
| S1 | 32 | >64 | INS 77DEGPSM | ||
| S2 | 8 | 16 | G91D | ||
| S3 | 8 | 16 | G91D | ||
| S23 | 8 | 32 | DEL 96ILK | ||
| S26 | 32 | >64 | INS 88RAKG | ||
| S40 | 64 | >64 | DEL 95RI | ||
| S42 | 8 | 32 | DEL 81S | ||
| S49 | 64 | 32 | INS 91KG | ||
| S53 | >64 | 64 | DEL 82M | ||
| S58 | >64 | >64 | INS 91RAG | ||
| S60 | 16 | 16 | INS 91RADR | ||
| S34 | >64 | >64 | DEL 95RI | GGA2160-2162UAU | |
| S36 | 16 | >64 | D139G | G2160U | |
| S44 | >64 | >64 | T64K | G91D | |
| S57 | >64 | >64 | T64K | G91D | |
| S61 | >64 | >64 | D94E | DEL 96ILKR | |
| S35 | 32 | >64 | T64K | G91D | C2164G |
| S38 | 16 | 64 | T64K | G91D | C2164G |
| S41 | 8 | 32 | T64K | G91D | C2164G |
| S51 | 32 | >64 | |||
| S52 | >64 | >64 | |||
| S59 | 32 | >64 | |||
INS, insertion; DEL, deletion. Position numbers for L4 and L22 are based on the H. influenzae numbering system, and those for 23S rRNA are based on the E. coli numbering system.
Alteration of ribosomal protein L22 alone by amino acid insertion, deletion, or substitution was detected in 11 isolates. For the strains with L22 mutations, the range of azithromycin MICs was 8 to >64 μg/ml and the range of clarithromycin MICs was 16 to >64 μg/ml (Table 4). Eight isolates had multiple mutations. One had a deletion in L22 and substitution of three bases in 23S rRNA. One isolate had changes in both L4 and 23S rRNA. Three isolates had alterations in ribosomal proteins L4 and L22. Three strains had mutations in L4, L22, and 23S rRNA. For strains with multiple mutations, the azithromycin MICs ranged from 8 to >64 μg/ml and the clarithromycin MICs ranged from 32 to >64 μg/ml (Table 4). The detected mutations were in a highly conserved region of ribosomal proteins. Among strains with multiple mutations, the 23S rRNA mutations were in positions 2160 to 2164 (E. coli numbering system). This region is not in the peptidyltransferase center but is proximal to the E site (25).
No mutation was found in the remaining three strains for which the azithromycin and clarithromycin MICs were high.
DISCUSSION
Three macrolide susceptibility groups were defined among clinical H. influenzae isolates, and the resistance mechanisms involved in the different susceptibility groups were studied. The hypersusceptible strains tested in this study were characterized by the absence of a macrolide efflux mechanism that was present in baseline and high-level macrolide-resistant strains. While the high-level macrolide-resistant strains lacked any known acquired macrolide resistance genes, most had alterations in ribosomal proteins L4 and L22 and/or ribosomal 23S rRNA. Macrolide-resistant S. pneumoniae and S. pyogenes strains with ribosomal alterations have been reported recently (1, 6, 15). H. influenzae laboratory mutants with high-level macrolide resistance, produced by exposure to azithromycin or clarithromycin, have been shown to be associated with mutations in 23S rRNA or ribosomal proteins L4 or L22 (4). By using transformation studies, Clark et al. have shown the importance of the alteration of ribosomal proteins L22 and L4 in the conferment of macrolide resistance (4).
Many gram-negative bacteria show diminished macrolide accumulation (24), which makes macrolide antibiotics of little value for treatment of infections caused by these species. For example, Enterobacteriaceae have a macrolide efflux pump called Acr, which is encoded by acrAB genes (24). Sanchez et al. recently described a gene cluster in H. influenzae with homology to the acrAB gene cluster of E. coli and have shown that inactivation of the acrAB gene cluster resulted in laboratory mutants that became susceptible to erythromycin, novobiocin, ethidium bromide, and crystal violet (18). The present study has confirmed the existence of a macrolide efflux mechanism in baseline clinical strains of H. influenzae and has demonstrated the existence of a small percentage of hypersusceptible clinical strains of H. influenzae which do not have a macrolide efflux mechanism. The protein(s) involved in macrolide efflux in the clinical strains studied is currently under investigation.
