Abstract
Background
The ability to manipulate the genetic networks underlying the physiological and behavioural repertoires of the adult honeybee worker (Apis mellifera) is likely to deepen our understanding of issues such as learning and memory generation, ageing, and the regulatory anatomy of social systems in proximate as well as evolutionary terms. Here we assess two methods for probing gene function by RNA interference (RNAi) in adult honeybees.
Results
The vitellogenin gene was chosen as target because its expression is unlikely to have a phenotypic effect until the adult stage in bees. This allowed us to introduce dsRNA in preblastoderm eggs without affecting gene function during development. Of workers reared from eggs injected with dsRNA derived from a 504 bp stretch of the vitellogenin coding sequence, 15% had strongly reduced levels of vitellogenin mRNA. When dsRNA was introduced by intra-abdominal injection in newly emerged bees, almost all individuals (96 %) showed the mutant phenotype. An RNA-fragment with an apparent size similar to the template dsRNA was still present in this group after 15 days.
Conclusion
Injection of dsRNA in eggs at the preblastoderm stage seems to allow disruption of gene function in all developmental stages. To dissect gene function in the adult stage, the intra-abdominal injection technique seems superior to egg injection as it gives a much higher penetrance, it is much simpler, and it makes it possible to address genes that are also expressed in the embryonic, larval or pupal stages.
Background
Due to its social attributes, its learning capabilities, and several facultative physiological and behavioural traits under social control, the honeybee provides unique opportunities as a model system [1,2]. The honeybee has a rich history as an experimental organism [3,4], and it is now receiving increased attention concerning general issues such as the molecular basis of learning and memory generation [5], the regulation of ageing [6], the regulatory anatomy of social systems [7,8], and the evolution of complex social systems through self-organization, emergence and multilevel natural selection [9,10]. The ability to manipulate the genetics underlying the physiological and behavioural repertoires of the adult honeybee worker will become instrumental for understanding the above issues. However, there is currently no established method for doing clear-cut reverse genetics on adult bees (but see [11,12]).
One recent and very promising method for targeted down-regulation of gene expression in a wide range of organisms is RNAi. RNAi is the process by which dsRNA inhibits the accumulation of homologous transcripts from cognate genes [13]. RNAi has evolved into a powerful tool for probing gene function in Drosophila, Tribolium, Caenorhabditis elegans and mice [14,15]. In honeybees, Beye et al. [12] showed that microinjection in preblastoderm eggs using dsRNAderived from a 300 bp stretch of the engrailed homeobox motif disrupts expression of the target gene during embryo development.
Here we extend the scope of the honeybee RNAi technique to include the adult stage. We show that adult honeybee gene expression exemplified by the gene coding for the lipoprotein vitellogenin can be specifically inhibited by microinjection of dsRNA into honeybee embryos as well as by intra-abdominal injection of adult bees. Furthermore, we demonstrate the superiority of the intra-abdominal injection technique with respect to efficacy and simplicity.
Results
The vitellogenin gene was chosen because its expression is unlikely to have a phenotypic effect until the adult stage in bees. The vitellogenin protein is produced by the adult insect fat body [16], which in honeybees consists of thin layers of cells spread against the body wall of the abdomen [17]. Vitellogenin is normally secreted into the hemolymph of reproductive females before transport into developing oocytes [16]. However, the sterile honeybee workers may have high hemolymph titers of this protein [18]. As shown by the control, vitellogenin mRNA is present in the fat body of bees < 24 hours old (Figure 1), while the level of corresponding protein in the hemolymph is undetectable at this age [19]. The expression then increases, and the level of mRNA is constant in the age interval of 7–15 days (Figure 1). During this period the individual vitellogenin titer may vary greatly, as shown in Figure 1. This variation may be due to individual differences in the rate at which vitellogenin is consumed for metabolic purposes [6]. Equal production rates would in this case give rise to variable titers. However, the phenomenon might also reflect individual differences in nutritional status, as the vitellogenin production is strongly dependent on the availability of proteinaceous food [20]. In any case, the vitellogenin titer is not an unambiguous assay for detecting RNAi. However, as the frequency of controls with negligible vitellogenin titers is low (7 % of all controls aged 5–15 days, N = 109), it is labor-saving to use it for detecting potential knockdown phenotypes.
