Abstract
The small GTPase Ran is required for the trafficking of macromolecules into and out of the nucleus. Ran also has been implicated in cell cycle control, specifically in mitotic spindle assembly. In interphase cells, Ran is predominately nuclear and thought to be GTP bound, but it is also present in the cytoplasm, probably in the GDP-bound state. Nuclear transport factor 2 (NTF2) has been shown to import RanGDP into the nucleus. Here, we examine the in vivo role of NTF2 in Ran import and the effect that disruption of Ran imported into the nucleus has on the cell cycle. A temperature-sensitive (ts) mutant of Saccharomyces cerevisiae NTF2 that does not bind to Ran is unable to import Ran into the nucleus at the nonpermissive temperature. Moreover, when Ran is inefficiently imported into the nucleus, cells arrest in G2 in a MAD2 checkpoint-dependent manner. These findings demonstrate that NTF2 is required to transport Ran into the nucleus in vivo. Furthermore, we present data that suggest that depletion of nuclear Ran triggers a spindle-assembly checkpoint-dependent cell cycle arrest.
INTRODUCTION
Efficient protein import into the nucleus requires both the small GTP-binding protein, Ran, and the homodimeric nuclear transport factor 2 (NTF2) protein. NTF2 was originally identified as an activity that stimulated the import of proteins into the nuclei of permeabilized mammalian cells (Moore and Blobel, 1994). The NTF2 protein binds specifically to the GDP-bound form of Ran (Paschal and Gerace, 1995; Clarkson et al., 1996; Stewart et al., 1998) and to nucleoporins (Paschal and Gerace, 1995; Clarkson et al., 1996) at distinct nonoverlapping sites. Recent studies demonstrate that NTF2 acts as a mediator of RanGDP import into the nucleus (Ribbeck et al., 1998; Smith et al., 1998; Steggerda et al., 2000). Once RanGDP is transported into the nucleus, the Ran nucleotide exchange factor, RCC1, converts RanGDP to RanGTP (Bischoff and Ponstingl, 1991). Since RCC1 is chromatin associated and exclusively nuclear (Ohtsubo et al., 1987), RanGDP to RanGTP exchange occurs only in the nucleus (Ohtsubo et al., 1989). The compartmentalization, of both the Ran exchange factor, within the nucleus (Ohtsubo et al., 1987), and the RanGAP (Ran GTPase activating protein), within the cytoplasm (Hopper et al., 1990), is thought to produce a Ran gradient that is hypothesized to drive vectorial nucleocytoplasmic transport.
ts Mutants of NTF2, RCC1, and Ran have well-characterized nuclear transport defects (Corbett and Silver, 1997; Wong et al., 1997; Mattaj and Englmeier, 1998). In addition, RCC1 mutants show a diverse range of cell cycle defects including chromosome instability, premature chromatin condensation and aberrant spindle formation (Dasso, 1993). These defects, until recently, were often considered to be the indirect consequence of the disruption of nucleocytoplasmic transport. However, the discovery of the centrosome-localized Ran binding protein RanBPM (Nakamura et al., 1998) has raised the possibility that the Ran GTPase system has a more direct role in cell cycle control (Sazer and Dasso, 2000). In fact, several recent studies demonstrated that in vitro RanGTP promotes microtubule formation when added to mitotic Xenopus egg extracts (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999). These observations indicate that Ran may influence cell cycle progression via a direct physical mechanism similar to the way tubulins and DNA replication factors mediate the events required for cell cycle progression.
In addition to proteins that are the physical components that mediate cell cycle events, surveillance mechanisms exist to ensure that critical events occur in the proper sequence in the course of the cell cycle. The spindle-assembly checkpoint ensures accurate nuclear division by monitoring the integrity of the mitotic spindle (Gardner and Burke, 2000; Straight and Murray, 1997). Characterization of the spindle-assembly checkpoint in S. cerevisiae has led to the isolation of eight checkpoint genes, MAD1,2,3, BUB1,2,3, MPS1, and CDC55 (Rudner and Murray, 1996). Disruption of any one of these genes results in the failure of cells to arrest in the presence of spindle damage (Hoyt et al., 1991; Li and Murray, 1991; Hardwick, 1998). Activation of the spindle-assembly checkpoint in yeast causes cells to arrest with condensed chromosomes before the onset of anaphase (Li and Murray, 1991; Hardwick et al., 1996). If Ran directly regulates microtubule assembly, and thus spindle assembly, one would expect perturbations in Ran to trigger a checkpoint-dependent response.
S. cerevisiae has been a useful model for studying cell cycle control and spindle formation in eukaryotic cells (Straight and Murray, 1997; Gardner and Burke, 2000). Unlike higher eukaryotes, budding yeast does not undergo nuclear envelope breakdown during mitosis; thus, any factor required for intranuclear spindle formation must be transported into the nucleus. If Ran is required for mitotic spindle formation in vivo, we would expect aberrant spindle formation in a subset of NTF2 mutants that are unable to bind to and import Ran into the nucleus. We would also predict that the resulting disruption of spindle formation would trigger the spindle-assembly checkpoint pathway and arrest cells before anaphase. Here, we investigate the in vivo role of NTF2 in Ran import and the effect that disruption of this import has on spindle formation and cell cycle progression.
MATERIALS AND METHODS
Yeast Strains and Plasmids
The wild-type (PSY580) and NTF2 deletion strains (ACY114, ACY115) used in this study have been described (Corbett and Silver, 1996). The MAD2 deletion strain, KH141, (MATa, MAD2::URA3, leu2,3–112, trp1–63, ade2, his3–11,15) was the generous gift of Andrew Murray. The yeast plasmids, pPS882 (CEN, LEU2, NTF2), pPS883 (CEN, URA3, NTF2), pPS919 (CEN, LEU2, ntf2–1), and pPS920 (CEN, LEU2, ntf2–2) used in this study have been described (Corbett and Silver, 1996). The bacterial expression plasmid for Ntf2p (pPS982) has been described (Wong et al., 1997). The NLS-green fluorescent protein (GFP) plasmid (pGADGFP) used for the NLS-GFP import assay has been described (Shulga et al., 1996). The plasmid for the expression of C-terminal fusion to the GFP has been described (Kahana et al., 1995). Fusion constructs to scRan (Gsp1p) were constructed by engineering an in-frame XhoI site at the termination site of the GSP1 coding region and a PstI 710 bp upstream of the 5′ start site of the GSP1 coding region (pAC410). The bacterial expression plasmids for Ntf2–1p (pAC41) and Ntf2–2p (pAC43) were generated by cloning an ∼480-bp PCR fragment containing a 5′-BamHI restriction enzyme site and a 3′-HindIII restriction enzyme site introduced by PCR amplification of pPS991 (ntf2–1ts) or pPS920 (ntf2–2ts) with the primers AC21 (5′CGGGATCCATGTCTCTCG ACTTTAACAC3′) and AC88 (5′CCCGAAGCTTCGCTATCGCCTTATACATCG3′). The PCR product was cloned into the T7-based expression vector pMW172 (Way et al., 1990). All procedures including yeast transformations, culture manipulations, and extract preparations were performed by standard methods (Adams et al., 1997).