The rates of susceptibility to azithromycin and clarithromycin differ depending on whether NCCLS breakpoints (16) or breakpoints derived from pharmacokinetic-pharmacodynamic parameters (9), which are based on correlation of serum concentrations of antibiotic with bacteriologic outcome of infections, are used. Jacobs et al. (10) have shown that when NCCLS breakpoints are used, the majority of the isolates that they tested could be categorized as being susceptible to azithromycin and clarithromycin (99.7 and 76.6%, respectively). However, with pharmacokinetic-pharmacodynamic breakpoints, virtually no strains were categorized as being susceptible, in agreement with the results of bacteriologic outcome studies of otitis media due to H. influenzae (5), reports of breakthrough bacteremia in patients infected with strains of S. pneumoniae with efflux-mediated macrolide resistance (13), and animal models of H. influenzae infection (14, 22). Interestingly, hypersusceptible strains without an efflux mechanism are categorized as being susceptible when pharmacokinetic-pharmacodynamic breakpoints are used, but no clinical or animal model studies have been conducted to evaluate such strains.
High-level macrolide-resistant clinical H. influenzae strains are rare, but most of the strains tested in our study were shown to have two resistance mechanisms: the macrolide efflux mechanism found in baseline strains and ribosomal mutations. The necessity for accumulation of two resistance mechanisms may explain the rarity of high-level macrolide-resistant H. influenzae strains, as baseline strains are intrinsically resistant to macrolides. However, the present study shows that resistance mechanisms generated in laboratory mutants (4) can be detected among clinical isolates.
In summary, a macrolide efflux mechanism, with or without additional ribosomal mutation, was detected in all of the baseline and high-level resistant strains of H. influenzae tested, while the strains defined as hypersusceptible had no detectable macrolide resistance mechanism. Demonstration of a macrolide resistance mechanism in the majority of baseline strains regarded as being susceptible by current NCCLS criteria, in conjunction with macrolide pharmacokinetic-pharmacodynamic data and the results of clinical studies, indicates that breakpoints for macrolide resistance require reevaluation.
Acknowledgments
This study was supported by a grant from GlaxoSmithKline, Collegeville, Pa.
REFERENCES
- 1.Bingen, E., R. Leclercq, F. Fitoussi, N. Brahimi, B. Malbruny, D. Deforche, and R. Cohen. 2002. Emergence of group A streptococcus strains with different mechanisms of macrolide resistance. Antimicrob. Agents Chemother. 46:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, S. B., H. R. Shinefield, B. Fireman, and R. Hiatt. 1992. Safety, immunogenicity, and efficacy in infancy of oligosaccharide conjugate Haemophilus influenzae type b vaccine in a United States population: possible implications for optimal use. J. Infect. Dis. 165(Suppl. 1):S139-S143. [DOI] [PubMed] [Google Scholar]
- 3.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, C. L., B. Bozdogan, M. Peric, M. R. Jacobs, and P. C. Appelbaum. 2002. In vitro selection of resistance in Haemophilus influenzae by amoxicillin-clavulanate, cefpodoxime, cefprozil, azithromycin, and clarithromycin. Antimicrob. Agents Chemother. 46:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dagan, R., C. E. Johnson, S. McLinn, N. Abughali, J. Feris, E. Leibovitz, D. J. Burch, and M. R. Jacobs. 2000. Bacteriologic and clinical efficacy of amoxicillin/clavulanate vs. azithromycin in acute otitis media. Pediatr. Infect. Dis. J. 19:95-104. [DOI] [PubMed] [Google Scholar]
