Abstract
Background
Methylation at CpG dinucleotides in genomic DNA is a fundamental epigenetic mechanism of gene expression control in vertebrates. Proteins with a methyl-CpG-binding domain (MBD) can bind to single methylated CpGs and most of them are involved in transcription control. So far, five vertebrate MBD proteins have been described as MBD family members: MBD1, MBD2, MBD3, MBD4 and MECP2.
Results
We performed database searches for new proteins containing an MBD and identified six amino acid sequences which are different from the previously described ones. Here we present a comparison of their MBD sequences, additional protein motifs and the expression of the encoding genes. A calculated unrooted dendrogram indicates the existence of at least four different groups of MBDs within these proteins. Two of these polypeptides, KIAA1461 and KIAA1887, were only present as predicted amino acid sequences based on a partial human cDNA. We investigated their expression by Northern blot analysis and found transcripts of ~8 kb and ~5 kb respectively, in all eight normal tissues studied.
Conclusions
Eleven polypeptides with a MBD could be identified in mouse and man. The analysis of protein domains suggests a role in transcriptional regulation for most of them. The knowledge of additional existing MBD proteins and their expression pattern is important in the context of Rett syndrome.
Background
Methylation at CpG dinucleotides in genomic DNA is a fundamental epigenetic mechanism of gene expression control in vertebrates [1-3]. Strong evidence exists for a correlation between DNA hypermethylation, hypoacetylation of histones, tightly packed chromatin, and transcriptional repression. Effects of DNA methylation are mediated through proteins which bind to symmetrically methylated CpGs. Such proteins contain a specific domain, the methyl-CpG-binding domain (MBD) which consists of ~70 residues in an α/β-sandwich fold built of three to four β-twisted sheets and a helix with a characteristic hairpin loop in the opposite layer [4-7]. Recently, a transcriptional repressor protein, Kaiso, lacking an MBD, has also been shown to bind to methylated CpG dinucleotides. This binding is mediated through zinc finger motifs [8,9]. Members of the MBD protein family are found in different animal species. So far, five vertebrate MBD proteins have been identified as members of the MBD protein family: MBD1, MBD2, MBD3, MBD4 and MECP2 (for review, see: [6,10]). Except for MBD4, all of them are associated with histone deacetylases (HDAC), and a transcriptional repression mechanism mediated by the recruitment of HDACs has been shown for MECP2, MBD1 and MBD2 [11-13].
One of these five MBD proteins, MECP2, is implicated in a human neurological disorder called Rett syndrome [14,15]. Symptoms of this syndrome are mental retardation, loss of speech and purposeful hand use, autism, ataxia, and stereotypic hand movements. The similar phenotype of conditional Mecp2 knockout mice and in vitro studies of functional consequences of MECP2 mutations indicate that the disorder is due to a loss of MECP2 function in the nervous system [16-21]. It remains unknown, why these patients present with a neurological phenotype, although MECP2 is ubiquitously expressed. It has been proposed that MECP2 is complemented by other MBD proteins in non-neural tissues and this hypothesis was tested for MBD2 by crossing Mecp2 knockout mice with Mbd2 knockout mice [19]. However, no evidence for functional redundancy of these two genes could be found in this way.
Here we report two new polypeptide sequences with an MBD as well as four MBD proteins in man and mouse that had not been mentioned as MBD protein family members up to date. Analysis of their amino acid sequence revealed additional domains associated with chromatin and point to a function in transcription control.
Results and Discussion
Human MBD proteins
We used a bioinformatics approach with the MBD of human MECP2 as query sequence to search for new members of the MBD protein family. Initial standard BLAST searches of the NCBI, Celera and SwissProt databases resulted only in five MBD proteins (MECP2, MBD1, MBD2, MBD3 and MBD4) which had previously been described and studied intensively. However the search of protein domain family databases (NCBI, Pfam, Smart and Prosite) revealed similarities to the MBD of MECP2 for four additional proteins, i.e. BAZ2A/TIP5, BAZ2B, CLLD8 and SETDB1 and for two cDNAs, KIAA1461 and KIAA1887. These databases use Hidden Markov Models (HMM) to detect motifs in amino acid sequences. An MBD has been described in CLLD8, SETDB1 and BAZ2A/TIP5 so far [22-24].