Ntf2p Purification and Immobilization
Yeast Ntf2p was purified from Escherichia coli as previously described for rat Ntf2p (Clarkson et al., 1996). Expression plasmids for Ntf2p (pPS982), Ntf2–1p (pAC41), or Ntf2–2p (pAC43) were transformed into E. coli BL21 (DE3). Transformants were inoculated into 2X tryptone-yeast extract medium containing 100 μg of ampicillin/ml and grown overnight at 30°C. It was not necessary to induce expression as the basal level of expression of the T7 polymerase yielded a large amount of Ntf2p. Bacteria were harvested by centrifugation and were stored at −80°C until required.
Ntf2p was isolated by thawing the cell pellet and resuspending it in 25% sucrose, 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were lysed by French press and treated with DNAse at 25°C for 30 min. The soluble fraction was isolated by centrifugation at 40,000 × g for 20 min and dialyzed overnight against 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1 mM PMSF (NTF2 buffer A). The lysate was clarified at 40,000 × g for 30 min at 4°C, was applied to DE52 ion exchange column (10 × 3 cm), and was washed with NTF2 buffer A. Ntf2p was eluted from the column with a gradient of 0 to 400 mM NaCl. Fractions containing Ntf2p were pooled, concentrated using a Centriprep-10 (Amicon, Charlotte, NC) concentrator, and applied to a column of Sephacryl SR100 preequilibrated in 20 mM Tris-HCl (pH 7.4), 50 mM NaCl, 2 mM MgCl2, 1 mM DTT, 0.1 mM PMSF (NTF2 buffer B). Fractions containing Ntf2p were collected and pooled.
Purified Ntf2p was cross-linked to cyanogen bromide (CNBr)-sepharose beads as previously described (Clarkson et al., 1996). Briefly, CNBr-sepharose beads (Pharmacia, Uppsala, Sweden) were swollen and washed in 1 mM HCl. Beads were transferred to coupling buffer (100 mM NaHCO3 (pH 8.3), 500 mM NaCl) and were added to 2–5 mg of Ntf2p in coupling buffer. Coupling was carried out at 4°C overnight. Residual active groups were blocked with 1 M Tris-HCl (pH 8.0) for 2 h at room temperature. Beads then were washed successively and extensively four times in coupling buffer and acid wash buffer (0.1 M sodium acetate (pH 4.0), 500 mM NaCl).
Binding Assays
Yeast cell extracts were prepared from cultures grown overnight at 30°C or at room temperature for ts strains in yeast extract-peptone-dextrose (YEPD) medium to confluency. Cells were harvested by centrifugation and washed once with water. Cells then were resuspended in one volume of PBSMT (1 × PBS, 2.5 mM MgCl2, 0.5% Triton X-100) supplemented with protease inhibitors (0.5 mM PMSF and 3 μg each of aprotinin, leupeptin, chymostatin, and pepstatin per milliliter). One volume of glass beads was added, and cells were lysed with 10–15 60-s pulses in a beadbeater (lysis was monitored by light microscopy to >70% lysis). The resulting lysate was clarified by centrifugation and assayed for protein concentration by using the Bio-Rad (Cambridge, MA) Protein Assay Kit.
Two milligrams of yeast lysate was incubated with 50 μl of Ntf2p-sepharose beads. Binding was carried out in PBSM (total volume, 500 μl) at 4°C for 1 h. Beads then were washed two times for 10 min in PBSM and one time for 10 min in PBSMT. Bound proteins were eluted with 100 μl of sample buffer and were resolved by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose for immunoblotting.
Immunoblot Analysis
Immunoblot analysis was performed essentially as described (Towbin et al., 1979) with the following modifications: following transfer, the nitrocellulose filter was blocked in TBST buffer (10 mM Tris, 140 mM NaCl, 0.05% Tween-20) plus 5% milk for 15 min. Filters to be used for the detection of scRan were blocked with TBS1/2T (TBS buffer containing 0.025% Tween-20) for 15 min. scRan-GFP was detected by incubation with a 1:5000 dilution (in TBST plus 5% milk) of an anti-GFP rabbit polyclonal antibody (the generous gift of P. Silver and J. Kahana, Harvard Medical School, Boston, MA). scRan was detected by incubation with a 1:1000 dilution (in TBS1/2T plus 5% milk) of an antiscRan rabbit polyclonal antibody (the generous gift of D. H. Wong and P. Silver, Harvard Medical School). Following several washes with TBST (or TBS1/2T for scRan), the filter next was incubated in a 1:5000 dilution of horseradish peroxidase-linked goat antirabbit polyclonal antiserum (Promega, Madison, WI) for 1 h at room temperature. The filter was again washed in TBST (or TBS1/2T for scRan) and detection was carried out with a 1:1 mixture of (chemiluminescent) ECL reagents (Amersham, Little Chalfont, UK) as recommended by the manufacturer.
Localization of scRan-GFP and Nuf2-GFP
The scRan-GFP and Nuf2-GFP (Kahana et al., 1995) fusion proteins were localized by directly viewing the GFP signal in living cells through a GFP-optimized filter (Chroma Technology, Brattleboro, VT) using an Olympus (Tokyo, Japan) BX60 epifluorescence microscope equipped with a Photometrics (Tucson, AZ) Quantix digital camera.
Indirect Immunofluorescence Microscopy
Ten-milliliter cultures were grown to log phase in YEPD overnight, then the cultures were split, and half was maintained for 3 h at 25°C and half was shifted to 37°C for 3 h. Cultures then were fixed with 600 μl of 37% formaldehyde for 30 min. Following fixation, cells were harvested by centrifugation and were washed once with 0.1 M potassium phosphate buffer (pH 6.5), once with P solution (1.2 M sorbitol, 0.1 M potassium phosphate buffer, pH 6.5), and were resuspended in 1 ml of P solution. DTT was added to 25 mM, and the cells were incubated at 30°C for 10 min with gentle agitation followed by the addition of 300 μg of zymolyase (United States Biological, Swampscott, MA). The digestion of the cell wall was monitored by microscopy. Digested cells were collected by centrifugation, were washed once with P solution, and were resuspended in 1 ml of P solution. Cells then were applied to Teflon-faced microscope slides precoated with 0.3% polylysine. After cells were adhered to slides, they were fixed with methanol at −20°C for 6 min and dried in cold acetone for 30 s. Cells were blocked by incubating for 15 min with PBS plus 0.5% bovine serum albumin (BSA). Antitubulin was diluted 1:100, and anti-Npl3p was diluted 1:1000 in PBS plus 0.5% BSA and incubated with the cells overnight at room temperature. Cells were washed several times with PBS plus 0.5% BSA and were incubated for 2 h with a 1:1000 dilution of either fluoroscein isothiocyanate (FITC)-labeled antimouse antibodies or Texas Red-labeled antimouse antibodies (Jackson ImmunoResearch, West Grove, PA) for antitubulin or with FITC-labeled antirabbit for Npl3p as indicated and with 4′,6-diamido-2-phenylindole (DAPI antibodies) (1 μg/ml).
Growth and Viability
NTF2, ntf2–1, and ntf2–2 cells were grown in YEPD overnight at 25°C, diluted to 2 × 106 cells/ml and shifted to 37°C. Growth was monitored by counting cells every 2 h using a hemacytometer. Viability was monitored every 2 h by plating 200 cells/plate onto YEPD plates. Plates were incubated at 25°C for 4 days, and colonies were counted.