Figure 1.
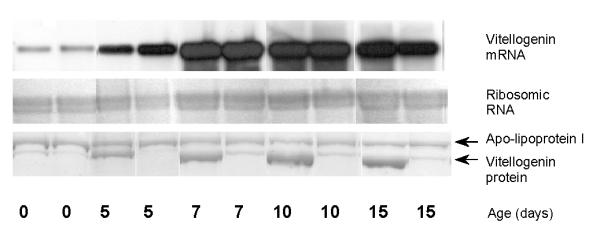
Wild-type control.Time course illustrating individual variation in vitellogenin mRNA expression vs. vitellogenin protein levels in wild-type controls. Total RNA loaded for Northern blot was 6 μg. An equal hemolymph volume of 0.1 μl was loaded for SDS-PAGE. Apo-lipoprotein I [40] and ribosomic RNA were included as controls.
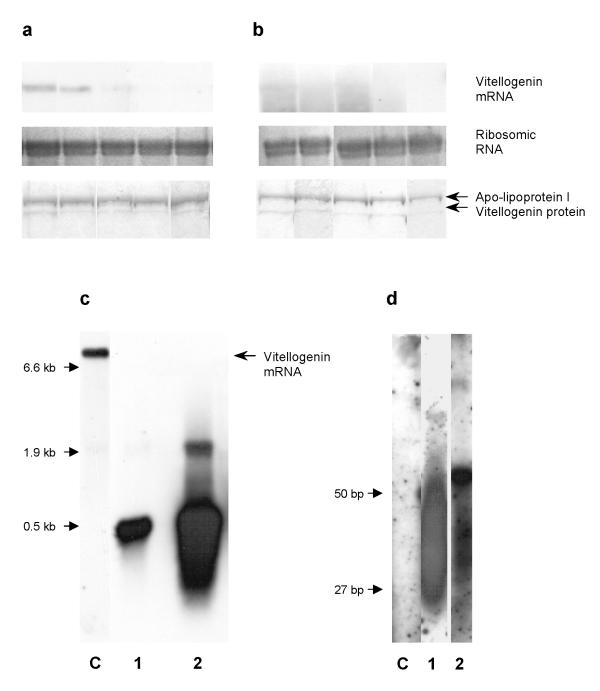
We found that 15 % (N = 70) of the adult bees reared from eggs injected with dsRNA had strongly reduced levels of vitellogenin mRNA. The disruption appeared to be incomplete in some individuals (Figure 2a), but the RNAi effect was detectable at emergence and persistent over 15 days (not shown). In the intra-abdominally injected group, 96 % (N = 30) showed loss of vitellogenin mRNA expression when sampled at 7 days old (Figure 2b). Furthermore, an unambiguous fragment with an apparent size similar to the template dsRNA (504 bp) was visualized in all adult injected bees with a mutant phenotype (Figure 2c). Samples enriched with short RNA show that vitellogenin mRNA fragments as small as 25 bp could be detected (Figure 2d). For all mutants, the level of vitellogenin in the hemolymph was almost undetectable (Figure 2a,2b). Reduced levels of vitellogenin mRNA was not encountered in any of the control individuals assayed in this manner (N = 96).
Figure 2.
Targeted vitellogenin disruption. (a) Individual samples of workers injected as embryo and (b) workers injected as adults. All bees were sampled as 7 days old. Total RNA loaded was 6 μg, and an equal 0.1 μl hemolymph volume was loaded for SDS-PAGE. (c) Visualization of ~500 bp fragment in a worker injected as adult. Lane 1: Control. Lane 2: 6 μg total RNA. Lane 3: 20 μg total RNA. (d) Pooled samples of 10 workers each enriched with small RNA fragments. Two oligonucleotides (27 and 50 bp) were included as markers. Lane 1: Control. Lane 2: Adult workers injected with dsRNA at the preblastoderm stage. Lane 3: Intra-abdominally injected workers.
