Abstract
The aminocoumarin resistance genes of the biosynthetic gene clusters of novobiocin, coumermycin A1, and clorobiocin were investigated. All three clusters contained a gyrBR resistance gene, coding for a gyrase B subunit. Unexpectedly, the clorobiocin and the coumermycin A1 clusters were found to contain an additional, similar gene, named parYR. Its predicted gene product showed sequence similarity with the B subunit of type II topoisomerases. Expression of gyrBR and likewise of parYR in Streptomyces lividans TK24 resulted in resistance against novobiocin and coumermycin A1, suggesting that both gene products are able to function as aminocoumarin-resistant B subunits of gyrase. Southern hybridization experiments showed that the genome of all three antibiotic producers and of Streptomyces coelicolor contained two additional genes which hybridized with either gyrBR or parYR and which may code for aminocoumarin-sensitive GyrB and ParY proteins. Two putative transporter genes, novA and couR5, were found in the novobiocin and the coumermycin A1 cluster, respectively. Expression of these genes in S. lividans TK24 resulted in moderate levels of resistance against novobiocin and coumermycin A1, suggesting that these genes may be involved in antibiotic transport.
The aminocoumarin antibiotics novobiocin, clorobiocin, and coumermycin A1 (Fig. 1A) are known as potent inhibitors of gyrase (18). Their equilibrium dissociation constants are in the range of 10 nM (10), i.e., their affinity for gyrase is considerably higher than that of modern fluoroquinolones. Novobiocin is licensed as an antibiotic for clinical use (Albamycin; Pharmacia-Upjohn) and is used for the treatment of infections with multiresistant gram-positive bacteria, e.g., Staphylococcus aureus.
FIG. 1.
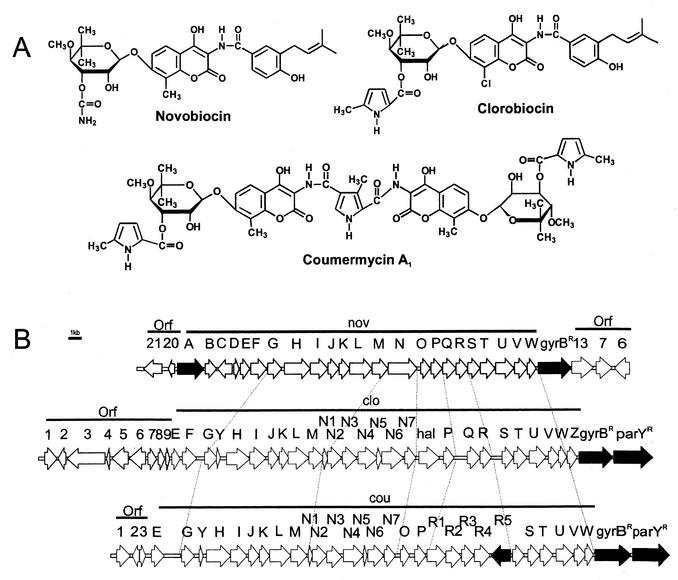
(A) Structures of the aminocoumarin antibiotics. (B) Gene clusters of novobiocin (top), clorobiocin (middle), and coumermycin A1 (bottom). Genes involved in aminocoumarin resistance are shown by solid arrows.
Novobiocin is produced by Streptomyces spheroides (synonym S. caeruleus [15]) NCIMB 11891, clorobiocin is produced by S. roseochromogenes var. oscitans DS12.976, and coumermycin A1 is produced by S. rishiriensis DSM 40489 (2). Obviously, these organisms must protect their gyrases from the inhibitory effect of aminocoumarin during antibiotic formation. Thiara and Cundliffe (29-31) reported that the principal resistance mechanism of the novobiocin producer S. sphaeroides is the de novo synthesis of a coumarin-resistant gyrase B subunit, which replaces the sensitive GyrB subunit in the active (GyrA)2(GyrB)2 heterotetramer. Thus, this novobiocin producer contains two gyrB genes, a constitutively expressed gyrBS, encoding the coumarin-sensitive protein, and the gyrBR gene, encoding the resistant protein and expressed in the presence of novobiocin. The promoter of gyrBR appears to be regulated by changes in the superhelical density of DNA (30).
Mitchell et al. (20) supplied evidence that additional genes may contribute to novobiocin resistance. They used the novobiocin producer S. niveus, which has recently been identified as a subjective synonym for S. spheroides (15).
We cloned and sequenced the novobiocin biosynthetic gene cluster (27), depicted in Fig. 1B. On its right border, the cluster contains the gyrBR resistance gene. Near the left border, the gene novA, encoding an ABC transporter, was identified. Méndez and Salas (19) suggested that novA is involved in transport of and possibly resistance against novobiocin. They classified the encoded protein as a type III ABC transporter, i.e., the ATP binding domain and the membrane domain are fused together on the same protein chain.