Determination of Yeast Cell DNA Content
Cells were prepared for the FACS by staining with propidium iodide (Epstein and Cross, 1992). Briefly, cells were ethanol fixed overnight at 4°C, washed, and resuspended in 1 ml of 50 mM sodium citrate, pH 7.0. Cells then were treated with 0.08 mg/ml Rnase A for 1 h at 50°C followed by 0.25 mg/ml proteinase K before incubation in 8 μg/ml propidium iodide. Each sample was analyzed with a FACS Caliber cytometer from Becton Dickinson (Franklin Lakes, NJ).
Construction and Analyses of MAD2Δ Strains
To construct the ntf2ts MAD2Δ, double mutants, KH141 (MAD2Δ) was mated with ACY115 (NTF2Δ) containing either pPS919 (ntf2–1ts) or pPS920 (ntf2–2ts). The resulting diploids were sporulated, and tetrads were dissected to isolate the double-mutant strains. Single and double mutants as well as a wild-type controls were grown overnight in YEPD at 25°C. Cells were diluted to 0.2 × 106 cells/ml in 50 ml of YEPD, with half grown at 25°C and half shifted to 37°C. Growth was monitored by measuring the OD600. To test the benomyl sensitivity of the yeast strains indicated, cells were grown overnight at 25°C, and 100,000, 10,000, 1,000, 100, and 10 cells were spotted on 10-μg/ml benomyl plates and incubated at 25°C. To test the nocodazole sensitivity of the yeast strains indicated, cells were grown overnight at 25°C and diluted to 0.2 × 106 cells/ml into YEPD containing 15 μg/ml nocodazole, and growth was monitored by OD600.
NLS-GFP Import Assay
The NLS-GFP import assay was performed as previously described (Shulga et al., 1996). Briefly, cells were grown to early-midlog phase in synthetic media containing 2% glucose at 25°C, were pelleted, were resuspended in 1 ml of 10 mM sodium azide and 10 mM 2-deoxy-d-glucose in glucose-free synthetic medium, and were incubated at 25°C for 45 min. The cells then were pelleted, were washed with 1 ml of ice-cold ddH20, were repelleted, were resuspended in 100 μl of glucose-containing synthetic medium prewarmed to 37°C and were incubated at 37°C. For scoring, 2-μl samples were removed every 2.5 min, and cells observed and counted through a GFP optimized filter (Chroma Technology) using an Olympus BX60 epifluorescence microscope. Cells were scored as “nuclear” if the nucleus was both brighter than the surrounding cytoplasm and a nuclear-cytoplasmic boundary was visible. At least 50 cells were counted at each time point.
RESULTS
NTF2 Imports Ran into the Nucleus In Vivo
To test whether Ntf2p imports Ran into the nucleus in vivo, we analyzed two previously identified ts NTF2 mutants, ntf2–1ts (M83T) and ntf2–2ts (D91G). Both of these mutations previously have been shown to disrupt protein import into the nucleus at the nonpermissive temperature, but neither has any effect on poly(A)+ RNA export (Corbett and Silver, 1996). The crystal structure of rat Ntf2p has been solved (Bullock et al., 1996). Yeast Ntf2p is 46% identical to rat Ntf2p, and rat Ntf2p functionally complements a yeast NTF2 deletion (Corbett and Silver, 1996). Given the functional conservation of the Ntf2 protein, we used the rat structure to model the yeast Ntf2–1ts and Ntf2–2ts mutant proteins. This analysis indicates that the Ntf2–1ts mutation lies in the Ntf2p dimerization domain and that the Ntf2–2ts mutation lies in a residue predicted to form a critical salt bridge between RanGDP and Ntf2p (Clarkson et al., 1996; Stewart et al., 1998; Kent et al., 1999).
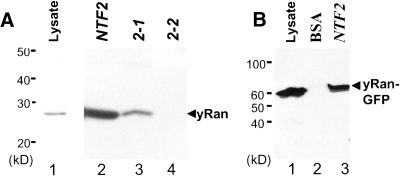
To analyze the effect that each of the two ts mutations in Ntf2p have on the interaction between Ntf2p and scRan (Gsp1p), we expressed and purified wild-type Ntf2p and the two mutant Ntf2 proteins from bacteria (Kent et al., 1996). Each of these proteins was covalently linked to CNBr-sepharose beads and incubated with yeast cell lysates, and the beads were analyzed by immunoblotting for bound scRan (Figure 1A). As previously reported, wild-type Ntf2 protein bound scRan very efficiently (Clarkson et al., 1997; Wong et al., 1997). The Ntf2–1 protein also bound to scRan although with decreased efficiency. In contrast, scRan binding to the Ntf2–2 protein was undetectable (Figure 1A, lane 4).
Figure 1.
Interaction of scRan with Ntf2p. Interactions were examined using bead-binding assays as described in Materials and Methods. Immunoblots detecting scRan are shown. (A) Interaction of scRan with Ntf2–1p and Ntf2–2p. Lane 1, yeast cell lysate; lane 2, wild-type Ntf2p beads; lane3, Ntf2–1p beads; and lane 4, Ntf2–2p beads. (B) Interaction of scRan-GFP with Ntf2p. Lane 1, yeast cell lysate; lane 2, BSA beads; and lane 3, Ntf2p beads.
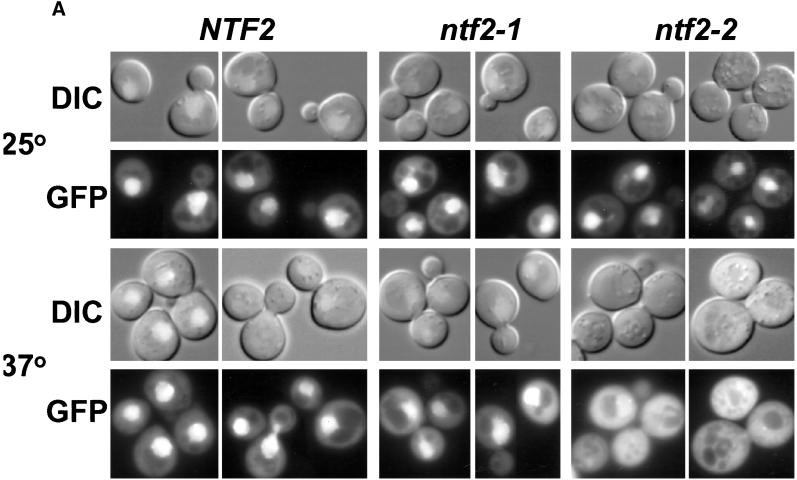
To follow the cellular localization of scRan in vivo, GFP (Chalfie et al., 1994) was fused to the C-terminus of scRan. Although scRan-GFP was unable to substitute for the endogenous scRan protein (our unpublished results), bead-binding assays demonstrate that the GFP fusion has no effect on the binding of scRan to Ntf2p (Figure 1B). Thus, scRan-GFP can be used as a tool to determine whether disruption of the interaction between Ntf2p and scRan has any effect on the localization of scRan in vivo. scRan-GFP localization was analyzed in yeast cells deleted for NTF2 and was maintained by plasmids encoding Ntf2p, Ntf2–1p, or Ntf2–2p (Corbett and Silver, 1996). These cells were grown at 25°C to log phase and then were shifted to 37°C for 3 h. In cells expressing wild-type Ntf2p, scRan was located throughout the cell but clearly was concentrated in the nucleus (Figure 2A), as demonstrated by costaining with DAPI (our unpublished results). A similar localization pattern was observed in the ntf2–1ts cells. In contrast, in the ntf2–2ts cells, scRan was diffusely localized throughout the cell with no concentration in the nucleus (Figure 2A). The steady-state level of scRan-GFP in each strain was approximately equal as determined by immunoblotting with anti-GFP (Figure 2B), indicating that the differences observed in the mutant strains were not due to differences in the level of scRan-GFP expression. Together these data indicate that the interaction between Ntf2p and scRan is required for the efficient localization of Ran to the nucleus.
