Abstract
Loss of axonal contact characterizes Schwann cells in benign and malignant peripheral nerve sheath tumors (MPNST) from neurofibromatosis type 1 (NF1) patients. Tumor Schwann cells demonstrate NF1 mutations, elevated Ras activity, and aberrant epidermal growth factor receptor (EGFR) expression. Using cDNA microarrays, we found that brain lipid binding protein (BLBP) is elevated in an EGFR-positive subpopulation of Nf1 mutant mouse Schwann cells (Nf1−/− TXF) that grows away from axons; BLBP expression was not affected by farnesyltransferase inhibitor, an inhibitor of H-Ras. BLBP was also detected in EGFR-positive cell lines derived from Nf1:p53 double mutant mice and human MPNST. BLBP expression was induced in normal Schwann cells following transfection with EGFR but not H-Ras12V. Furthermore, EGFR-mediated BLBP expression was not inhibited by dominant-negative H-Ras, indicating that BLBP expression is downstream of Ras-independent EGFR signaling. BLBP-blocking antibodies enabled process outgrowth from Nf1−/− TXF cells and restored interaction with axons, without affecting cell proliferation or migration. Following injury, BLBP expression was induced in normal sciatic nerves when nonmyelinating Schwann cells remodeled their processes. These data suggest that BLBP, stimulated by Ras-independent pathways, regulates Schwann cell-axon interactions in normal peripheral nerve and peripheral nerve tumors.
All axons of the peripheral nervous system are ensheathed or myelinated by a neural crest-derived population of glia known as Schwann cells. Although elaborate interactions between Schwann cell membranes and axons must occur in developing peripheral nerves, little is known about the molecules that cause or maintain Schwann cell process outgrowth around axons. The intimate interaction of Schwann cells with axons is disrupted only in pathological situations, such as after nerve injury and in tumors of the peripheral nervous system.
Neurofibromatosis type 1 (NF1) is an autosomal dominant disorder affecting approximately 1 in 3,500 individuals worldwide. NF1 predisposes patients to the formation of complex benign peripheral nerve tumors called neurofibromas, in which many Schwann cells have lost contact with axons. At least 95% of NF1 patients exhibit neurofibromas, and approximately 4% of NF1 patients develop lethal malignant peripheral nerve sheath tumors (MPNST). Schwann cells are thought to be the primary tumorigenic cell type, as Schwann cells in neurofibromas (32, 48) and MPNST (35) lose both copies of the NF1 gene, consistent with the hypothesis that NF1 acts as a tumor suppressor gene.
Mice with homozygous mutations in the Nf1 gene are not viable, and Nf1 heterozygous mice do not develop neurofibromas (10, 28). However, neurofibromas do form in chimeric Nf1 mutant mice (14) and mice lacking Nf1 specifically in Schwann cells (59). In vitro, Nf1 mutant embryonic mouse Schwann cells are angiogenic and invasive with an altered proliferation rate (30, 31). However, unlike neurofibroma Schwann cells within tumors, they maintain interaction with neurons in vitro. Omission of serum in the culture medium promotes the development of hyperproliferative Nf1−/− Schwann cells (Nf1−/− TXF) that do not adhere normally to axons and yet retain expression of Schwann cell markers including GFAP, p75NGFR, and S100 (30, 45). Furthermore, these cells have increased expression of the epidermal growth factor receptor (EGFR), a characteristic of MPNST cells and a subpopulation of Schwann cells in neurofibromas (17). Based on these results, we propose that Nf1−/− TXF cells model at least a subset of cells in human neurofibromas and possibly MPNST.
Nf1−/− TXF cell defects must be caused at least in part by loss of Nf1 function. The only proven function of the NF1 gene product, neurofibromin, is its ability to downregulate activated Ras by converting Ras-GTP to Ras-GDP via its GTPase-activating protein (GAP) domain (13). Cultured embryonic mouse Schwann cells mutated at Nf1, neurofibroma Schwann cells, and MPNST cell lines with reduced neurofibromin have elevated Ras-GTP (5, 18, 31, 49), as do neurofibroma extracts (24). Elevated Ras-GTP in Schwann cells does not by itself cause disruption of axon-glial interaction, however, as Nf1−/− Schwann cells do not lose contact with axons (31). Exposure of cells to farnesyltransferase inhibitors (FTI), drugs that inhibit Ras function by reducing lipid addition to certain Ras molecules (46, 51), has been used as a test for a Ras-dependent phenotype. FTI treatment reduces growth of Nf1−/− TXF cells and MPNST cells but is incapable of reversing other phenotypes of Nf1−/− mouse Schwann cells, including cell invasion (30, 57). These studies implicate Ras as well as additional molecular pathways downstream of NF1.
To identify genes altered as a consequence of NF1 loss that may be relevant to peripheral nerve tumorigenesis, we utilized cDNA microarray technology to compare gene expression between wild-type and Nf1 mutant mouse Schwann cells. We identified one cDNA, encoding the brain lipid binding protein (BLBP)/brain fatty acid binding protein (B-FABP)/fatty acid binding protein 7 (6, 50), that was strikingly overexpressed in the Nf1−/− TXF cells but not normalized by a Ras inhibitor, FTI. BLBP has been implicated elsewhere in neuron-radial glial cell signaling in the developing central nervous system (2, 20, 34) and is a marker for brain tumors (22). Our results show that aberrant BLBP expression prevents Nf1−/− TXF Schwann cells from forming a stable association with axons and support a role for BLBP in tumorigenesis in the peripheral nervous system.