Recently our group has also cloned and sequenced the core regions of the biosynthetic gene clusters of coumermycin A1 (35) and of clorobiocin (23) (Fig. 1B). The present study was undertaken in order to identify and compare putative aminocoumarin resistance genes in the three aminocoumarin clusters.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this study are listed in Table 1. S. lividans TK24 was cultured at 28°C and 170 rpm for 2 to 4 days in baffled shake flasks in HA medium containing 1.0% malt extract, 0.4% yeast extract, 0.4% glucose, and 1.0 mM CaCl2 (pH 7.3). For preparing protoplasts of S. lividans TK24, CRM medium containing 10.3% sucrose, 2.0% tryptic soy broth, 1.0% MgCl2, 1.0% yeast extract, and 0.75% glycine (pH 7.0) was used. Streptomyces protoplasts were prepared and transformed as described before (14). Regeneration of protoplasts was carried out on R2YE medium (14). For selection of thiostrepton-resistant strains of S. lividans TK24, HA agar plates containing 50 μg of thiostrepton per ml were used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| Streptomyces lividans TK24 | Streptomycin resistant, no plasmids | 14 |
| Escherichia coli XL1 Blue MRF′ | Tetr, host strain for cloning experiments | Stratagene |
| Plamids and cosmids | ||
| Litmus 28 | Cloning vector, Ampr | NE BioLabs |
| Litmus 38 | Cloning vector, Ampr | NE BioLabs |
| pBluescript SK(−) | Cloning vector, Ampr | Stratagene |
| pGEM-3Zf(−) | Cloning vector, Ampr | Promega |
| pGEM-11Zf(+) | Cloning vector, Ampr | Promega |
| pUWL201 | E. coli-Streptomyces shuttle vector, Ampr TsrrermE∗p promoter | 27 |
| 10-9C | Cosmid isolated from S. spheroides genomic DNA library | 27 |
| p10-9CE2 | 4.3-kb EcoRI fragment from 10-9C containing gyrBR in pBluescript SK(−) | This work |
| 4-2H | Cosmid isolated from S. rishiriensis genomic DNA library | 35 |
| pMS63 | 11.8-kb NotI-HindIII fragment from novobiocin biosynthetic gene cluster containing novA in pBluescript SK(−) | This work |
| pZW10 | 12.75-kb BglII fragment (bp 16013 to 28761 from AF 235050) from coumermycin A1 biosynthetic gene cluster in pBluescript SK(−) | This work |
DNA isolation and manipulation.
Standard methods were used for DNA isolation and manipulation in Escherichia coli XL1 Blue MRF′ (25). DNA fragments were isolated from agarose gels with a QIAquick gel extraction kit according to the instructions of the manufacturer.
Construction of plasmids for heterologous gene expression in S. lividans TK24.
From the novobiocin biosynthetic gene cluster, gyrBR and novA were cloned for heterologous expression in S. lividans TK24. A 2.3-kb ApaI-PstI fragment from plasmid p10-9CE2 containing gyrBR was isolated and ligated into the same sites of Litmus 38. The resulting construct was digested with ApaI and EcoRI, and the 2.3-kb fragment was introduced into pGEM-11Zf(+). The insert of the plasmid obtained was excised with HindIII and EcoRI and cloned into the corresponding sites of pUWL201 to give the expression plasmid pGES1+.
novA was isolated from pMS63 by digestion with SnaBI and PstI (2.05 kb) and cloned into the EcoRV and PstI sites of pBluescript SK(−). After digestion with HindIII and PstI, novA was ligated into the corresponding sites of pUWL201. The resulting expression plasmid was named pTES3.
From the coumermycin A1 biosynthetic gene cluster, the three genes gyrBR, parYR, and couR5 were cloned for expression experiments. The 2.2-kb PvuII-MluI fragment of the cosmid 4-2H containing gyrBR was ligated into the EcoRV and BssHII sites of Litmus 28. The insert was excised with HindIII and SpeI and cloned into pUWL201 to give the expression plasmid pGES2. Cosmid 4-2H was also digested with BamHI and XhoI, and a 2 kb-fragment containing the C terminus of parYR was cloned into the same sites of pGEM-11Zf(+) to give pGES31. A fragment encoding the N-terminal region of ParYR was amplified by PCR with cosmid 4-2H as template. The synthetic oligonucleotides used for the amplification were primer gyrBX-1 (GCCCCTCTAGACGCGTGCGTGACCCAAAG) and primer gyrBX-2 (GATGACCTCGATGTGGTCGCAGGCAC); an XbaI site (underlined) was introduced into primer gyrBX-1. The PCR fragment was cloned into the XbaI and BamHI sites of pGES31. The plasmid containing the complete parYR gene was digested with HindIII and EcoRI, and the resulting fragment was ligated into the corresponding sites of pUWL201, forming the expression plasmid pGES3.
To generate a construct for coexpression of gyrBR and parXR, pGEM-11Zf(+) was digested with NsiI and treated with Klenow fragment, creating a blunt end. The vector was then digested with NotI and ligated with a 3.44-kb PvuII-NotI fragment isolated from cosmid 4-2H to give pGES41. Subsequently, a 0.87-kb NotI-XhoI fragment was also isolated from cosmid 4-2H and ligated into the corresponding sites of pGES41. The insert containing gyrBR and parYR was excised with HindIII and EcoRI and ligated into pUWL201 generating the expression construct pGES4.
couR5 was cloned into pGEM-3Zf(+) as a 1.56-kb SmaI-AccI fragment from pZW10. Then the insert was excised with HindIII and PstI and cloned into the same sites of pUWL201 to give the expression plasmid pTES4.
The constructs were introduced into S. lividans TK24, a Streptomyces strain which is very closely related to S. coelicolor and which is commonly used for expression experiments by protoplast transformation. Transformants were selected by thiostrepton resistance, and the presence of the intact expression construct was confirmed by plasmid isolation and restriction analysis (data not shown).
Novobiocin and coumermycin A1 susceptibility testing.
About 106 spores of S. lividans TK24 containing the expression plasmids pGES1+, pGES2, pGES3, pGES4, pTES3, or pTES4 were plated on minimal medium (14) containing 20 μg of thiostrepton per ml and different concentrations of novobiocin or coumermycin A1, respectively. Growth was determined after 6 days of incubation at 30°C.