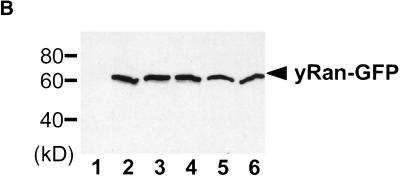
Figure 2.
Localization of scRan. (A) NTF2, ntf2–1ts, and ntf2–2ts cells were transformed with a 2μ plasmid encoding scRan-GFP (pAC410). Transformants were grown in liquid media lacking uracil to log phase at 25°C, were split, and were grown at 25°C or 37°C for 3 h. scRan-GFP was viewed directly in living cells. (B) Levels of scRan-GFP are the same in NTF2, ntf2–1ts, and ntf2–2ts transformants grown at 25°C and 37°C. Cells were lysed as described in Materials and Methods, 10 μg of each lysate was resolved on SDS-polyacrylamyide gel, transferred to nitrocellulose, and detected with anti-GFP. Lane 1, vector alone (no GFP) 37°C; lanes 2–4, 25°C (lane 2, NTF2; lane 3, ntf2–1ts; and lane 4, ntf2–2ts); lanes 5 and 6, 37°C (lane 5, ntf2–1ts; lane 6, ntf2–2ts).
If Ntf2p is required to import Ran into the nucleus, one would predict that overexpression of scRan would suppress the ntf2–2 phenotype. In fact, consistent with previous reports demonstrating that the overexpression of scRan can complement an NTF2 deletion (Paschal et al., 1997), the overexpression of scRan suppresses the ntf2–2ts phenotype (our unpublished results).
Cell Cycle Arrest in ntf2–1ts and ntf2–2ts Mutant Cells
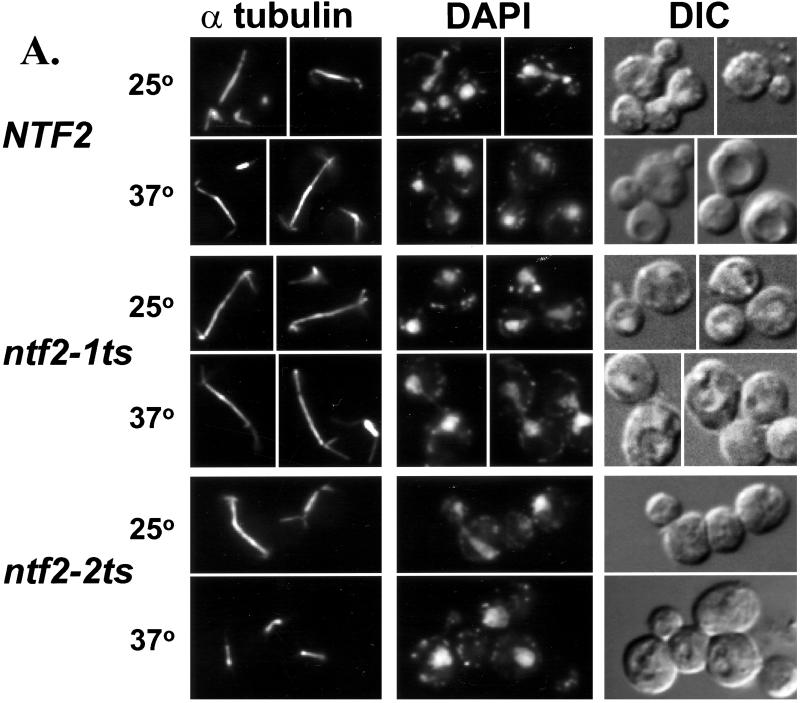
Ran has been implicated in a variety of cell cycle processes including mitotic spindle formation (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999). The NTF2 alleles described above provide excellent tools to test whether changes in the level of nuclear Ran affect microtubule morphology and/or cell cycle arrest in vivo. To observe the microtubules in the mutant NTF2 cells, the NTF2, ntf2–1ts, and ntf2–2ts cells were immunostained with an antitubulin antibody to visualize microtubules. At both the permissive and nonpermissive temperature wild-type and ntf2–1ts cells displayed a normal distribution of microtubule morphologies. In contrast, at the nonpermissive temperature the ntf2–2ts cells, which are unable to efficiently import Ran into the nucleus, accumulate in mitosis as large budded cells with short spindles (Figure 3A). DAPI staining of these cells shows that the majority of these cells have not segregated their chromatin. Approximately 60% of the ntf2–2ts cells display this phenotype as compared with only 18% of the ntf2–1ts cells and <1% of the wild-type cells, both of which efficiently import Ran into the nucleus.
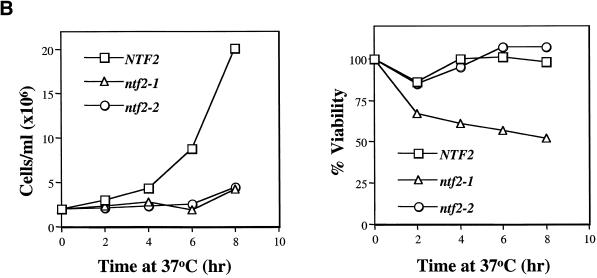
Figure 3.
Cell cycle arrest in the ntf2–2ts mutant. (A) Visualization of spindles in NTF2 mutants. NTF2, ntf2–1ts, and ntf2–2ts cells were grown to log phase at 25°C, were split, and were shifted at 25°C or 37°C for 3 h. Cells were stained with antitubulin, to visualize microtubules, and with DAPI, to visualize DNA, as described in Materials and Methods. (B) Growth and viability of NTF2 mutants at 37°C. Cells were grown overnight at 25°C, diluted to 2 × 106 cells/ml, and shifted to 37°C. Samples were removed every 2 h, counted, and 200 cells were plated onto YEPD at 25°C. The results are plotted as cell number versus time at 37°C (left panel) or percent of viability versus time at 37°C (right panel).
To determine whether the ntf2–2ts cells undergo a reversible mitotic arrest, the growth and viability of NTF2, ntf2–1ts, and ntf2–2ts cells were analyzed following a shift to 37°C. As previously shown (Corbett and Silver, 1996), both the ntf2–1ts and ntf2–2ts mutant cells cease growth rapidly when shifted to 37°C (Figure 3B, left panel). The viability of ntf2–1ts cells at the nonpermissive temperature is significantly reduced compared with wild-type NTF2 cells. In contrast, the viability of ntf2–2ts cells is similar to the NTF2 cells (Figure 3B, right panel). These data indicate that the Ntf2–2 mutation causes a specific and reversible cell cycle arrest, rather than a loss of viability.