To test whether the observed ~500 bp fragment was spanning parts upstream of the sequence in the original template dsRNA, we designed a new probe for Northern blotting using the clone AmR9, located ~1.5 kb upstream of the original AP4a5 (see Methods). We found that the new probe did not hybridize to the fragment observed in the adult injected bees (Figures 3). Given that the hybridization of lanes 2 and 3 in Figure 3 are comparable, the amount of dsRNA still present in the abdomen can be tentatively estimated to 0.1–0.2 μg (total RNA in the bee abdomen is ~60 μg).
Figure 3.
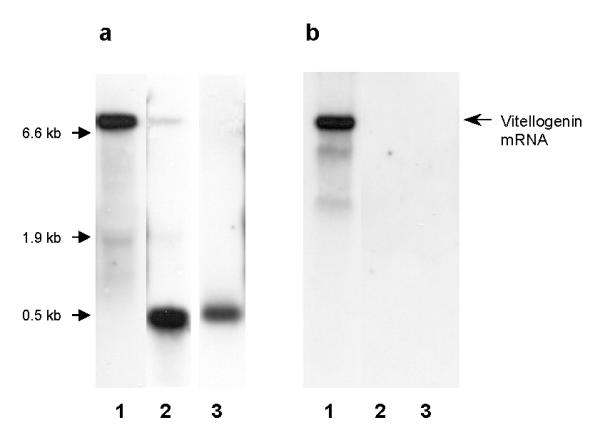
Sequence dependent Northern blot. Hybridization prepared with (a) AP4a5, and (b) AmR9. Lane 1: Control. Lane 2: Sample from an intra-abdominally injected worker, 6 μg total RNA. Lane 3: Original dsRNA template, 0.5 Ηg.
Discussion
Vitellogenin is a female specific glucolipoprotein yolk precursor produced by all oviparous animals [21]. Invertebrate and vertebrate vitellogenins constitute a multigene superfamily together with insect apolipophorin II/I, human apolipoprotein B (apoB-100), and the large subunit of mammalian microsomal triglyceride transfer protein (MTP) [22]. It is generally synthesized in large amounts directly prior to yolk deposition and is of fundamental importance in reproduction [21]. As vitellogenin is also synthesized by the functionally sterile workers [18], it has been suspected for some time that vitellogenin has functions other than reproduction in honeybees. It was recently shown that nurse bees use vitellogenin to produce royal jelly [23]. Furthermore, vitellogenin serves important functions related to immune function and longevity (GVA, ZLPS, KN, A Hagen, K Schrøder, Ø Mikkelsen, TBL Kirkwood and SWO, submitted). As the amount of vitellogenin mRNA in the honeybee worker is considerable, our results suggest that RNAi can be used to also knock down abundantly expressed genes.
It cannot be excluded that the observed high efficacy of the intra-abdominal injection technique for inhibiting vitellogenin expression is due to the structure and function of the honeybee fat body. When dsRNA is injected into the honeybee abdomen, the fat body tissue is readily exposed. Furthermore, the fat body plays an immunological role that includes uptake of potentially harmful macromolecules from the hemolymph, in many ways analogous to the mammalian liver [24]. The observed efficacy may thus not be representative for genes being expressed in other organs or tissues in the adult bee. However, it is promising that this does not seem to be the case in Drosophila melanogaster. Dzitoyeva et al. [25] demonstrated that intra-abdominal injections of homologous dsRNA in this species silence lacZ transgene expression in the gut as well as in the optic and antennal lobes, and that the method is potent in silencing endogenous GM06434 mRNA in the central nervous system. Even though the scope of this technique should turn out to be more restricted in honeybees, our results suggest that a number of important genes being expressed in the fat body can be targeted this way.