Southern hybridization.
A 0.9-kb BglII fragment (bp 1012 to 1923 of AF205853) containing gyrBR and a 1.33-kb BglII-PvuII fragment (bp 3271 to 4600 of AF205853) containing a part of the parYR gene from S. rishiriensis were labeled with the Digoxigenin High Prime DNA labeling and detection starter kit II (Roche Applied Science, Mannheim, Germany) and used as probes for Southern blot analysis on Hybond-N nylon membranes (Amersham Biosciences, Freiburg, Germany). Genomic DNA from Streptomyces spheroides NCIMB 11891, S. roseochromogenes var. oscitans DS12.976, S. rishiriensis DSM 40489, and S. coelicolor A3(2) was isolated as described before (14).
Sequence analysis.
Double-stranded sequencing was performed by the dideoxynucleotide chain termination method on a LI-COR automatic sequencer (MWG-Biotech AG, Ebersberg, Germany).
Database searches were performed in the GenBank database with the BLAST 2.0 program. DNASIS (version 2.1, 1995; Hitachi Software Engineering) was used for computer-aided sequence analyses.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper were deposited in the GenBank nucleotide sequence database under accession numbers AF205854, AF205853, and AY136281.
RESULTS
Identification and sequence analysis of type II topoisomerase genes.
Our previous sequencing of the biosynthetic gene clusters of novobiocin, clorobiocin, and coumermycin A1 (23, 27, 35) revealed the presence of a gyrBR gene towards the right end of all three clusters (Fig. 1), each presumably coding for a coumarin-resistant GyrB subunit. We now extended the sequencing at the borders of the clusters (Fig. 1B). This revealed that both the coumermycin A1 and the clorobiocin clusters contained a gene immediately downstream of gyrBR which showed, on average, 44% identity and 57% similarity (amino acid level) to gyrBR of the same organism, suggesting that it may encode a type II topoisomerase B subunit containing a well-conserved ATP binding site. We suggest the name parYR for these two genes. No parYR homologue was found in the novobiocin cluster.
The promoter regions of gyrBR showed about 75% sequence identity (nucleotide level) between the three clusters, indicating that expression of gyrBR in all these strains may be regulated in a similar way, as described by Thiara and Cundliffe (30). The intergenic region between gyrBR and parYR comprised 79 bp and 80 bp in S. rishiriensis and S. roseochromogenes, respectively. Both organisms contained a well-conserved ribosome binding site (AGGAG) 8 bp upstream of the parYR start codon. No evidence was found for the presence of a transcription terminator or for sequence similarity to common bacterial promoters. This may indicate that gyrBR and parYR are transcribed as a single operon.
The sequence of gyrBR of S. spheroides identified in our study was not identical to that reported previously by Thiara and Cundliffe (EMBL Z17304). However, the sequence published by these authors and their restriction map given for the DNA region surrounding the gyrBR gene (31) were in excellent agreement with the gyrBR region of S. rishiriensis. We reconfirmed the identity of the strains used in our laboratory by ordering new strain samples from the respective culture collections and by identification of the antibiotics produced by all strains by high-pressure liquid chromatography and by mass spectroscopy in comparison to authentic standards. This procedure confirmed the identity of all our strains. The most plausible explanation for these findings is that the sequence which Thiara and Cundliffe (31) examined was derived from S. rishiriensis rather than from S. spheroides.
In the three clusters, the gyrBR genes were equal in size (coding for proteins of 677 amino acids) and showed a sequence identity of approximately 92% on the amino acid level. They showed, on average, 75% identity to the gyrB gene (SCO3874) of S. coelicolor A3(2), the genome of which has recently been sequenced, and 41% identity to the gyrB genes of other gram-positive bacteria, such as Bacillus subtilis, Staphylococcus aureus, and Clostridium perfringens (Fig. 2B).
FIG. 2.
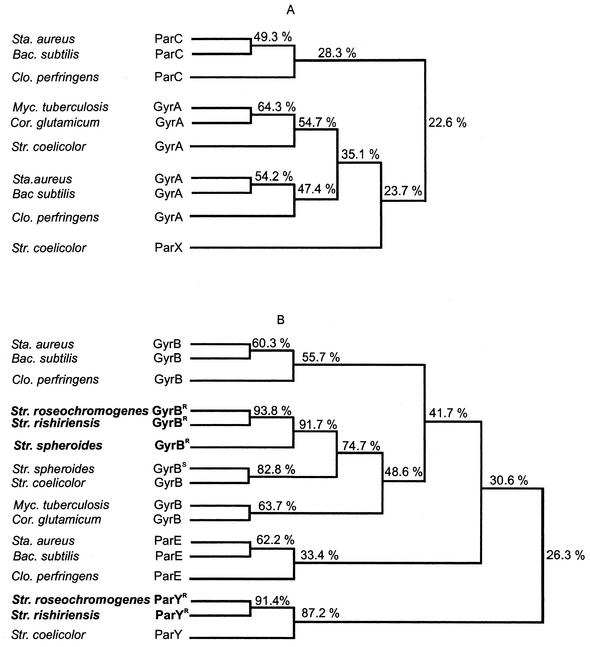
Phylogenetic trees based on sequence similarities of the genes coding for the A subunits (A) and the B subunits (B) of type II topoisomerases in gram-positive bacteria. Resistance genes of the aminocoumarin antibiotic producers are shown in boldface. The phylogeny was constructed with DNASIS for Windows, version 2 (Hitachi, San Bruno), scoring with a gap penalty of 5.0, a K-tuple of 2.0, a fixed gap penalty of 10.0, and a floating gap penalty of 10.0. The number of top diagonals and window size were both set 5. Bacterial strains: Sta., Staphylococcus; Bac., Bacillus; Clo., Clostridium; Myc., Mycobacterium; Cor., Corynebacterium; Str., Streptomyces.