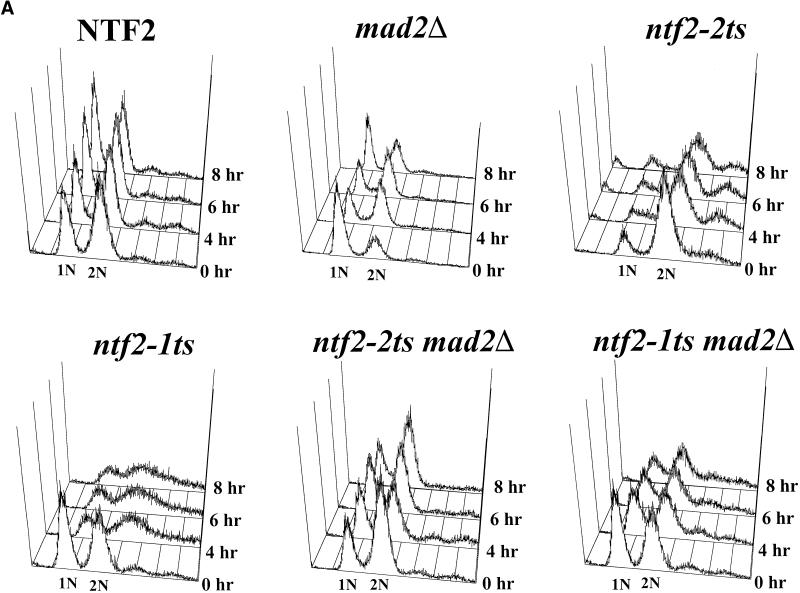
To further characterize the arrest phenotype observed in the NTF2 mutants, we utilized FACS to determine whether the DNA had replicated in the arrested cells. NTF2, ntf2–1ts, and ntf2–2ts cells were grown overnight at room temperature, diluted, shifted to 37°C, and samples taken at 0, 4, 6, and 8 h. Cells were stained with propidium iodide, and the DNA content was analyzed by flow cytometry. Wild-type cells showed an equal distribution of cells with 1N and 2N DNA content, whereas, the majority of the ntf2–2ts cells contained 2N DNA content (Figure 4A). Thus, ntf2–2ts mutant cells arrest as large budded cells with duplicated DNA. In the ntf2–1ts mutant cells, following a shift to 37°C, there was a broad peak between 1N and 2N DNA content, suggesting that many of the cells lag in their DNA replication, which is consistent with a general slowdown in cell cycle progression in this mutant. This confirms that, unlike the ntf2–2ts cells, the ntf2–1ts cells do not arrest at a specific point in the cell cycle.
Figure 4.
Analysis of the cell cycle arrest in ntf2–2ts mutant cells. (A) DNA content in NTF2 mutants. NTF2, ntf2–1ts, ntf2–2ts, MAD2Δ, ntf2–2ts MAD2Δ, and ntf2–1ts MAD2Δ cells were grown overnight at 25°C, were diluted to 2 × 106 cells/ml in YEPD, and were shifted to 37°C, and samples were taken at 0, 4, 6, and 8 h. The DNA content of individual NTF2, ntf2–1ts, and ntf2–2ts cells was determined by flow cytometry. (B) Spindle pole body localization in NTF2 mutants. NTF2, ntf2–1ts and ntf2–2ts cells were transformed with a plasmid encoding Nuf2-GFP (Kahana et al., 1995) to visualize spindle pole bodies. Transformed cells were grown to log phase at 25°C and then shifted to 37°C for 3 h. GFP-labeled spindle pole bodies were visualized by direct fluorescent microscopy.
FACS analysis indicated that the arrest in the ntf2–2ts mutant cells occurs after DNA replication. To determine whether the arrest is pre- or post-M phase, the spindle pole bodies were visualized with Nuf2-GFP (Kahana et al., 1995). In both the wild-type NTF2 and ntf2–1ts cells, spindle pole bodies show normal cell cycle localization (Figure 4B) with the majority of large budded cells containing duplicated and separated spindle pole bodies. In contrast, 75% of the ntf2–2ts cells arrest as large budded cells with spindle pole bodies that have duplicated but have not yet migrated to the poles (Figure 4B) indicating a G2 arrest.
The ntf2–2 Cell Cycle Arrest is MAD2 Dependent
To determine whether the cell cycle defects observed in the ntf2–2ts cells are checkpoint dependent, we crossed cells expressing either the ntf2–2ts allele or the ntf2–1ts allele of NTF2 with a mitotic arrest-deficient 2 mutant, MAD2Δ (Li and Murray, 1991). The resulting diploids were sporulated, and tetrads were dissected to produce wild-type NTF2; ntf2–2ts, ntf2–1ts, and MAD2Δ single mutants, as well as the ntf2–1ts MAD2Δ and ntf2–2ts MAD2Δ double mutants.
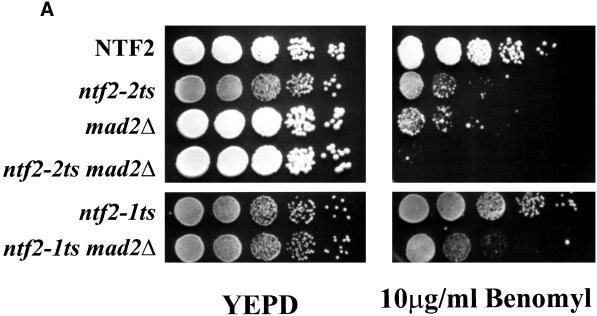
MAD2Δ cells exhibit wild-type growth under normal conditions; however, growth is compromised upon exposure to the microtubule-destabilizing drug benomyl (Li and Murray, 1991). The growth of each of the single and double mutants was first tested on plates containing 10 μg/ml benomyl. As expected, the MAD2Δ mutant is sensitive to benomyl (Figure 5A). Surprisingly, unlike the ntf2–1ts, the ntf2–2ts also exhibited a benomyl-sensitive phenotype (Figure 5A), indicating that the ntf2–2ts mutation has an allele-specific effect on microtubule function. In addition, the ntf2–2ts MAD2Δ double mutant exhibited a hypersensitivity to benomyl suggesting a compounding of this defect.
Figure 5.
The ntf2–2ts MAD2Δ double mutant is hypersensitive to microtubule-destabilizing drugs. (A) ntf2–2ts is a unique benomyl-sensitive allele of NTF2. NTF2, ntf2–2ts, ntf2–1ts, ntf2–2ts MAD2Δ, and ntf2–1ts MAD2Δ cells were grown at 25°C overnight, and 100,000, 10,000, 1000, 100, and 10 cells were spotted onto YEPD (left) and 10 μg/ml benomyl plates (right). Plates were incubated at 25°C for 4 days. (B) The ntf2–2ts MAD2Δ double mutant is hypersensitive to nocodazole. NTF2, MAD2Δ, ntf2–2ts, and ntf2–2ts MAD2Δ were grown at 25°C overnight, were diluted to 0.2 × 106 cells/ml into YEPD containing 15 μg/ml nocodazole, and growth was monitored by OD600.