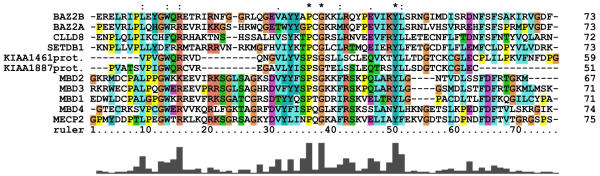
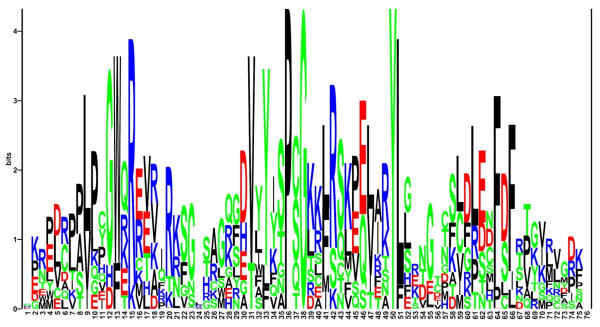
Nine of the eleven MBD-containing protein sequences could also be detected by screening the Sequence Similarity DataBase (SSDB) [25] at GenomeNet. The cDNAs for KIAA1461 and KIAA1887 were not found in the KEGG database that underlies the SSDB. These results are summarized in Tab 1. MBD amino acid sequences of the five previously published and the six newly described human polypeptides were aligned (Fig. 1). A sequence logo derived from the alignment of all eleven sequences is shown in Fig. 2. These analyses implicate a small number of highly conserved and apparently essential amino acids within the domain. At three positions identical amino acids are present and five positions with conservative substitutions can be found.
Table 1.
Summary of the human MBD polypeptides
| Accession No. a) | Name | Search method / databases b) | Domains c) | MBD position d) |
| P51608 | MECP2 | a, b, c, d, e, f, g, ac, bc, cc | 96–149 | |
| Q9UIS9 | MBD1 | a, b, c, d, e, f, g, ac, bc, cc | zf-CXXC | 7–59 |
| Q9UBB5 | MBD2 | a, b, c, d, e, f, g, ac, bc, cc | 151–204 | |
| O95983 | MBD3 | a, b, c, d, e, f, g, ac, bc, cc | 8–60 | |
| Q9Z2D7 | MBD4 | a, b, c, d, e, f, g, ac, bc, cc | HhH-GPD | 82–135 |
| Q9UIF9 | BAZ2A/TIP5 | e, f, g | AT_hook, DDT, PHD, bromodomain | 526–577 |
| Q9UIF8 | BAZ2B | e, f, g | DDT, PHD, bromodomain, | 549–600 |
| Q96T68 | CLLD8 | e, f, g | SET | 162–216 |
| Q15047 | SETDB1 | e, f, g | SET | 597–653 |
| Q9P267 | KIAA1461 | e, f | PWWP | 21–79 |
| Q96Q00 | KIAA1887 | e, f | 304–456 |
a) Accession numbers according to the SwissProt database. b) a = BLASTP, b = TBLASTN, c = TBLASTX, d = PSI-BLAST, e = Pfam, f = Ncbi, g = SSDB, ac = BLASTP in Celera database, bc = TBLASTN in Celera database, cc = TBLASTX in Celera database. c) domain nomenclature according to the Pfam database, all sequences contain an MBD in addition. d) position in amino acid sequence
Figure 1.
Alignment of the human methyl-CpG-binding proteins according to Swissprot. ClustalX alignment of the human MBDs. Columns are colored by conservation and property [50]. Residue conservation above each column indicates: "*" completely conserved; ":" favored substitutions; "." weakly favored substitutions. A quality graph is depicted below the alignment.
Figure 2.
Sequence logo of the eleven human MBD sequences. The height of the letters corresponds to the frequency of the amino acid at its position. The size of each stack stands for the information present at this position, measured in bits. Top letters represent the consensus sequence. Grey bars indicate gaps in some of the aligned sequences.