The two parYR genes coded for proteins of 702 amino acids with 91% sequence identity. A very similar gene exists in the genome of S. coelicolor (87% identity, amino acid level), SCO5822, which has been annotated as a putative DNA gyrase subunit B in the database and which has not yet been functionally identified. This gene of S. coelicolor will be referred to as parY hereafter.
Many bacteria contain two type II topoisomerases, i.e., a gyrase encoded by the genes gyrA and gyrB and a topoisomerase IV encoded by the genes parC and parE (34). It is not possible to distinguish gyrase and topoisomerase IV unambiguously by characteristic sequence motifs, although sequence comparison with functionally identified topoisomerases in related organisms usually allows classification into one of the two groups. This was, however, not possible for the parY genes; in a sequence comparison to the known gyrB and parE genes of gram-positive bacteria (Fig. 2B), the parY genes formed a group on their own, with only around 26% amino acid sequence identity to the products of the gyrB and parE genes.
Expression of gyrBR and parYR genes.
In order to investigate the role of the gyrBR and the parYR genes in aminocoumarin resistance, we expressed gyrBR and parYR of S. rishiriensis in Streptomyces lividans TK24, with gyrBR from S. spheroides as the comparison. All three genes were cloned separately into the Streptomyces expression vector pUWL201 (see Materials and Methods), which contains the constitutive ermE*p promoter for foreign gene expression and a thiostrepton resistance marker. Table 2 shows the growth inhibition caused by different antibiotic concentrations in the transformants. In a control strain transformed with the empty vector pUWL201, complete growth inhibition was achieved with 100 μg of novobiocin and 50 μg of coumermycin A1 per ml. The higher antibacterial activity of coumermycin A1 compared to novobiocin has been reported previously (12, 22).
TABLE 2.
Growth of S. lividans TK24 harboring resistance genes from S. spheroides (Ss) and S. rishiriensis (Sr)
| Gene(s) | Growtha with novobiocin at (μg/ml):
|
Novobiocin MIC (μg/ml) | Growth with coumermycin A1 at (μg/ml):
|
Coumermycin A1 MIC (μg/ml) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 50 | 100 | 250 | 500 | 600 | 750 | 0 | 50 | 100 | 250 | 300 | 500 | |||
| Control (pUWL201) | +++++ | + | 0 | 0 | 0 | 0 | 0 | 100 | +++++ | 0 | 0 | 0 | 0 | 0 | 50 |
| Ss gyrBR | +++++ | +++++ | +++++ | +++++ | ++++ | +++ | + | >750 | +++++ | +++++ | ++++ | +++ | ++ | 0 | 500 |
| Sr gyrBR | +++++ | +++++ | +++++ | +++++ | ++++ | +++ | + | >750 | +++++ | +++++ | ++++ | +++ | ++ | 0 | 500 |
| Sr parYR | +++++ | +++++ | +++++ | ++++ | 0 | 0 | 0 | 500 | +++++ | +++++ | +++ | + | 0 | 0 | 300 |
| Sr gyrBR + parYR | +++++ | +++++ | +++++ | +++++ | ++++ | +++ | + | >750 | +++++ | +++++ | ++++ | +++ | ++ | 0 | 500 |
| Ss novA | +++++ | +++++ | ++ | + | 0 | 0 | 0 | 500 | +++++ | ++++ | + | 0 | 0 | 0 | 250 |
| Sr couR5 | +++++ | ++++ | ++ | 0 | 0 | 0 | 0 | 250 | +++++ | ++++ | + | 0 | 0 | 0 | 250 |
+++++, more than 3,000 colonies; ++++, 1,000 to 3,000 colonies; +++, 500 to 1,000 colonies; ++, 100 to 500 colonies; +, 10 to 100 colonies; 0, no growth observed. MIC, minimum concentration causing complete growth inhibition under the conditions of this experiment.
The gyrBR genes from the novobiocin producer S. spheroides and from the coumermycin A1 producer S. rishiriensis clearly provided resistance, with complete inhibition achieved with 500 μg of coumermycin A1 and >750 μg of novobiocin per ml. Both genes provided equally effective protection against novobiocin and against coumermycin A1, proving that they encode general aminocoumarin resistance rather than specific protection from either of these two structurally different antibiotics.
Likewise, the newly discovered parYR gene clearly provided resistance against both novobiocin and coumermycin A1, although the resistance level was somewhat lower than that produced by the gyrBR genes, with complete inhibition achieved with 300 μg of coumermycin A1 and 500 μg of novobiocin per ml (Table 2).
In an additional experiment, we placed the entire sequence comprising both gyrBR and parYR from S. rishiriensis into the pUWL201 expression vector. Upon expression in S. lividans TK24, this construct led to the same level of resistance as expression of gyrBR alone (Table 2). Since gyrBR and parYR are presumably transcribed as a single operon (see above), this result indicates that the effects of gyrBR and parYR expression are not additive or synergistic under the experimental conditions of our study.
A attempt to purify GyrBR and ParYR and to investigate their activity in vitro remained unsuccessful. Expression of fusion proteins of GyrBR and ParYR with N-terminal His6 tags from the pRSET B vector system in E. coli yielded high expression, but only in the form of insoluble inclusion bodies. Varying the cultivation temperature between 37°C and 15°C, the inducer concentrations (50 μM to 500 μM), and the time of induction did not help to alleviate this problem.