Both spindle checkpoint mutants and microtubule assembly mutants exhibit benomyl sensitivity; however, checkpoint mutants are often more sensitive to the drug nocodazole than are mutants in genes that are directly involved in microtubule stability (Straight and Murray, 1997). To distinguish between an effect of the ntf2–2ts mutant, and thus the depletion of nuclear Ran, on microtubule stability and the spindle checkpoint, we tested NTF2, MAD2Δ, the ntf2–2ts single mutants and the ntf2–2ts MAD2Δ double mutant for their ability to grow in the presence of nocodazole. The ntf2–2ts mutant grows only slightly more slowly than wild-type cells in nocodazole (Figure 5B); this is most likely due to the slower growth of the ntf2–2ts at 25°C (compare to Figure 6A) rather than to a specific nocodazole sensitivity of this mutant. This suggests that the benomyl sensitivity observed for the ntf2–2ts mutant is due to a disruption of microtubule stability rather than to perturbation of the spindle checkpoint. Consistent with this is the nocodazole hypersensitivity exhibited by the ntf2–2ts MAD2Δ double mutant (Figure 5B). In this double mutant the depletion of Ran from the nucleus, resulting from the ntf2–2ts mutation, destabilizes microtubules mimicking the effects of the addition of benomyl. The addition of nocodazole deals a double microtubule-destabilizing blow to the cell that cannot be sensed in the absence of the MAD2 spindle-assembly checkpoint pathway. Therefore, the ntf2–2ts MAD2Δ double mutant displays hypersensitivity to nocodazole.
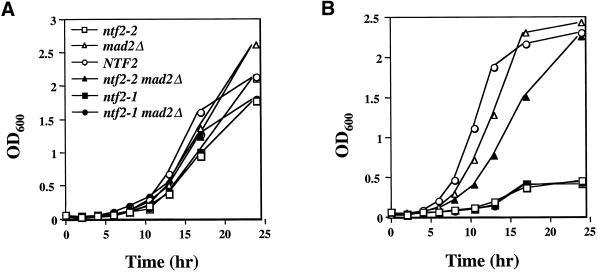
Figure 6.
MAD2Δ suppresses the temperature sensitivity of the ntf2–2ts but not of the ntf2–1ts allele. Cells were grown overnight at 25°C, and 0.2 × 106 cells/ml were inoculated into 50 ml of YEPD, split, and (A) grown at 25°C and (B) grown at 37°C. Growth was monitored by OD600.
To determine whether the ts arrest observed in the ntf2 mutants was MAD2 dependent, we tested the growth of the single and double mutants at 25°C and 37°C. While all strains grew well at the permissive temperature (Figure 6A), both the ntf2–2ts and ntf2–1ts strains displayed the previously observed ts phenotype (Corbett and Silver, 1996) and were unable to grow at 37°C (Figure 6B). As expected, the ntf2–1ts MAD2Δ double mutant also was unable to grow at 37°C. However, the MAD2Δ suppressed the ntf2–2ts phenotype allowing the ntf2–2ts MAD2Δ double mutant to grow at 37°C (Figure 6B). This is consistent with a role for Ran in microtubule stability. The destabilization of the microtubules by the depletion of Ran from the nucleus cannot be sensed in the absence of the MAD2 pathway in the double mutant. This results in the inability of the cells to arrest. Thus, the cells progress through the cell cycle and are able to grow at 37°C. Since Ran is not completely excluded from the nucleus, the residual nuclear Ran that is present is sufficient for the cell survival over the time course of this experiment. This demonstrates that the ts growth defect observed in the ntf2–2ts mutant is dependent on the MAD2 spindle-assembly checkpoint pathway.
To rule out the possibility that the MAD2Δ suppresses all nuclear transport mutants with cell cycle defects, MAD2Δ was crossed with a mutant in importin-α, srp1–31ts, which arrests as large budded cells with 2N DNA content at 37°C (Loeb et al., 1995). The srp1–31ts MAD2Δ double mutant maintains the ts phenotype observed for the srp1–31ts single mutant. Furthermore, the double mutant arrests as large budded cells at 37°C, which is similar to the srp1–31ts single mutant (our unpublished results). This indicates that the MAD2Δ is not a general suppressor of nuclear transport mutants with cell cycle defects. This finding, in combination with the inability of the MAD2Δ to suppress the ntf2–1ts mutation, suggests that the suppression of the ntf2–2ts allele by MAD2Δ is specific and is consistent with the hypothesis that the ntf2–2ts mutant triggers a spindle-assembly checkpoint arrest.
To determine whether a MAD2Δ suppresses the ntf2–2ts cell cycle defect, the MAD2Δ mutant and each of the double mutants (ntf2–1 MAD2Δ and ntf2–2 MAD2Δ) were subjected to FACS analysis. The single and double mutants were grown at room temperature, were diluted, and were shifted to 37°C. Samples were taken at 0, 4, 6, and 8 h, and DNA content was analyzed by FACS analysis. The MAD2Δ cells were close to the stationary phase when diluted but rapidly reached log phase with an equal distribution of cells with 1N and 2N DNA very much like wild-type cells grown at 37°C (see Figure 4A). The ntf2–1ts MAD2Δ phenotype was somewhat improved compared with the ntf2–1ts alone; however, there was still a broadening of the peaks, suggesting a slow down in DNA replication indicative of the single ntf2–1ts mutant that is not rescued by the MAD2Δ (see Figure 4A). The DNA content of the ntf2–2ts MAD2Δ double mutant was more similar to that observed for wild-type cells than for ntf2–2ts single mutants (see Figure 4A). This indicates allele-specific rescue of the ntf2–2ts cell cycle phenotype by the MAD2Δ.