Alternatively, it may also be that the capacity of the fat body for taking up potentially harmful macromolecules from the hemolymph is indeed the key for targeting other tissues by intra-abdominal injection of dsRNA. If this dsRNA is processed into small interfering RNA (siRNA) or another mobile signal [26] that is exported to the hemolymph, this would make the fat body a source of mobile silencing agents that probably could be more easily taken up by other tissues. The results by Dzitoyeva et al. [25], and the fact that microinjections in the adult honeybee brain using dsRNA homologous to biogenic amine receptors results in incomplete knockdown of the target gene (T Farooqui, K Robinson, H Vaessin and BH Smith, submitted), are consistent with this explanation. If this suggestion is correct, the intra-abdominal injection technique may be superior to injecting dsRNA directly into a target tissue, both because of the actual mechanism, and the fact that the technique allows injection of much higher amounts of dsRNA without harming the individual.
The 'degradative PCR mechanism' suggested by Lepardi et al. [27], states that new dsRNA generated by siRNA is extended towards the 5', and siRNA from the 3' is able to produce full-length products. According to this, one would expect to detect fragments spanning upstream regions of the AP4a5 sequence if the observed ~500 bp fragment resulted from cleavage of new dsRNA. However, the clone AmR9, located ~1.5 kb upstream of AP4a5, did not hybridize to the fragment. Furthermore, the fragment was not found in knockdown adult bees reared from injected eggs where much less dsRNA template was introduced. These observations suggest that some of the original dsRNA template injected into adult bees was present even after 15 days. This persistence of injected dsRNA as high molecular weight material is in agreement with the results of Parrish et al. [28] who tested the fate of 32P labeled dsRNA in C. elegans. However, their experiments lasted only 12 hours, and to the best of our knowledge we are the first to report the presence of template dsRNA even after 15 days, and the activation of RNAi more than 21 days after delivery of dsRNA. It should be noted that the dsRNA template found in adult workers might be slightly smaller than the dsRNA injected into the bees (Figure 3a), but whether this is of biological relevance is not clear.
We do not have a clear answer to why the template dsRNA appears so stable. However, the RNAi machinery is probably tuned to silence transposons and protect against virus attacks [15]. Introducing a comparably huge amount of dsRNA probably challenges the RNAi machinery to deal with a situation far beyond what it is designed for. Thus, what we observe may just be due too low capacity of the Dicer enzyme system, which produces 21–23 bp dsRNAs that target the selective destruction of homologous RNAs [29]. This suggests that it might be possible to ensure a consistent inhibition of gene function by injection of a great excess of dsRNA since it is not itself toxic to bees.
If our results with vitellogenin turn out to be representative for large number of genes being expressed in various tissues of the adult honeybee, the intra-abdominal injection technique provides a way to do functional genomic studies on adult honeybees by very simple means. The technique can be considered to be conditional to the extent that we can expose the adult bee to the dsRNA at our choice. It allows groups of genes to be simultaneously rendered ineffective without the need for time-consuming crosses, and it may even allow sequential targeted disruption that might turn out to be crucial for understanding certain types of regulatory structures.
The technique can easily be extended to include delivery of siRNA. High-pressure injection of siRNA into the tail vein of postnatal mice has recently proven to be highly efficient for specific inhibition of transgene expression in a variety of organs [30]. As use of siRNA seems to be a means to target specific isoforms in Drosophila [31], and to ensure a moderate longevity of RNAi in mice [30] delivery of siRNA by intra-abdominal injection may provide additional possibilities for temporal and spatial control of RNAi in adult workers.