Obviously, gyrBR and parYR had been actively expressed from the pUWL201 constructs in S. lividans TK24, as demonstrated by the resulting aminocoumarin resistance. We therefore tried to purify the GyrBR and ParYR proteins from these strains by affinity chromatography on novobiocin-Sepharose and elution with different concentrations of KCl and urea (26, 29). This resulted in successful purification of the native coumarin-sensitive gyrase B subunit of S. lividans TK24, which could be reconstituted with the GyrA subunit to an enzymatically active, supercoiling enzyme. However, we did not succeed in isolating the coumarin resistance enzymes. Apparently, these proteins did not bind to the novobiocin-Sepharose and eluted together with the bulk of the proteins. Attempts to determine gyrase activity in crude extracts without purification were unsuccessful.
Hybridization experiments with gyrBR and parYR.
Thiara and Cundliffe (30) reported that the novobiocin producer S. spheroides is able to express a novobiocin-resistant GyrBR protein in addition to the novobiocin-sensitive GyrBS protein. It was likely that the same strategy was employed by the producers of clorobiocin and coumermycin A1. The same scenario may be encountered for the parY genes, i.e., we speculate that the genomes of S. roseochromogenes and S. rishiriensis may contain an additional parYS gene besides the parYR genes located in the clorobiocin and coumermycin A1 biosynthetic gene clusters.
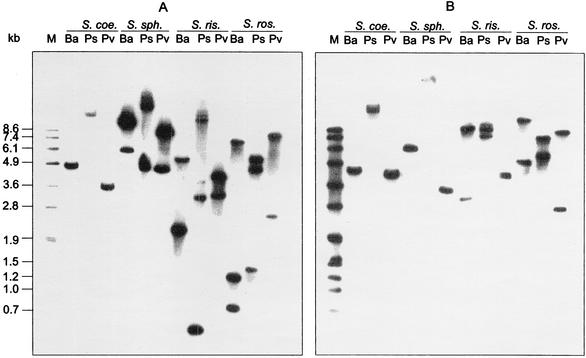
In order to investigate the number of gyrB and parY genes in these organisms, we conducted Southern hybridization experiments with gyrBR and parYR from S. rishiriensis as probes. The high sequence similarity within the gyrB genes and within the parY genes, as well as the sequence differences between gyrB and parY (see Fig. 2B), suggested that selective hybridization for each group was possible. Therefore, genomic DNA from the producers of novobiocin, clorobiocin, and coumermycin A1 as well as from the completely sequenced organism S. coelicolor A3(2) was digested with different enzymes and hybridized to the two probes consecutively. The resulting blots are shown in Fig. 3. As expected, the gyrBR probe of S. rishiriensis hybridized with the gyrB gene from S. coelicolor, resulting in a single band in each restriction digest. The observed bands coincided with the fragment size calculated from the genomic sequence (BamHI, 4,496 bp; PstI, 11,668 bp; PvuII, 3,453 bp). In contrast, two bands hybridizing with gyrBR were detected in S. spheroides, similar to the genes gyrBR and gyrBS described by Thiara and Cundliffe (31). Likewise, two gyrB genes were detected in both S. roseochromogenes and S. rishiriensis. The gyrBR gene of S. rishiriensis contains a PstI restriction site, and the gyrBR gene of S. roseochromogenes contains both a PstI and a BamHI site. This led to the appearance of three bands in the respective lanes (Fig. 3A).
FIG. 3.
Southern blotting of genomic DNA of S. coelicolor (S. coe.), S. spheroides (S. sph.), S. rishiriensis (S. ris.), and S. roseochromogenes (S. ros.). Genomic DNA was digested with BamHI (Ba), PstI (Ps), and PvuII (Pv). gyrBR (A) and parYR (B) from S. rishiriensis were used as hybridization probes (see Materials and Methods). In blot B, S. rishiriensis DNA digested with PvuII showed two bands at 4.1 and 4.3 kb, which are not clearly resolved in this figure.
With the parYR probe from S. rishiriensis, the parY gene of S. coelicolor was again detected, with the correct fragment sizes as calculated from the genomic sequence (BamHI, 4,183 bp; PstI, 13,021 bp; PvuII, 3,965 bp). Also in S. spheroides, a single gene hybridizing to the parYR probe was detected. This suggests that the novobiocin producer, like S. coelicolor, also contains just one parY gene in its genome, since the novobiocin biosynthetic gene cluster does not contain a parYR resistance gene.
In both S. roseochromogenes and S. rishiriensis, two bands hybridizing with parYR were detected, suggesting that these organisms contain an additional parY gene besides the parYR genes identified in the biosynthetic gene clusters of clorobiocin and coumermycin A1.
Expression of putative transporter genes novA and couR5.
The gene novA, located near the left border of the novobiocin biosynthetic gene cluster, showed sequence similarity to type III ABC transporters (19), such as the ABC transporters found in the biosynthetic gene clusters of complestatin (4) and chloroeremomycin (33). The characteristic Walker motifs within the C-terminal part of the protein are especially well preserved. Méndez and Salas (19) suggested that the novA gene product may be involved in the transport of and/or resistance against novobiocin, but no experimental evidence has been provided so far. Our sequencing of the clorobiocin and the coumermycin A1 clusters revealed no similar ABC transporter within or near these clusters (Fig. 1B). However, the coumermycin A1 cluster contained the gene couR5, which showed sequence similarity to transmembrane efflux proteins presumably involved in antibiotic transport, e.g., to the putative actinorhodin transporter from S. coelicolor (8) and to the putative tetracenomycin resistance protein from S. glaucescens (11).