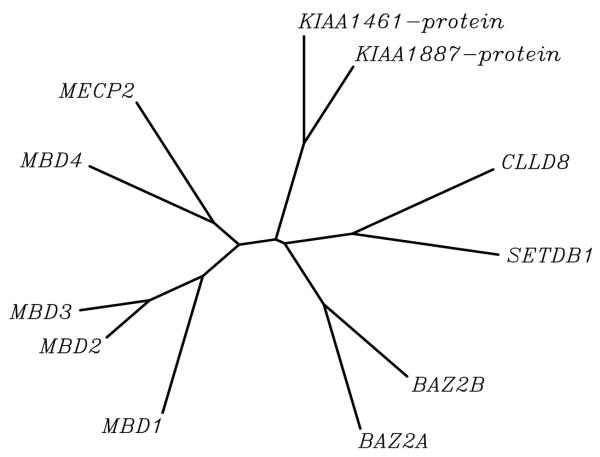
A phylogenetic tree of the MBD amino acid sequences of all eleven polypeptides is shown in Fig. 3. Four major MBD subsets are indicated there. The MBDs of the originally described proteins (MBD1, MBD2, MBD3, MBD4 and MECP2) are found as one group besides a second (BAZ2A/TIP5, BAZ2B) and third subset (CLLL8 and SETDB1) which are joined by a very short branch. KIAA1461 and KIAA1887 appear in a fourth branch. MBDs of the original five proteins are more similar to each other than to the novel ones, which explains why BLAST analyses with the MECP2 MBD query failed to identify the second, third or fourth class.
Figure 3.
Unrooted dendrogram depicting methyl-CpG-binding domains of the human protein family. Tree representation of the similarity between the human MBD proteins. The left branch clusters the original 5 MBD proteins. Branch lengths are proportional to the amount of inferred evolutionary change.
Domain analysis
An analysis of the amino acid sequences revealed that the MBD was the only domain shared by all eleven sequences. The MBD of MECP2, MBD1, MBD2, MBD4 and BAZ2A/TIP5 mediates binding to DNA, in case of MECP2, MBD1 and MBD2 preferentially to methylated CpG [13,24,26-28]. MBD4 has a special role acting as a DNA repair enzyme that reverses spontaneous CpG to TpG base exchanges, thereby maintaining methylated CpG motifs. It binds preferably to m5CpG/GpT mismatches [29,30].
In case of human MBD3 and SETDB1 the MBD has been shown to mediate protein-protein interactions [23,31]. Xenopus MBD3 is exceptional in its binding to methylated CpG which can be explained by the difference of an amino acid residue within the MBD (Lys30) important for DNA binding [31]. It remains to be determined whether the MBDs of BAZ2B, CLLD8, KIAA1461 and KIAA1887 mediate DNA binding or protein-protein interactions. Additional domains found in seven of the eleven polypeptides indicate that they are associated with chromatin and function in epigenetic mechanisms of gene regulation. Some of the proteins are already known to be involved in transcriptional repression, and the domains of the remainder strongly suggest a comparable function.
MECP2 recruits the Sin3A co-repressor complex and MBD2 the NuRD co-repressor complex, which itself contains MBD3. Both complexes contain HDACs, and MBD1 is also associated with HDAC activity although the identity of the deacetylase remains unknown [13]. Within the C-terminal part of MECP2, a histidine and proline-rich region is present which is conserved in certain neural-specific transcription factors [32].
BAZ2A/TIP5 is part of the NoRC, nucleolar remodeling complex, which represses rDNA transcription by recruiting histone methyltransferases, HDACs and DNA methyltransferases [33].
BAZ2B has a domain structure similar to BAZ2A/TIP5, both contain a DDT (DNA binding homeobox and Different Transcription factors) and a tandem PHD-bromodomain. The PHD domain is a C4HC3 zinc-finger-like motif and the bromodomain consists of 110 amino acids and is found in many chromatin-associated proteins that can interact specifically with acetylated lysine. Tandem PHD-bromodomains have been found in several transcriptional co-repressors [34]. The DDT domain is exclusively associated with nuclear domains in other proteins and was found in different transcription and chromatin remodeling factors [35]. An AT_hook motif (which allows binding to the minor groove of AT-rich DNA regions) was found in BAZ2A/TIP5 but not in BAZ2B.
SETDB1 is a H3-K9 histone methyltransferase [23], its mouse homologue, ESET, has furthermore been shown to interact with the mSin3A/B co-repressor complex [36]. The SET domain is a signature motif for lysine-specific histone methyltransferases [37,38]. This domain is also present in CLLD8 to which no function has yet been assigned.