Conclusions
As we observe substantial RNAi in adults obtained from eggs injected with dsRNA at the preblastoderm stage, this injection method seems to provide a means to disrupt gene function in the embryonic, larval and pupal stages. However, intra-abdominal injection of dsRNA in adults is highly superior to egg injection for specific disruption of gene function in the adult stage. The intra-abdominal injection technique is much more efficient. It is substantially simpler, and it makes it possible to specifically address gene function in adults even in those cases where the focal gene is also expressed in the embryonic, larval or pupal stages. The number of (partially) sequenced honeybee genes is steadily increasing [32-34]. It is thus encouraging that we apparently do not need to wait for the development of sophisticated transformation techniques before we can start doing advanced reverse genetics on this fascinating model system.
Methods
Preparation of dsRNA
Primers were designed from the sequence of Apis mellifera vitellogenin (GenBank accession number: AJ517411), clone AP4a5 (MD Piulachs, J Cruz, X Bellés, KRG, AR Barchuk and ZLPS, submitted). The AP4a5 primer sequences were fused with the T7 promoter sequence (underlined) as 5'-TAATACGACTCACTATAGGGCGAACGACTCGACCAACGACTT-3', and 5'-TAATACGACTCACTATAGGGCGAAACGAAAGGAACGGTCAATTCC-3' PCR reactions were performed according to standard procedures using AP4a5 as template. PCR product was purified using the QIAquick™ PCR purification kit (QIAGEN).
RNA was prepared using the Promega RiboMax™ T7 system (Promega), where sense and antisense strands were transcribed from DNA template in the same reaction. RNA was phenol-chloroform extracted and isopropanol precipitated. RNA was resuspended in nuclease free water, heated at 95°C for 1 min and left to cool at room temperature. The product was divided in two batches and diluted with nuclease free water to a final concentration of 5 and 10 μg/μl, respectively.
Injections of eggs and subsequent rearing of adults
Honeybee eggs 0–6 hours old were collected from specifically designed hives kept in a flight room, allowing frequent collection of eggs of defined age with only slight disturbance of the colony [35,36]. Injections of dsRNA (10 μg/μl, N = 500) or nuclease free water (controls, N = 300) were performed as described [12]. We did not include individuals injected with nonsense dsRNA in the control group as a previous study, [12], strongly suggests that RNAi is highly specific in honeybees [12]. The average amount injected into each embryo was 300 ρl. The eggs were incubated at 35°C and 80 % RH for 65 hours before reintroduction of the surviving embryos into empty cells of flight room colonies with removable comb sections. The coordinates of the manipulated eggs were documented. dsRNA injected eggs (N = 398) and controls (N = 273) were inserted in separate sections. After 18 days the sections of comb were taken out and all brood not located at the coordinates of the introduced eggs were removed. The combs were incubated at 35°C and 80 % RH for 2 days. Bees emerging during this time interval were collected and paint marked before introduction into a queenright nursing colony (N = 103 for dsRNA injected bees, N = 136 for the control).
Injections of adult bees
Newly emerged workers were immobilized to a piece of styrophor with two crossed needles at 8°C. In this position they were injected with 1 μl dsRNA solution (5 μg/μl, N = 50) or 1 μl nuclease free water (N = 40). Injections were made dorsally between the 5th and 6th abdominal segment with a Hamilton micro syringe, needle G 30 (Becton Dickinson). Individuals showing sign of hemolymph leakage after withdrawal of the needle were discarded (N = 3). The bees stayed in the fixed position for one hour before introduction into the nursing colony.
Sampling of adult bees
Bees were collected at ages 0, 5, 7, 10 an 15 days and anaesthetized on ice before hemolymph collection. For each worker, 1 μl hemolymph was dissolved in 10 μl Tris-buffer in duplicate (20 mM Tris, 150 mM NaCl, 5 mM EDTA, pH 7.5,1 mM phenylmethylsulfonyl fluorid, 5 mM benzamidin, 0.7 μM pepstatin, 8 μM chymostanin, 10 μM leupeptin and 0.8 μM aprotinin, Sigma). The abdomen was then separated from the thorax with forceps and the intestine removed. The abdominal segments with adhering fat-body tissue were rinsed in nuclease free water and frozen in 1 ml TRIZOL reagent (Invitrogen).