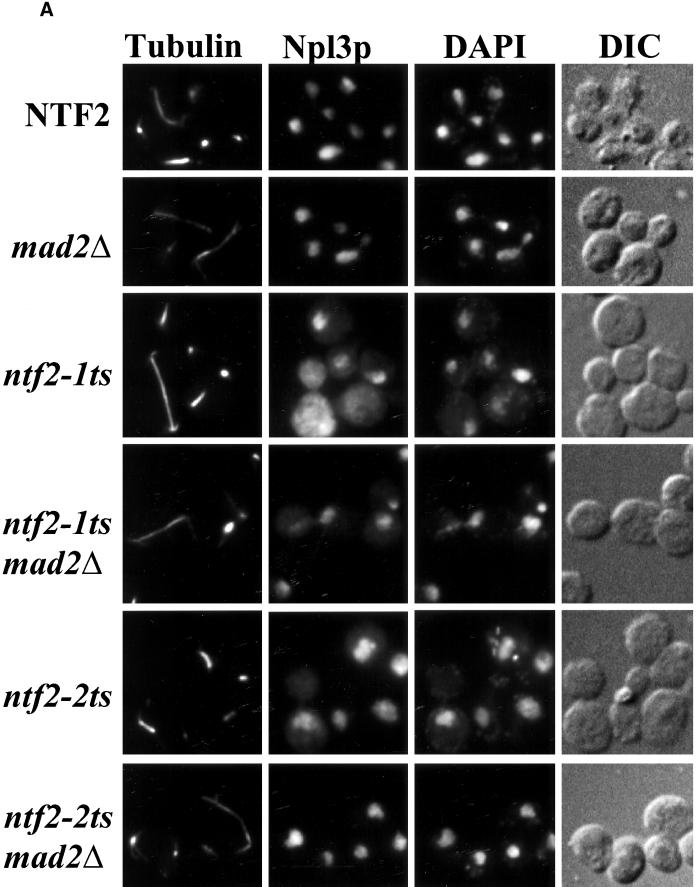
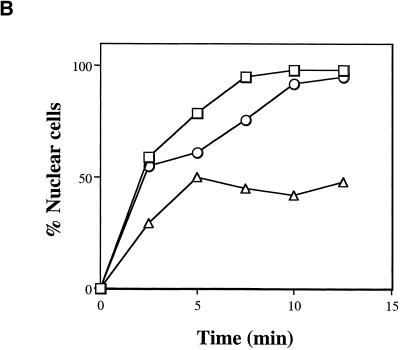
To further analyze the cell cycle phenotype and to assess protein import in the ntf2–2ts MAD2Δ double mutant, cells were grown at 37°C and costained for tubulin, to observe microtubules, and for the nuclear protein Npl3p, to observe endogenous nuclear protein localization. The MAD2Δ strain showed no defect in protein import or spindle morphology (Figure 7A). As expected, both the ntf2–1ts and ntf2–2ts cells showed mislocalization of Npl3p, however, the ntf2–2ts mutant appeared to have a less profound protein import defect than the ntf2–1ts strain (Figure 7A). To further quantify this apparent difference, we performed an NLS-GFP import assay (Shulga et al., 1996) to measure the relative nuclear protein import rates in the wild-type, ntf2–1ts, and ntf2–2ts strains. Consistent with the mislocalization of Npl3p observed in ntf2–1ts and ntf2–2ts cells, the relative rate of NLS-GFP import in ntf2–1ts cells is significantly slower than in ntf2–2ts cells (Figure 7B). These data indicate that nuclear protein import is more compromised in the ntf2–1ts than in the ntf2–2ts allele and suggest that the cell cycle defect observed in ntf2–2ts cells does not correlate with protein import.
Figure 7.
The ntf2–2ts cell cycle arrest is MAD2 dependent. (A) MAD2Δ suppresses the ntf2–2ts cell cycle arrest. NTF2, MAD2Δ, ntf2–1ts, ntf2–1ts MAD2Δ, ntf2–2ts, and ntf2–2ts MAD2Δ cells were grown overnight at 25°C and were shifted to 37°C for 3 h. Cells were stained with an antitubulin antibody followed by antimouse Texas Red to visualize microtubules, anti-Npl3p was followed by staining with antirabbit FITC to analyze nuclear protein localization and with DAPI to visualize DNA. (B) ntf2–1ts cells exhibit a more severe protein import defect than ntf2–2ts cells. A standard import assay (Shulga et al., 1996) was used to analyze nuclear import rates. Results are plotted as the percent cells showing nuclear signal versus time for NTF2 (□), ntf2–1ts (▵), and ntf2–2ts (○).
The most important question is whether the MAD2Δ is able to suppress the spindle defect observed in the ntf2–2ts cells. Examination of the ntf2–2ts MAD2Δ double mutant revealed wild-type spindle morphology, suggesting that deletion of MAD2 alleviates the cell cycle arrest phenotype observed in the ntf2–2ts single mutant (Figure 7A). This demonstrates that the G2 arrest observed in ntf2–2ts is MAD2 dependent. Taken together, these results indicate that depletion of Ran from the nucleus in ntf2–2ts cells results in a G2 arrest that is independent of nuclear transport and is dependent on the spindle-assembly checkpoint monitored by MAD2.
DISCUSSION
Ran has long been suspected to be involved in the cell cycle (Sazer and Dasso, 2000). However, it has been difficult to distinguish a direct effect of Ran on cell cycle progression from an effect on nuclear transport. Several recent in vitro studies were able to address this question in a cell-free system using eunucleated sperm DNA and mitotic Xenopus extracts (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999). These experiments demonstrated that RanGTP was required for microtubule nucleation in this system. How do we determine the significance of these studies in a cellular system? We were able to ask this question in yeast because yeast undergoes mitosis without nuclear envelope breakdown. Therefore, any factor required for mitotic spindle formation must be imported into the nucleus. We have utilized two ts mutants of the Ran import factor NTF2, ntf2–1ts and ntf2–2ts, to separate the roles of nuclear Ran in nuclear transport and cell cycle control.
It had previously been shown that both the ntf2–1ts and ntf2–2ts mutants have protein import defects (Corbett and Silver, 1996). Here we show that unlike the Ntf2–1 protein, the Ntf2–2 protein cannot interact with Ran and is unable to efficiently import Ran into the nucleus. These results support previous studies indicating that Ntf2p imports Ran into the nucleus (Ribbeck et al., 1998; Smith et al., 1998; Steggerda et al., 2000). However, these results also suggest that Ntf2p has additional roles in nuclear transport since the ntf2–1ts mutant has a profound protein import defect despite its ability to efficiently mediate Ran import.
The identification of an NTF2 mutant that is unable to import Ran into the nucleus provided a tool to determine whether or not nuclear Ran is involved in mitotic spindle formation in vivo. Here we show that the Ran import defective ntf2–2ts allele is a unique microtubule assembly-defective allele of NTF2 and that this cell cycle defect is enhanced by a deletion of the MAD2 checkpoint gene. In addition, the ntf2–2ts but not the ntf2–1ts phenotype is suppressed by the MAD2Δ. These results suggest a link between nuclear Ran and the mitotic spindle checkpoint, and they support a role for nuclear Ran in cell cycle progression that is distinct from nuclear transport.
We cannot completely rule out protein import defects in our analysis of Ran in cell cycle progression. It is possible that the ntf2–2ts allele specifically disrupts the nuclear import of other proteins, such as tubulin, that are involved in spindle formation, and affects microtubule assembly resulting in the observed G2 arrest. The level of tubulin in nuclei of ntf2–2ts cells grown at 37°C is similar to wild-type cells grown at 37°C. In contrast, consistent with the severe protein import defect observed, ntf2–1ts cells are significantly reduced in the level of tubulin in nuclei but are able to form mitotic spindles (Quimby and Corbett, unpublished observations). This suggests that insufficient nuclear tubulin does not cause the microtubule defect observed in ntf2–2ts cells but does not eliminate the possibility that some other spindle assembly protein is not imported efficiently. However, taken together, the degree of protein import defect observed in ntf2–2ts cells and the single amino acid substitution that specifically disrupts the interaction with Ran suggest that the Ntf2–2 mutation specifically disrupts the import of Ran, which leads to the observed cell cycle defect.
There are two possible explanations for our observations on the role of Ran in mitotic spindle-assembly. One, nuclear Ran may be directly required for proper spindle formation, and, as Ran is depleted, spindles are physically disrupted. The resultant disrupted spindles are sensed by the MAD2 spindle checkpoint mechanism causing cells to arrest. Second, the cellular checkpoint system may directly sense the level of nuclear Ran to ensure that the nucleocytoplasmic transport system is intact before the cell progresses through mitosis. Our result demonstrating that ntf2–2ts is benomyl but not nocodazole sensitive in combination with the in vitro studies suggesting a direct role for Ran in spindle formation support the former hypothesis. If Ran is directly involved in spindle formation, how do we explain the suppression of the ntf2–2ts phenotype by disruption of the MAD2 checkpoint? One possibility is that the spindle assembly checkpoint directly senses the level of Ran in the nucleus and triggers the checkpoint at a level that is slightly above the critical level required for proper spindle formation. In this model, the single mutant (ntf2–2ts) should arrest at 37°C because the spindle-assembly checkpoint is intact and senses a reduction in the level of nuclear Ran. Moreover, the double mutant (ntf2–2ts MAD2Δ) should continue through the cell cycle due to a disruption of the spindle-assembly checkpoint, and cells should appear normal because the low level of Ran that is present is capable of supporting mitotic spindle formation. In fact this is what we observe, which is consistent with a model in which Ran plays a direct role in mitotic spindle formation.