- 6.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doern, G. V., A. B. Brueggemann, G. Pierce, H. P. Holley, Jr., and A. Rauch. 1997. Antibiotic resistance among clinical isolates of Haemophilus influenzae in the United States in 1994 and 1995 and detection of beta-lactamase-positive strains resistant to amoxicillin-clavulanate: results of a national multicenter surveillance study. Antimicrob. Agents Chemother. 41:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotfried, M. H. 2001. Epidemiology of clinically diagnosed community-acquired pneumonia in the primary care setting: results from the 1999-2000 respiratory surveillance program. Am. J. Med. 111(Suppl. 9A):25S-29S. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs, M. R. 2001. Optimisation of antimicrobial therapy using pharmacokinetic and pharmacodynamic parameters. Clin. Microbiol. Infect. 7:589-596. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, M. R., S. Bajaksouzian, A. Zilles, G. Lin, G. A. Pankuch, and P. C. Appelbaum. 1999. Susceptibilities of Streptococcus pneumoniae and Haemophilus influenzae to 10 oral antimicrobial agents based on pharmacodynamic parameters: 1997 USA surveillance study. Antimicrob. Agents Chemother. 43:1901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 12.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonks, J., J. Garau, L. Gomez, M. Xercavins, A. Ochoa De Echaguen, I. F. Gareen, P. T. Reiss, and A. A. Medeiros. 2002. Failure of macrolide antibiotic treatment in patients with bacteremia due to erythromycin resistant Streptococcus pneumoniae. Clin. Infect. Dis. 35:556-564. [DOI] [PubMed] [Google Scholar]
- 14.Mitten, M. J., J. Meulbroek, M. Nukkala, L. Paige, K. Jarvis, A. Oleksijew, A. Tovcimak, L. Hernandez, J. D. Alder, P. Ewing, Y. S. Or, Z. Ma, A. M. Nilius, K. Mollison, and R. K. Flamm. 2001. Efficacies of ABT-773, a new ketolide, against experimental bacterial infections. Antimicrob. Agents Chemother. 45:2585-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai, K., P. C. Appelbaum, T. A. Davies, L. M. Kelly, D. B. Hoellman, A. T. Andrasevic, L. Drukalska, W. Hryniewicz, M. R. Jacobs, J. Kolman, J. Miciuleviciene, M. Pana, L. Setchanova, M. K. Thege, H. Hupkova, J. Trupl, and P. Urbaskova. 2002. Susceptibilities to telithromycin and six other agents and prevalence of macrolide resistance due to L4 ribosomal protein mutation among 992 pneumococci from 10 central and Eastern European countries. Antimicrob. Agents Chemother. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Pfaller, M. A., A. F. Ehrhardt, and R. N. Jones. 2001. Frequency of pathogen occurrence and antimicrobial susceptibility among community-acquired respiratory tract infections in the respiratory surveillance program study: microbiology from the medical office practice environment. Am. J. Med. 111(Suppl. 9A):4S-12S. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez, L., W. Pan, M. Vinas, and H. Nikaido. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 179:6855-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokol, W. 2001. Epidemiology of sinusitis in the primary care setting: results from the 1999-2000 respiratory surveillance program. Am. J. Med. 111(Suppl. 9A):19S-24S. [DOI] [PubMed] [Google Scholar]
- 20.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tessier, P. R., M. K. Kim, W. Zhou, D. Xuan, C. Li, M. Ye, C. H. Nightingale, and D. P. Nicolau. 2002. Pharmacodynamic assessment of clarithromycin in a murine model of pneumococcal pneumonia. Antimicrob. Agents Chemother. 46:1425-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wondrack, L., M. Massa, B. V. Yang, and J. Sutcliffe. 1996. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob. Agents Chemother. 40:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann, R. A., C. L. Thomas, and J. Wower. 1990. Structure and function of rRNA in decoding domain and at the peptidyltransferase center, p. 331-347. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evaluation. American Society for Microbiology, Washington, D.C.