SDS-PAGE and western blotting
Individual samples of 2.0–5.0 μg hemolymph protein were subjected to one-dimensional SDS-electrophoresis using 7 % polyacrylamide gels [37]. N = 70 and N = 100 for workers stemming from eggs injected with dsRNA and nuclease free water, respectively. Likewise, N = 30 and N = 20 for bees injected with dsRNA and nuclease free water as adults. The identity of the vitellogenin band was confirmed by transferring a set of samples (N = 14) to nitrocellulose paper and probe with diluted antiserum (1:75 000) raised in rabbit as described [38] Bound antibodies were detected by biotinylated secondary goat antirabbit and Vectastain ABC-AmP™ Detection System (Vector).
Northern blot hybridization
Total RNA was extracted from the abdomen using TRIZOL reagent (Invitrogen). Individual samples of total RNA (6 μg) was electrophoresed on 1. 2 % denaturing formaldehyde agarose gel and visualized by ethidium-bromide with UV light. The gel was washed in 10× SSC before the RNA was transferred to Biodyne B membrane (Pall) in 20X SSC. Prehybridization was performed in hybridization buffer (5X SSC, 5 % (w/v) Dextran, 1 % (w/v) SDS, 5 % liquid block (Amersham, RPN 3540) at 65°C for 2.5 hours. Fluorescent-labeled (Amersham, RPN 3540) fragments (500 Ηg) of thevitellogeninclones AP4a5 or AmR9 (MD Piulachs, J Cruz, X Bellés, KRG, AR Barchuk and ZLPS, submitted) were added in the prehybridization buffer and hybridization was allowed to proceed over night. The final washes were performed at 65°C in 5X SSC, 0.1 % (w/v) SDS for 15 min and 1X SSC, 0.1 % (w/v) SDS for 15 min. The probe was visualized by Gene Images detection module (Amersham, RPN 3510). All workers injected with dsRNA (as embryos or adults), and subsequently assayed for hemolymph vitellogenin (cf. methods above), were assayed in this manner. In the case of the controls, the sample size was somewhat reduced as the SDS-PAGE indicated high vitellogenin titers for most bees (N = 82 and N = 14 for workers injected as embryos and adults, respectively).
Samples enriched with small RNA fragments were prepared by filtration with Microcon filtration units YM-100 (Millipore) as described [39]. Individual samples of knockdown phenotypes were selected based on the previous screen for RNAi. Low-molecular weight RNA (3 μg) was then electrophoresed through a 2.5 % agarose gel as pooled samples of total RNA stemming from 10 individuals. Samples were then blotted to a Hybond XL-membrane (Amersham, RPN 203S) and visualized as previously described.
Authors' contributions
GVA designed and coordinated the study. She produced dsRNA and carried out injections, in vitro rearing and sampling of bees. She participated in the Northern analysis and drafted the manuscript. ZLPS provided vitellogenin clones and sequence information. KRG optimized, coordinated and participated in the Northern analysis. KN performed the SDS-PAGE, the western blotting and extracted total RNA. SWO participated in coordination of the study and in writing of the manuscript.
Acknowledgments
Acknowledgements
We appreciate the help of M Beye and M Hasselmann in producing vitellogenin dsRNA, and of AM Nascimento on 33P hybridizations for visualizing small RNA fragments by Northern blot. We also thank A Hagen for technical assistance. We are grateful to K Hartfelder and MJA da Rocha for generous support and critical discussions. We thank G Hunt and K Hartfelder for helpful comments on the manuscript. GVA's contribution was financially supported by the Norwegian Research Council, project no. 133680/110.
Contributor Information
Gro V Amdam, Email: gro.amdam@ihf.nlh.no.
Zilá LP Simões, Email: zlpsimoe@usp.br.