Our results not only indicate that nuclear Ran is required for proper mitotic spindle formation in vivo, but also demonstrate that the role of Ran is conserved through evolution. How do our results obtained in a closed mitotic system relate to vertebrate cells that undergo open mitosis? In most vertebrate cells the mitotic spindle is initiated in the cytoplasm in the absence of RanGTP. Our findings are consistent with a model in which the rapid release of RanGTP from the nucleus upon nuclear envelope breakdown may serve as a signal for the spindle to proceed through mitosis. In fact, it has been shown that microtubule dynamics differ markedly in interphase versus mitotic cells (Saxton et al., 1984; Verde et al., 1990). In our model, cells would take advantage of the nuclear compartmentalization of RanGTP to prevent premature spindle progression. These results suggest a mechanism where in higher eukaryotes the release of nuclear RanGTP serves as a catalyst for early events in mitotic spindle assembly.
In any model, Ran is likely to interact directly with some specific spindle-associated protein in mitotic cells. One candidate protein is RanBPM, which was identified in a two-hybrid screen with Ran and has been shown to induce ectopic microtubule nucleation (Nakamura et al., 1998). If nuclear Ran interacts with RanBPM and acts as some type of signal for the cell to progress through mitosis, the depletion of Ran would halt this process and arrest cell division. If excess Ran is added to mitotic cell extracts, more would be available to interact with RanBPM forming ectopic microtubules, as was seen in the in vitro experiments (Nakamura et al., 1998). Another candidate Ran-interacting protein is Mad2p or one of the components of the Mad2p complex. Most likely, Ran interacts with a variety of proteins during mitosis, either individually or as a complex, and identification of these proteins will be the next step in defining the role of Ran in the cell cycle.
ACKNOWLEDGMENTS
We are grateful to Dr. Alec Hodel for helpful guidance with FACS analysis, to Dr. Pam Silver for many of the plasmids and antibodies used in this study, and to Drs. Mary Dasso and Patrizia Fanara for critical reading of the manuscript. B.B.Q. is a recipient of a National Institute of Health (NIH) fellowship (5F32GM19681–01). C.W. is supported by the Georgia Industrial Fellowship for Teachers (GIFT) program. A.H.C. is supported by a grant from NIH (GM58728) and a Biomedical Career Award from the Burroughs Wellcome Foundation.
Abbreviations used:
- BSA
bovine serum albumin
- CNBr
cyanogen bromide
- DTT
dithiothreitol
- FACS
fluorescence activated cell sorter
- FITC
fluoroscein isothiocyanate
- GFP
green fluorescent protein
- NTF2
nuclear transport factor 2
- PMSF
phenylmethylsulfonyl fluoride
- ts
temperature sensitive
- YEPD
yeast extract-peptone-dextrose
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Bullock TL, Clarkson WD, Kent HM, Stewart M. The 1.6Å resolution crystal structure of nuclear transport factor 2 (NTF2) J Mol Biol. 1996;260:422–431. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen W, Ward W, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Clarkson WD, Corbett AH, Paschal BM, Kent HM, McCoy AJ, Gerace L, Silver PA, Stewart M. Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J Mol Biol. 1997;272:716–730. doi: 10.1006/jmbi.1997.1255. [DOI] [PubMed] [Google Scholar]
- Clarkson WD, Kent HM, Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions on nucleoporins p62 and Nsp1p. J Mol Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. RCC1 in the cell cycle: the regulator of chromosome condensation takes on new roles. Trends Biol Sci. 1993;13:96–101. doi: 10.1016/0968-0004(93)90161-f. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Gardner RD, Burke DJ. The spindle checkpoint: two transitions, two pathways. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- Hardwick KG. The spindle checkpoint. Trends Genet. 1998;14:1–4. doi: 10.1016/S0168-9525(97)01340-1. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Traglia HM, Dunst RW. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kent HM, Clarkson WD, Bullock TL, Stewart M. Crystallization and preliminary X-ray diffraction analysis of nuclear transport factor 2. J Struct Biol. 1996;116:325–328. doi: 10.1006/jsbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- Kent HM, Moore MS, Quimby BB, Baker AM, McCoy AJ, Murphy GA, Corbett AH, Stewart M. Engineered mutants in the switch II loop of Ran define the contribution made by key residues to the interaction with nuclear transport factor 2 (NTF2) and the role of this interaction in nuclear protein import. J Mol Biol. 1999;289:565–577. doi: 10.1006/jmbi.1999.2775. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;55:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Loeb JDJ, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink GR. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Masuda H, Horri J, Ki K, Yokoyama N, Ohba T, Nishitani H, Miyata T, Tanaka M, Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J Cell Biol. 1998;143:1041–1052. doi: 10.1083/jcb.143.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamara M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Kai R, Furuno N, Sekiguchi T, Havashida H, Kuma K, Miyata T, Fukushige S, Murotu T, Matsubara K, Nishimoto T. Isolation and characterization of the active cDNA of the human cell cycle gene (RCC1) involved in the regulation of onset of chromosome condensation. Genes Dev. 1987;1:585–593. doi: 10.1101/gad.1.6.585. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Fritze C, Guan T, Gerace L. High levels of the GTPase Ran/TC4 relieve the requirement for nuclear protein transport factor 2. J Biol Chem. 1997;272:21534–21539. doi: 10.1074/jbc.272.34.21534. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Lippowsky G, Kent HM, Stewart M, Görlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Dasso M. The Ran decathlon: multiple roles of Ran. J Cell Sci. 2000;113:1111–1118. doi: 10.1242/jcs.113.7.1111. [DOI] [PubMed] [Google Scholar]
- Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Brownwell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- Steggerda SM, Black BE, Paschal BM. Monoclonal antibodies to NTF2 inhibit nuclear protein import by preventing nuclear translocation of the GTPase Ran. Mol Biol Cell. 2000;11:703–719. doi: 10.1091/mbc.11.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Kent H, McCoy A. Structural basis for the molecular recognition between Nuclear Transport Factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J Mol Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- Straight AF, Murray AW. The spindle assembly checkpoint in budding yeast. Methods Enzymol. 1997;283:425–440. doi: 10.1016/s0076-6879(97)83035-2. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some application. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Labbé J-C, Dorée M, Karsenti E. Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature. 1990;343:233–238. doi: 10.1038/343233a0. [DOI] [PubMed] [Google Scholar]
- Way M, Pope B, Gooch J, Hawkins M, Weeds AG. Identification of a region in segment 1 of gelsolin critical for actin binding. EMBO J. 1990;9:4103–4109. doi: 10.1002/j.1460-2075.1990.tb07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wong DH, Corbett AH, Kent HM, Stewart M, Silver PA. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]