The predicted protein sequence of KIAA1461 harbors a PWWP motif [39] named after the conserved amino acids Pro-Trp-Trp-Pro. It was first described in the WHSC1 protein, encoded by a gene within the Wolf-Hirschhorn syndrome critical region. The PWWP domain of Dnmt3b, a DNA methyltransferase, has been recently shown to bind to DNA. A common feature of PWWP containing proteins is the presence of additional domains known to be associated with chromatin [40]. Furthermore KIAA1461 has been shown to interact with the KIAA1549 protein in a yeast-two-hybrid experiment (http://www.kazusa.or.jp/huge/ppi). This interaction partner has not been studied in detail so far, but contains a serine-rich stretch as well as a helix-turn-helix motif (PS00622, LuxR family) according to Prosite (http://www.expasy.org/prosite). Helix-turn-helix motifs can be found in many transcription regulation proteins.
In the predicted protein sequence of KIAA1887, only a proline-rich extension (http://www.ebi.ac.uk/interpro) but no protein motif as such could be found.
The co-existence of MBDs and domains involved in chromatin modification, present in many of the identified polypeptides, could also point to a connection between the latter mechanism and methylated DNA. Interestingly, a very recent study [41] has shown that Mecp2 is associated with a H3-K9 methyltransferase activity, indicating a link between DNA methylation and histone methylation.
MBD proteins in other species
In the mouse, homologues were found for all human MBD proteins. Sequence identity scores range from 63.8% to 94.0% (Tab. 2) indicating a conserved function in both species. Human and mouse MBD1, MBD2, MBD3, MBD4 and MECP2 are curated orthologues [27,42-44].
Table 2.
Mouse homologues of human MBD proteins
| Human protein | Mouse protein | % identity a) |
| Human protein | Mouse protein | % identity a) |
| MBD1 (645 aa) | Mbd1 (713 aa) | 66.8 % in 698 aa |
| MBD2 (478 aa) | Mbd2 (454 aa) | 93.2 % in 454 aa |
| MBD3 (331 aa) | Mbd3 (362 aa) | 85.1 % in 355 aa |
| MBD4 (647 aa) | Mbd4 (631 aa) | 63.8 % in 647 aa |
| BAZ2A (2009 aa) | Baz2a (1972 aa) | 80.1 % in 2039 aa |
| BAZ2B (2034 aa) | ENSMUSP00000028367 (2065 aa) | 82.6 % in 2027 aa |
| CLLD8 (746 aa) | ENSMUSP00000022552 (701 aa) | 65.2 % in 715 aa |
| SETDB1 (1446 aa) | ESET (1457 aa) | 91.0 % in 1465 aa |
| KIAA1461 (1544 aa) | ENSMUSP00000036847 (1518 aa) | 94.0 % in 1521 aa |
| KIAA1887 (1040 aa) | ENSMUSP00000026476 (101 aa) | 91.1 % in 101 aa |
Comparison of the human MBD proteins and their mouse homologues. Identity scores were calculated with the LALIGN program (http://www.ch.embnet.org/software/LALIGN_form.html). The numbers of amino acid (aa) residues are indicated. a) The number of aligned amino acids can exceed the residue number of the sequences due to gaps inserted by the algorithm.
Homology searches in the ENSEMBL database revealed the following murine homologues of BAZ2B, CLLD8, KIAA1461 protein and KIAA1887 protein. The mouse homologue of BAZ2B, the ENSEMBL protein ENSMUSP00000028367 (gene ENSMUSG00000026987) shows a 82.6 % amino acid sequence identity. The predicted mouse gene ENSMUSG00000021980 coding for ENSEMBL protein ENSMUSP00000022552 has a 65.2 % amino acid sequence identity to the human CLLD8. We furthermore found the predicted gene ENSMUSG00000036792 coding for the ENSEMBL protein ENSMUSP00000036847 as mouse homologue of human KIAA1461 protein with a 94.0 % amino acid sequence identity. For the KIAA1887 protein, the mouse gene sequence ENSMUSG00000025409 (ENSEMBL protein ENSMUSP00000026476) with 91.1 % sequence identity was present in the database. The latter database entry however consists of only 101 amino acids. The existence of translated proteins remains to be determined for all four mouse genes.