Karina R Guidugli, Email: karina@rge.fmrp.usp.br.
Kari Norberg, Email: kari.norberg@ihf.nlh.no.
Stig W Omholt, Email: stig.omholt@ihf.nlh.no.
References
- Beye M, Poch A, Burgtorf C, Moritz RFA, Lehrach H. A gridded genomic library of the honeybee (Apis mellifera): A reference library system for basic and comparative genetic studies of a hymenopteran genome. Genomics. 1998;49:317–320. doi: 10.1006/geno.1998.5253. [DOI] [PubMed] [Google Scholar]
- RobinsonB GE, Aronstein K, Evans JE, Fahrbach SE, Johnston SK, Maleszka R, Page RE, Robertson HM, Weaverc DB. Proposal for the Sequencing of a New Target Genome: White Paper for a Honey Bee Genome Project. The Honey Bee Genome Sequencing Consortium. 2002. http://www.nhb.org/bee-r/HoneyBeeWhitePaper.pdf
- Robinson KO, Ferguson HJ, Cobey S, Vaessin H, Smith BH. Sperm-mediated transformation of the honeybee, Apis mellifera. Insect Molec Biol. 2000;9:625–634. doi: 10.1046/j.1365-2583.2000.00225.x. [DOI] [PubMed] [Google Scholar]
- Seeley TD. The wisdom of the hive. The social physiology of honey bee colonies. Harvard, Harvard University Press. 1995.
- Menzel R, Giurfa M. Cognitive architecture of a mini-brain: the honeybee. TICS. 2001;5:62–71. doi: 10.1016/S1364-6613(00)01601-6. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Omholt SW. The regulatory anatomy of honeybee lifespan. J theor Biol. 2002;216:209–228. doi: 10.1006/jtbi.2002.2545. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Fahrbach SE, Winston ML. Insect societies and the molecular biology of social behavior. Bioessays. 1997;19:1099–1108. doi: 10.1002/bies.950191209. [DOI] [PubMed] [Google Scholar]
- Pankiw T, Page RE. Response thresholds to sucrose predict foraging division of labor in honeybees. Behav Ecol Sociobiol. 2000;47:265–267. doi: 10.1007/s002650050664. [DOI] [Google Scholar]
- Page RE, Erber J. Levels of behavioral organization and the evolution of division of labor. Naturwissenschaften. 2002;89:91–106. doi: 10.1007/s00114-002-0299-x. [DOI] [PubMed] [Google Scholar]
- Gadau J, Page RE, Werren JH, SchmidHempel P. Genome organization and social evolution in Hymenoptera. Naturwissenschaften. 2000;87:87–89. doi: 10.1007/s001140050016. [DOI] [PubMed] [Google Scholar]
- Fiala A, Muller U, Menzel R. Reversible downregulation of protein kinase a during olfactory learning using antisense technique impairs long-term memory formation in the honeybee, Apis mellifera. J neuroscience. 1999;19:10125–10134. doi: 10.1523/JNEUROSCI.19-22-10125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beye M, Haertel S, Hagen A, Hasselmann M, Omholt SW. Specific developmental gene silencing in the honey bee using a homeobox motif. Insect Molec Biol. 2002;11:527–532. doi: 10.1046/j.1365-2583.2002.00361.x. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. RNAi: nature abhors a double-strand. Current Opinion in Genetics & Development. 2002;12:225–232. doi: 10.1016/S0959-437X(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Haunerland NH, Shirk PD. Regional and functional differentiation in the insect fat body. Annu Rev Entomol. 1995;40:121–145. [Google Scholar]
- Snodgrass RE. Anatomy of the honeybee. London, Cornell University Press. 1956.