In order to investigate the possible role of novA and couR5 in aminocoumarin resistance, we expressed both genes in S. lividans TK24 with the same expression vector, pUWL201, and the same experimental procedure for resistance determination as described for the gyrBR and parYR genes.
The results are shown in Table 2. The expression of both novA from S. spheroides and couR5 from S. rishiriensis in S. lividans TK24 provided resistance against novobiocin and coumermycin A1, but the level of resistance was lower than that observed upon expression of gyrBR or parYR. This suggests that novA and couR5 are involved in the transport of novobiocin and coumermycin A1, presumably in the sequestration of these antibiotics into the medium, but that the principal resistance mechanism of the aminocoumarin antibiotic producers appears to be the synthesis of coumarin-resistant topoisomerases rather than efflux mechanisms.
Ritchie and coworkers (20) reported the cloning of two DNA fragments from the novobiocin producer which conveyed a low level of resistance to novobiocin. However, the restriction map given for these fragments is not in agreement with the sequence of novA.
DISCUSSION
The principal resistance mechanism of the novobiocin producer S. spheroides has been reported to be the de novo synthesis of an aminocoumarin-resistant GyrB subunit, i.e., GyrBR, in the presence of novobiocin (30). By cloning of the biosynthetic gene clusters of novobiocin, clorobiocin, and coumermycin A1, we have now shown that a gyrBR gene is contained in the biosynthetic gene clusters of all three aminocoumarin producers. Unexpectedly, the biosynthetic gene clusters of clorobiocin and coumermycin A1 were found to contain an additional gene with obvious sequence similarity to genes encoding type II topoisomerase B subunits. This new gene was termed parYR.
Many organisms contain two type II topoisomerases, i.e., gyrase and topoisomerase IV (3, 34). Gyrase is encoded by the genes gyrA and gyrB, and their gene products form the heterotetramer (GyrA)2(GyrB)2. Gyrase is unique within the topoisomerases by its ability to introduce negative supercoils into DNA, energetically driven by ATP hydrolysis. Its biological function is to control DNA supercoiling and to relieve topological stress arising during the transcription and replication processes.
Topoisomerase IV is encoded by the genes parC and parE. Their gene products correspond functionally to GyrA and GyrB, and they also form an active (ParC)2(ParE)2 heterotetramer. This complex functions as a decatenating enzyme and resolves the interlinked daughter chromosomes following DNA replication. It has been suggested that topoisomerase IV is an important target, in some organisms even the principal target, of the fluoroquinolones and the aminocoumarin antibiotics (7, 9).
However, several organisms do not contain a topoisomerase IV, e.g., Deinococcus radiodurans, Campylobacter jejuni, and the actinobacteria Mycobacterium tuberculosis, Mycobacterium leprae, and Corynebacterium glutamicum. In the entire class of actinobacteria, 118 putative gyrB genes but no parE genes have been identified (www.seasquirt.mbio.co.jp). Expression of the gyrase of Mycobacterium smegmatis (17) revealed that this enzyme exhibits, besides supercoiling activity, pronounced decatenating activity and is therefore likely to fulfill both functions in the organism.
Streptomycetes belong to the actinobacteria. The genomic sequence of Streptomyces coelicolor A3(2) (1) contains a gyrA gene and an adjacent gyrB gene (SCO3873 and SCO3874, respectively) and in addition two further genes (SCO5836 and SCO5822) with sequence similarity to the A and B subunits of type II topoisomerases, respectively. These genes are separated by a stretch of 13 kb in the genome and have been annotated in the database as DNA gyrase-like protein, subunit A and probable DNA gyrase subunit B, respectively. Their function has not been investigated so far. We will use the names parX (SCO5836) and parY (SCO5822) for these genes hereafter. Homologues of parX and parY apparently exist in the genome of Streptomyces avermitilis (http://avermitilis.ls.kitasato-u.ac.jp), although their sequence is not available to the public. These genes (SAV2423 and SAV2442) have been annotated as putative subunits of topoisomerase IV, but no experimental evidence on their function exists.
The deduced parY gene product of S. coelicolor shows 88% sequence identity with the products of the parYR resistance genes of S. rishiriensis and S. roseochromogenes identified in this study (Fig. 2). Sequence comparison with the gyrB and the parE genes of the most closely related, high-GC-content, gram-positive bacteria shows that the parY genes can be placed neither into the gyrB nor into the parE group (Fig. 2B). A very similar picture arises from sequence comparison of the gyrA, parC, and parX sequences (Fig. 2A).
Expression of parYR from S. rishiriensis in S. lividans TK24 resulted in novobiocin and coumermycin A1 resistance, similar to that observed upon expression of gyrBR. This suggests that ParYR is able to function as a coumarin-resistant gyrase B subunit, in clear contrast to ParE of E. coli. In E. coli, complementation experiments showed that gyrase (consisting of GyrA and GyrB) can partly substitute for topoisomerase IV, but topoisomerase IV (encoded by ParC and ParE) cannot substitute for gyrase, since topoisomerase IV does not possess the unique supercoiling activity of gyrase (3, 24).