DNA methylation as a mechanism of gene expression regulation exists also in plants. In our database searches we detected plant MBD proteins as well. The Pfam database contains polypeptides from Arabidopsis thaliana and Triticum aestivum. BLAST analyses revealed additional proteins in Zea Mays, Hordeum vulgare and Lycopersicon esculetum. Entries for MBD containing proteins from plants over C. elegans to mouse and human are present in Pfam.
Expression patterns of MBD genes
Expression analyses had been carried out previously for all genes of the mouse/human MBD family except for KIAA1461 and KIAA1887 (only the abundance of KIAA1887 ESTs in different tissues has been reported [45]). The results of published Northern blot experiments are summarized in Tab. 3. Since expression levels of MBD4 were too low to be detected by Northern blots, only results of RT-PCR studies in three tissues are shown. However the presence of MBD4 EST sequences from numerous tissues points to a ubiquitous expression (http://www.ncbi.nlm.nih.gov/UniGene).
Table 3.
Expression patterns of the mouse/human MBD genes
| Tissue | MECP2a) | MBD1b) | MBD2b) | MBD3b) | MBD4b) | CLLD8c) | BAZ2Ad) | BAZ2Bd) | SETDB1e) | KIAA1461f) | KIAA1887g) |
| Brain | + | + | + | + | + | (+) | + | (+) | + | (+) | (+) |
| Heart | + | + | + | + | + | + | + | + | + | + | |
| Kidney | + | + | + | + | + | + | - | + | (+) | + | |
| Liver | + | (+) | (+) | + | + | (+) | - | + | (+) | + | |
| Lung | + | + | + | + | + | + | - | + | (+) | (+) | |
| Skeletal muscle | + | + | + | + | (+) | + | + | + | + | + | |
| Spleen | + | + | + | + | + | - | + | + | |||
| Testis | + | + | + | + | + | + | + | + | + | ||
| ES cells | - | (+) | + | + | |||||||
| Placenta | + | (+) | + | + | + | + | + | ||||
| Pancreas | + | + | + | + | + | + | + |
The table shows the expression of the eleven MBD genes in major tissues as detected by Northern blot. Empty spaces denote lacking information for that tissue. (+) indicates very low or doubtful expression, – no expression. a) [44,51]b) [27] for these genes, comprehensive expression data was only available from mouse. c) [22] Expression in additional tissues has been reported. d) [46] Expression in additional tissues has been reported. e) [52] SETDB1 was originally called KIAA0067. f) Northern blot results of this study. g) [45] and northern blot results of this study
We performed Northern blot analyses for KIAA1461 and KIAA1887. Strong signals of ~8 kb were detected for KIAA1461 in skeletal muscle, heart, pancreas, kidney and placenta. A faint band could be detected in brain, lung and liver. For KIAA1887 a strong band of ~5 kb was present in heart, kidney, liver, skeletal muscle, placenta and pancreas, weaker signals could be seen for brain and lung tissue (Fig. 4).
Figure 4.
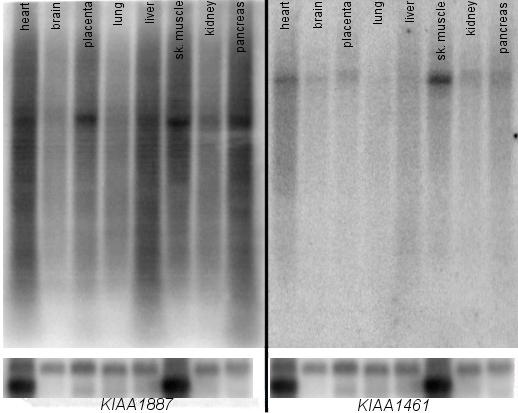
Northern blot of KIAA1461 and KIAA1887. Human multiple tissue Northern blot showing the expression of the KIAA1461 and KIAA1887 genes. The calculated sizes of the transcripts are ~5 kb for KIAA1887 and ~8 kb for KIAA1461. A β-actin probe was hybridized as loading control, shown at the bottom.