- Engels W. Extraoocytäre Komponenten des Eiwachstums bei Apis mellifera. Insectes Soc. 1968;15:271–288. [Google Scholar]
- Barchuk AR, Bitondi MMG, Simões ZLP. Effects of juvenile hormone and ecdysone on the timing of vitellogenin appearance in hemolymph of queen and worker pupae of Apis mellifera. J Insect Sci. 2002;2:1. doi: 10.1673/031.002.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitondi MMG, Simões ZLP. The relationship between level of pollen in the diet, vitellogenin and juvenile hormone titres in Africanized Apis mellifera workers. J Apic Res. 1996;35:27–36. [Google Scholar]
- Byrne BM, Gruber M, Ab G. The evolution of egg yolk proteins. Prog Biophys Mol Biol. 1989;53:33–69. doi: 10.1016/0079-6107(89)90005-9. [DOI] [PubMed] [Google Scholar]
- Babin PJ, Bogerd J, Koogman FP, Van Marrewijk WJA, Van der Horst DJ. Apolipophorin II/I, apoprotein B, vitellogenin and microsomal triglyseride transfer protein genes are derived from a common ancestor. J Molec Evol. 1999;49:150–160. doi: 10.1007/pl00006528. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Norberg K, Hagen A, Omholt SW. Social exploitation of vitellogenin. Proc Natl Acad Sci USA. [DOI] [PMC free article] [PubMed]
- Bulet P. Drosophila antimicrobial peptides. M S-Med Sci. 1999;15:23–29. [Google Scholar]
- Dzitoyeva S, Dimitrijevic N, Manev H. Intra-abdominal injection of double-stranded RNA into anesthetized adult Drosophila trigger RNA interference in the central nervous system. Molecular Psychiatry. 2001;6:665–670. doi: 10.1038/sj.mp.4000955. [DOI] [PubMed] [Google Scholar]
- Boutla AB, Kalantidis K, Tavernarakis N, Tsagris M, Tabler M. Induction of RNA interference in Caenorhabditis elegans by RNAs derived from plants exhibiting post-transcriptional gene silencing. Nucleic Acids Research. 2002;30:1688–1694. doi: 10.1093/nar/30.7.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepardi C, Wei Q, Paterson BM. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- Parrish S, Fleenor J, SiQun X, Mello C, Fire A. Functional anatomy of a dsRNA trigger: Different requirements for the two trigger strands in RNA interference. Molecular Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role of the bidentate ribonuclease in the initial step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nature Genetics. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- Celotto AM, Graveley BR. Exon-specific RNAi: A tool for dissecting the functional relevance of alternative splicing. RNA. 2002;8:718–724. doi: 10.1017/S1355838202021064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Wheeler DE. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc Natl Acad Sci USA. 1999;96:5575–5580. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield CW, Band MR, Bonaldo MF, Kumar CG, Liu L, Pardinas J, Robertson HM, Soares B, Robinson GE. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 2002;12:555–566. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Winningham KM, Hoffman DR. The bee venom protease allergen contains a CUB domain. J Allergy Clin Immunol. 2002;109:S79. doi: 10.1016/j.jaci.2004.07.043. [DOI] [PubMed] [Google Scholar]
- Omholt SW, Hagen A, Elmholdt O, Rishovd S. A laboratory hive for frequent collection of honey bee eggs. Apidologie. 1995;26:297–304. [Google Scholar]
- Omholt SW, Rishovd S, Hagen A, Elmholdt O, Dalsgard B, Fromm S. Successful production of chimeric honey bee larvae. J Exp Zool. 1995;272:410–412. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Jensen PV, Børgesen LW. Regional and functional differentiation in the fat body of pharaoh's ant queens, Monomorium pharaonis (L.). Arth Struc Dev. 2000;29:171–184. doi: 10.1016/S1467-8039(00)00021-9. [DOI] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: Cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- Shipman BA, Ryan RO, Schmidt JO, Lae JH. Purification and properties of a very high density lipoprotein from the hemolymph of the honeybee Apis mellifera. Biochemistry. 1987;26:1885–1889. doi: 10.1021/bi00381a015. [DOI] [PubMed] [Google Scholar]