The function of the ParY proteins in comparison to topoisomerase IV of other organisms needs further investigation. From the results of our study, it appears possible that streptomycetes may possess two type II topoisomerases, (GyrA)2(GyrB)2, involved in supercoiling, and (ParX)2(ParY)2, involved in both supercoiling and decatenation. The coumarin antibiotic producers contain an additional gene coding for a coumarin-resistant GyrBR protein which can replace the coumarin-sensitive GyrB protein in the A2B2 heterotetramer. Likewise, the coumarin-resistant ParYR protein may replace the coumarin-sensitive ParY protein in the (ParX)2(ParY)2 heterotetramer in the producers of clorobiocin and coumermycin. The absence of a parYR resistance gene in the producer of novobiocin may be related to the lower affinity of novobiocin to topoisomerase IV compared to the affinity of clorobiocin and coumermycin A1 (12, 22).
Gyrase B mutations which convey resistance to aminocoumarins mostly involve Arg 136 in E. coli or corresponding arginine residues in other organisms (5, 6, 32). Arg 136 has been shown to be of principal importance in the binding of aminocoumarin drugs to gyrase (13, 16). However, Arg 136 is not conserved even in the aminocoumarin-sensitive GyrBS proteins of S. coelicolor and S. spheroides, or in the ParY protein of S. coelicolor (Table 3). On the contrary, only the aminocoumarin-resistant ParYR proteins of S. rishiriensis and S. roseochromogenes contain an arginine residue in this position (Table 3). Likewise, nearly all amino acids shown by X-ray studies to be involved in the binding of novobiocin and/or clorobiocin to gyrase were well conserved in the deduced GyrBR and ParYR sequences (Table 3). It is therefore not clear why aminocoumarins cannot bind to these enzymes. Computer modeling of the three-dimensional structure of these proteins may help to elucidate the reason.
TABLE 3.
Alignment of amino acids involved in aminocoumarin binding by E. coli gyrase, with corresponding positions of type II topoisomerase B subunits of different bacteriaa
| Species | Protein | Size (aa) | Position | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | GyrB | 804 | Val 43 | Asn 46 | Glu 50 | Asp 73 | Gly 77 | Ile 94 | Val 120 | Arg 136 | Gly 164 | Thr 165 |
| Novobiocin interaction | + | + | + | + | − | − | − | + | − | − | ||
| Clorobiocin interaction | + | + | + | + | + | − | − | + | − | + | ||
| Staphylococcus aureus | GyrB | 644 | Ile 51 | Asn 54 | Glu 58 | Asp 81 | Gly 85 | lle 102 | Ser 128 | Arg 144 | Gly 172 | Thr 173 |
| S. pneumoniae | GyrB | 648 | Ile 50 | Asn 53 | Glu 57 | Asp 80 | Gly 84 | Val 101 | Ser 127 | Lys 143 | Gly 171 | Thr 172 |
| S. coelicolor | GyrB | 687 | Val 70 | Asn 73 | Glu 77 | Asp 100 | Gly 104 | Val 121 | Val 147 | Thr 163 | Gly 191 | Thr 192 |
| S. spheroides | GyrBR | 677 | Leu 63 | Asn 66 | Glu 70 | Asp 93 | Gly 97 | Val 114 | Leu 140 | Thr 156 | Gly 184 | Thr 185 |
| S. rishiriensis | GyrBR | 677 | Leu 63 | Asn 66 | Glu 70 | Asp 93 | Gly 97 | Val 114 | Leu 140 | Thr 156 | Gly 184 | Thr 185 |
| S. roseochromogenes | GyrBR | 677 | Leu 63 | Asn 66 | Glu 70 | Asp 93 | Gly 97 | Val 114 | Leu 140 | Thr 156 | Gly 184 | Thr 185 |
| S. coelicolor | ParY | 707 | Ile 60 | Asn 63 | Glu 67 | Asp 90 | Gly 94 | Val 111 | Ala 137 | Leu 153 | Gly 197 | Thr 198 |
| S. rishiriensis | ParYR | 702 | Ile 56 | Asn 59 | Glu 63 | Asp 86 | Gly 90 | Ala 107 | Ala 132 | Arg 148 | Gly 192 | Thr 193 |
| S. roseochromogenes | ParYR | 702 | Ile 56 | Asn 59 | Glu 63 | Asp 86 | Gly 90 | Ala 107 | Ala 132 | Arg 148 | Gly 193 | Thr 194 |
Resistance proteins of the aminocoumarin producers are shown in bold face. Boldface amino acids are positions of known mutations conferring resistance to aminocoumarins in E. coli, Staphylococcus aureus (28), or Streptococcus pneumoniae (21). The interaction of E. coli gyrase with aminocoumarins has been described (16, 32).
Acknowledgments
We thank H. Simon (Jena University) for help in the purification of gyrase from Streptomyces spp. We are grateful to Zhao-Xin Wang and Florence Pojer for sequencing of the coumermycin and clorobiocin biosynthetic gene clusters, respectively.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to L. Heide and S.-M. Li).