Taken together, MBD1, MBD2, MBD3 and MECP2 as well as SETDB1, CLLL8, BAZ2A, KIAA1461 and KIAA1887 show a broad tissue distribution. The expression of BAZ2B is more restricted according to northern blot results [46]. It is of note that CLLL8, BAZ2A, KIAA1461 and KIAA1887 show a very low expression in brain.
Conclusions
In this study we present additional four proteins and two cDNAs with a methyl-CpG-binding domain in mouse and man. Transcripts of SETDB1, BAZ2A, CLLL8, KIAA1461 and KIAA1887 are found in all adult tissues studied. Among the six proteins described here as new members of the MBD protein family, CLLL8, BAZ2B, KIAA1461 and KIAA1887 show a low expression in brain. MECP2 is found preferentially in mature neurons of the brain [47,48], and the cell types that express CLLD8, BAZ2A/TIP5, SETDB1 as well as the predicted KIAA1461 and KIAA1887 in brain are not known.
Rett syndrome is caused by mutations in MECP2. Even though MECP2 is ubiquitously expressed, the phenotype of the syndrome is restricted to the brain. This could be explained by a greater need of long lived, non-dividing neuronal cells for a special chromatin state that involves MECP2 and tightly suppresses transcription of undesired genes. Another explanation would be that the loss of function of MECP2 in non-neural tissues is compensated by another protein with similar properties. MBD2 has been studied in this respect, but no evidence for a genetic interaction of the two genes was found by combining Mbd2- and Mecp2-null mice [19]. Based on gene expression studies of Mecp2 knockout mice and biochemical evidence, it has been suggested that the essential function of Mecp2 in the brain might not be transcriptional [49]. In view of this aspect and the protein-protein interaction property of the methyl-CpG-binding domain of human MBD3 and SETDB1, functional compensation would not necessarily require a DNA binding property.
Almost all presented polypeptides are known or predicted to be involved in mechanisms of gene expression regulation. In order to understand the higher-order interplay of MBD proteins and associated complexes, it will be a major task to identify interacting proteins as well as regulated targets of all components. This will help to solve the question whether some of the polypeptides can functionally complement MECP2 in tissues other than the brain.
Methods
BLAST-based and database searches
Non-redundant GenBank, high throughput genomic sequences and expressed sequence tag (EST) databases were searched using BLASTP and TBLASTN of the NCBI web tools (http://www.ncbi.nlm.nih.gov/blast) applying default conditions. The human MECP2 chain A (MBD) was used as a query input. TBLASTX was carried out applying the web tools at the European Bioinformatics Institute (http://www.ebi.ac.uk/blast2/).
Using the MBD of human MECP2 as query, the NCBI (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein), Pfam (release 7) (http://www.sanger.ac.uk/Software/Pfam), Prosite (release 17.17) (http://www.expasy.ch/prosite/) and Smart (version 3.4) (http://smart.embl-heidelberg.de/) databases were screened for proteins with this domain. The resulting polypeptides were sorted according to their species of origin and additional identified domains within their sequence.
SSDB at GenomeNet (http://ssdb.genome.ad.jp) was searched with pfm:MBD as query motif. BLASTP and BLASTN searches were carried out against the Celera database (http://www.celera.com) with standard settings.
Homologues of KIAA1461, CLLL8 and KIAA1887 were identified in the ENSEMBL database.
Alignment and Phylogeny
Alignments and phylogenetic trees were computed using the ClustalW program at GenomeNet (http://clustalw.genome.ad.jp/) with standard settings and the ClustalX program (version 1.8) [50] for graphical representation.
The sequence logo was constructed by means of the plogo script (http://www.cbs.dtu.dk/~gorodkin/appl/plogo.html). File formats were converted with the GCG package (version 10.2) when necessary.
Sequence comparison between mouse and man was carried out using the LALIGN algorithm at EMBNET (http://www.ch.embnet.org/software/LALIGN_form.html) with default settings.
Cell lines. RNA and genomic DNA isolation, cDNA synthesis
CCRF-CEM and lymphoblastoid cells were grown according to manufacturer's instructions (DSMZ, Braunschweig, Germany and the Coriell Institute for Medical Research, Camden, Netherlands) and harvested at 95 % confluency. Genomic DNA was isolated with the QIAGEN Blood Maxi-KIT (QIAGEN, Hilden, Germany). Total RNA was isolated using the Trizol reagent (Invitrogen, Karlsruhe, Germany). A reverse transcriptase reaction was performed in the presence of 50 ng/μl oligo(dT) and 2 mM dA/C/G/TTP with 10 U/μl SSII reverse transcriptase (Invitrogen, Karlsruhe, Germany). The resulting cDNA was purified with the QIAquick PCR purification kit (QIAGEN, Hilden, Germany). cDNA (200 ng) was used in subsequent 50- μl PCR amplifications with 10 pmol gene-specific primers. Standard PCR conditions and respective annealing temperatures were used.
Northern blotting
A cDNA fragment of KIAA1461 was PCR-amplified with exon specific primers KIAA1461for (5'-CTAGACCATGGGAAAAATGT-3') / KIAA1461rev (5'-ACTTGGAGACTGCTCCTCTA-3') and human genomic DNA as template using standard conditions. For KIAA1887 a cDNA fragment was PCR-amplified with exon 6 specific primers KIAA1887for (5'-CAGACCCCCTACTGTATTTC-3') / KIAA1887rev (5'-CAAAAGGTTAAAGCTTCCAT-3') and cDNA from a lymphoblastoid cell line as template using standard conditions. A cDNA fragment of β-actin amplified with primers β-actinfor (5'-TGAACCCTAAGGCCAACCGTG-3') / β-actinrev (5'-GCTCATAGCTCTTCTCCAGGG-3') was used as a loading control. These probes were radioactively labeled with 32P-dCTP in a random prime reaction and hybridized in ExpressHyb solution (CLONTECH, Palo Alto, CA, USA) to a human multiple tissue northern blot (CLONTECH, Palo Alto, CA, USA) for 16 h at 65°C. Washing was performed in 2 × SSC / 0.1% SDS at 65°C for 10 min. Signals were detected with a PhosphorImager (Amersham Biosciences, Freiburg, Germany).
Abbreviations
MBD: Methyl-CpG-binding domain
HDAC: histone deacetylase
Authors' contributions
TCR: carried out the database searches, comparative analyses and molecular genetic experiments
HHR: revised the manuscript and participated in the study coordination
UAN: devised the study, supervised and coordinated it and drafted the manuscript
All authors read and approved the manuscript.
Acknowledgments
Acknowledgements
We thank Ralph Schulz and Bettina Lipkowitz for excellent technical assistance and Ulf Gurok and Antje Krause for critical reading and suggestions. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) (SFB 577, Teilprojekt C3).
Contributor Information
Tim C Roloff, Email: rolof_t@molgen.mpg.de.
H Hilger Ropers, Email: ropers@molgen.mpg.de.
Ulrike A Nuber, Email: nuber@molgen.mpg.de.
References
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Singal R, Ginder GD. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- El Osta A, Wolffe AP. DNA methylation and histone deacetylation in the control of gene expression: basic biochemistry to human development and disease. Gene Expr. 2000;9:63–75. doi: 10.3727/000000001783992731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki I, Shimotake N, Fujita N, Nakao M, Shirakawa M. Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. EMBO J. 1999;18:6653–6661. doi: 10.1093/emboj/18.23.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield RI, Smith BO, Nan X, Free A, Soteriou A, Uhrin D, Bird AP, Barlow PN. The solution structure of the domain from MeCP2 that binds to methylated DNA. J Mol Biol. 1999;291:1055–1065. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Ng HH, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–1406. doi: 10.1128/MCB.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van dV, I, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Percy AK. Rett syndrome: clinical correlates of the newly discovered gene. Brain Dev. 2001;23:S202–S205. doi: 10.1016/s0387-7604(01)00350-3. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Wolffe AP. Functional consequences of Rett syndrome mutations on human MeCP2. Nucleic Acids Res. 2000;28:4172–4179. doi: 10.1093/nar/28.21.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo S, Nomura Y, Segawa M, Fujita N, Nakao M, Dragich J, Schanen C, Tamura M. Functional analyses of MeCP2 mutations associated with Rett syndrome using transient expression systems. Brain Dev. 2001;23:S165–S173. doi: 10.1016/s0387-7604(01)00345-x. [DOI] [PubMed] [Google Scholar]