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 2.Berger, J., and A. D. Batcho. 1978. Coumarin-glycoside antibiotics. J. Chromatogr. Libr. 15:101-158. [Google Scholar]
- 3.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 4.Chiu, H. T., B. K. Hubbard, A. N. Shah, J. Eide, R. A. Fredenburg, C. T. Walsh, and C. Khosla. 2001. Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc. Natl. Acad. Sci. USA 98:8548-8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contreras, A., and A. Maxwell. 1992. gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol. Microbiol. 6:1617-1624. [DOI] [PubMed] [Google Scholar]
- 6.del Castillo, I., J. L. Vizan, M. C. Rodriguez-Sainz, and F. Moreno. 1991. An unusual mechanism for resistance to the antibiotic coumermycin A1. Proc. Natl. Acad. Sci. USA 88:8860-8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Moreno, M. A., J. L. Caballero, D. A. Hopwood, and F. Malpartida. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769-780. [DOI] [PubMed] [Google Scholar]
- 9.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 10.Gormley, N. A., G. Orphanides, A. Meyer, P. M. Cullis, and A. Maxwell. 1996. The interaction of coumarin antibiotics with fragments of DNA gyrase B protein. Biochemistry 35:5083-5092. [DOI] [PubMed] [Google Scholar]
- 11.Guilfoile, P. G., and C. R. Hutchinson. 1992. Sequence and transcriptional analysis of the Streptomyces glaucescens tcmAR tetracenomycin C resistance and repressor gene loci. J. Bacteriol. 174:3651-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooper, D. C., J. S. Wolfson, G. L. McHugh, M. B. Winters, and M. N. Swartz. 1982. Effects of novobiocin, coumermycin A1, clorobiocin, and their analogs on Escherichia coli DNA gyrase and bacterial growth. Antimicrob. Agents Chemother. 22:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kampranis, S. C., N. A. Gormley, R. Tranter, G. Orphanides, and A. Maxwell. 1999. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemistry 38:1967-1976. [DOI] [PubMed] [Google Scholar]
- 14.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, UK.
- 15.Lanoot, B., M. Vancanneyt, I. Cleenwerck, L. Wang, W. Li, Z. Liu, and J. Swings. 2002. The search for synonyms among streptomycetes by with SDS-PAGE of whole-cell proteins. Emendation of the species Streptomyces aurantiacus, Streptomyces cacaoi subsp. cacaoi, Streptomyces caeruleus and Streptomyces violaceus. Int. J. Syst. Evol. Microbiol. 52:823-829. [DOI] [PubMed] [Google Scholar]
- 16.Lewis, R. J., O. M. Singh, C. V. Smith, T. Skarzynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 17.Manjunatha, U. H., M. Dalal, M. Chatterji, D. R. Radha, S. S. Visweswariah, and V. Nagaraja. 2002. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 30:2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxwell, A. 1999. DNA gyrase as a drug target. Biochem. Soc. Trans. 27:48-53. [DOI] [PubMed] [Google Scholar]
- 19.Méndez, C., and J. A. Salas. 2001. The role of ABC transporters in antibiotic-producing organisms: drug secretion and resistance mechanisms. Res. Microbiol. 152:341-350. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell, J. I., P. G. Logan, K. E. Cushing, and D. A. Ritchie. 1990. Novobiocin-resistance sequences from the novobiocin-producing strain Streptomyces niveus. Mol. Microbiol. 4:845-849. [DOI] [PubMed] [Google Scholar]
- 21.Munoz, R., M. Bustamante, and A. G. de la Campa. 1995. Ser-127-to-Leu substitution in the DNA gyrase B subunit of Streptococcus pneumoniae is implicated in novobiocin resistance. J. Bacteriol. 177:4166-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng, H., and K. J. Marians. 1993. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268:24481-24490. [PubMed] [Google Scholar]
- 23.Pojer, F., S.-M. Li, and L. Heide. 2002. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901-3911. [DOI] [PubMed] [Google Scholar]
- 24.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Staudenbauer, W. L., and E. Orr. 1981. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 9:3589-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steffensky, M., A. Mühlenweg, Z.-X. Wang, S.-M. Li, and L. Heide. 2000. Identification of the novobiocin biosynthetic gene cluster of Streptomyces sphaeroides NCIB 11891. Antimicrob. Agents Chemother. 44:1214-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stieger, M., P. Angehrn, B. Wohlgensinger, and H. Gmunder. 1996. GyrB mutations in Staphylococcus aureus strains resistant to cyclothialidine, coumermycin, and novobiocin. Antimicrob. Agents Chemother. 40:1060-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiara, A. S., and E. Cundliffe. 1988. Cloning and characterization of a DNA gyrase B gene from Streptomyces sphaeroides that confers resistance to novobiocin. EMBO J. 7:2255-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiara, A. S., and E. Cundliffe. 1989. Interplay of novobiocin-resistant and -sensitive DNA gyrase activities in self-protection of the novobiocin pro-ducer Streptomyces sphaeroides. Gene 81:65-72. [DOI] [PubMed] [Google Scholar]
- 31.Thiara, A. S., and E. Cundliffe. 1993. Expression and analysis of two gyrB genes from the novobiocin producer Streptomyces sphaeroides. Mol. Microbiol. 8:495-506. [DOI] [PubMed] [Google Scholar]
- 32.Tsai, F. T., O. M. Singh, T. Skarzynski, A. J. Wonacott, S. Weston, A. Tucker, R. A. Pauptit, A. L. Breeze, J. P. Poyser, R. O'Brien, J. E. Ladbury, and D. B. Wigley. 1997. The high-resolution crystal structure of a 24-kDa gyrase B fragment from E. coli complexed with one of the most potent coumarin inhibitors, clorobiocin. Proteins 28:41-52. [PubMed] [Google Scholar]
- 33.van Wageningen, A. M., P. N. Kirkpatrick, D. H. Williams, B. R. Harris, J. K. Kershaw, N. J. Lennard, M. Jones, S. J. Jones, and P. J. Solenberg. 1998. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5:155-162. [DOI] [PubMed] [Google Scholar]
- 34.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430-440. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Z. X., S. M. Li, and L. Heide. 2000. Identification of the coumermycin A1 biosynthetic gene cluster of Streptomyces rishiriensis DSM 40489. Antimicrob. Agents Chemother. 44:3040-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]