- Nan X, Bird A. The biological functions of the methyl-CpG-binding protein MeCP2 and its implication in Rett syndrome. Brain Dev. 2001;23:S32–S37. doi: 10.1016/s0387-7604(01)00333-3. [DOI] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/S0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Mabuchi H, Fujii H, Calin G, Alder H, Negrini M, Rassenti L, Kipps TJ, Bullrich F, Croce CM. Cloning and characterization of CLLD6, CLLD7, and CLLD8, novel candidate genes for leukemogenesis at chromosome 13q14, a region commonly deleted in B-cell chronic lymphocytic leukemia. Cancer Res. 2001;61:2870–2877. [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC – a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Genuardi M, Karbowski M, Yeung AT, Matsumoto Y, Bellacosa A. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J Cell Physiol. 2000;185:473–480. doi: 10.1002/1097-4652(200012)185:3<473::AID-JCP19>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem. 2002;277:35434–35439. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- Vacca M, Filippini F, Budillon A, Rossi V, Mercadante G, Manzati E, Gualandi F, Bigoni S, Trabanelli C, Pini G, et al. Mutation analysis of the MECP2 gene in British and Italian Rett syndrome females. J Mol Med. 2001;78:648–655. doi: 10.1007/s001090000155. [DOI] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., III Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T, Copley R, Bork P. DDT – a novel domain in different transcription and chromosome remodeling factors. Trends Biochem Sci. 2001;26:145–146. doi: 10.1016/S0968-0004(00)01769-2. [DOI] [PubMed] [Google Scholar]
- Yang L, Mei Q, Zielinska-Kwiatkowska A, Matsui Y, Blackburn ML, Benedetti D, Krumm A, Taborsky GJ, Jr, Chansky HA. An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and mSin3 transcription corepressors mSin3A/B. Biochem J. 2002 doi: 10.1042/BJ20020854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/S1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, Altherr MR, den Dunnen JT. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat Struct Biol. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2002 doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm Genome. 1999;10:906–912. doi: 10.1007/s003359901112. [DOI] [PubMed] [Google Scholar]
- Quaderi NA, Meehan RR, Tate PH, Cross SH, Bird AP, Chatterjee A, Herman GE, Brown SD. Genetic and physical mapping of a gene encoding a methyl CpG binding protein, Mecp2, to the mouse X chromosome. Genomics. 1994;22:648–651. doi: 10.1006/geno.1994.1442. [DOI] [PubMed] [Google Scholar]
- D'Esposito M, Quaderi NA, Ciccodicola A, Bruni P, Esposito T, D'Urso M, Brown SD. Isolation, physical mapping, and northern analysis of the X-linked human gene encoding methyl CpG-binding protein, MECP2. Mamm Genome. 1996;7:533–535. doi: 10.1007/s003359900157. [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Ohara O. Prediction of the coding sequences of unidentified human genes. XXI. The complete sequences of 60 new cDNA clones from brain which code for large proteins. DNA Res. 2001;8:179–187. doi: 10.1093/dnares/8.4.179. [DOI] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Nezu J, Shimane M. A novel family of bromodomain genes. Genomics. 2000;63:40–45. doi: 10.1006/geno.1999.6071. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- LaSalle JM, Goldstine J, Balmer D, Greco CM. Quantitative localization of heterogeneous methyl-CpG-binding protein 2 (MeCP2) expression phenotypes in normal and Rett syndrome brain by laser scanning cytometry. Hum Mol Genet. 2001;10:1729–1740. doi: 10.1093/hmg/10.17.1729. [DOI] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy JF, Sedlacek Z, Bachner D, Delius H, Poustka A. A complex pattern of evolutionary conservation and alternative polyadenylation within the long 3"-untranslated region of the methyl-CpG-binding protein 2 gene (MeCP2) suggests a regulatory role in gene expression. Hum Mol Genet. 1999;8:1253–1262. doi: 10.1093/hmg/8.7.1253. [DOI] [PubMed] [Google Scholar]
- Nomura N, Nagase T, Miyajima N, Sazuka T, Tanaka A, Sato S, Seki N, Kawarabayasi Y, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified human genes. II. The coding sequences of 40 new genes (KIAA0041-KIAA0080) deduced by analysis of cDNA clones from human cell line KG-1 (supplement). DNA Res. 1994;1:251–262. doi: 10.1093/dnares/1.5.251. [DOI] [PubMed] [Google Scholar]