MATERIALS AND METHODS
Cell culture.
Nf1 heterozygous C57BL/6 mice were mated to obtain Nf1 wild-type (+/+), heterozygous (+/−), and homozygous null (−/−) embryos, as determined by PCR genotyping (10). Mouse Schwann cells were isolated from embryonic day 12.5 dorsal root ganglia (DRG) as previously described (31) and cultured on poly-l-lysine-coated plates in Dulbecco modified Eagle medium (DMEM) plus 10% fetal bovine serum. Nf1−/− TXF cells appeared as hyperproliferative colonies in Nf1−/− cell cultures without serum (30) and were grown in serum-free N2 medium supplemented with 5 ng of recombinant human GGF2 (Cambridge Neuroscience)/ml and 2 μM forskolin (Calbiochem).
For coculture of neurons and Schwann cells, DRG neurons (DRGN) were isolated from wild-type embryos (E12.5) as described previously (31). Wild-type or Nf1−/− TXF mouse Schwann cells were preincubated in suspension at 37°C with 10 μM Cell Tracker green 5-chloromethyl fluorescein diacetate (Molecular Probes) for 1 h followed by a 30-min preincubation with affinity-purified rabbit anti-mouse BLBP antibodies (20) or rabbit immunoglobulin G (IgG) at a concentration of 80 μg/ml. Labeled cells were then seeded onto DRGN cultures and incubated for 48 h before fixing.
Mouse Schwann cells preincubated with rabbit IgG or anti-BLBP antibodies as described above were also plated on LabTek chamber slides (Nalge-Nunc International), coated with poly-l-lysine alone or poly-l-lysine plus laminin, and fixed after being incubated for 48 h.
Nf1:p53 mouse tumor cell lines were established from Nf1:p53 compound heterozygous mouse tumors (53). These cells were cultured in DMEM with 10% fetal bovine serum. MPNST cells were grown as described previously (18). Cell lines derived from malignant triton tumors include 38-1-3-7, 40-1-7, 67C-4, 33-2-20, 67A-4, 39-2-11, 61E-4, 37-3-8-17, 32-7-33, and 61C-4; the 32-5-30-2 cell line was created from MPNST; and cell lines 32-5-24, 38-2-17-8, and 41-2-9 were derived from unclassified tumors.
Microarray analysis.
mRNA was isolated (MicroFastTrack kit 2.0; Invitrogen) from 2 to 4 million wild-type (Nf1+/+) or Nf1 mutant (Nf1+/−, Nf1−/−, and Nf1−/− TXF) Schwann cells at 80 to 90% confluency. Prior to being sent to Incyte Genomics, samples were quantitated with a spectrophotometer, and actin reverse transcription-PCR (RT-PCR) (see RT-PCR below) was conducted to ensure the quality of the samples. Nf1+/+ mRNA was used as a template in a reverse transcriptase reaction to synthesize Cy3-labeled control cDNA; samples (200 ng each) isolated from Nf1+/− cells, Nf1−/− cells, Nf1−/− TXF cells, or Nf1−/− TXF cells treated with 1 μM FTI (L-745,832; Merck Research Laboratories) (33) for 2 days (30) were used to create four separate Cy5-labeled cDNA targets. Each Cy5 target was hybridized simultaneously with the Cy3 control to the MouseGEM 1.0 cDNA microarray. Fluorescent signal intensity data were exported to the University of Cincinnati Bioinformatics Core Facility and analyzed with GEMTools 2.4 (Incyte Genomics) and GeneSpring 3.2 (Silicon Genetics) computer software.
RT-PCR analysis.
mRNA isolated from mouse Schwann cells (MicroFastTrack 2.0 kit) was used as a template to create double-stranded cDNA (Superscript preamplification system; Gibco-BRL). Both oligo(dT) primers and random hexamers were used in the reverse transcriptase reaction. Duplicate samples omitting reverse transcriptase were used to control for genomic DNA contamination. Mouse β-actin primers (sense primer, 5′-TGT GAT GGT GGG AAT GGG TCA-3′; antisense primer, 5′-TTT GAT GTC ACG CAC GAT TTC C-3′) were included in each reaction mixture as a positive control for cDNA. Mouse β-actin and mouse BLBP (sense primer, 5′-AGA CCC GAG TTC CTC CAG TT-3′; antisense primer, 5′-ATC ACC ACT TTG CCA CCT TC-3′) were PCR amplified in the same reaction for 40 cycles under the following cycling conditions: 94°C, 1 min; 54°C, 1 min; and 72°C, 2 min. Each reaction mixture contained 1 μl of cDNA, 0.7 μM primers, 2.0 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1× PCR buffer, and 1.0 U of Taq polymerase (Gibco-BRL).
Total RNA was isolated from human MPNST cells with Trizol reagent (Life Technologies). Human BLBP (sense primer, 5′-CGC TCC TGT CTC TAA AGA GGG G-3′; antisense primer, 5′-TGG GCA AGT TGC TTG GAG TAA C-3′) was amplified in a single-tube RT-PCR system (Stratagene) from DNase-treated samples (500 ng). The glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) (sense primer, 5′-AAC ATC ATC CCT GCC TCT CTA CTG-3′; antisense primer, 5′-TTG ACA AAG TGG TCG TTG AGG-3′) was amplified in a separate reaction to confirm the integrity of each sample.
Quantitative real-time PCR of BLBP and GAPDH was conducted with the SYBR Green master mix in the ABI Prism 7700 sequence detection system. Amplification conditions included an initial denaturing step at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Cycle threshold values were obtained where fluorescence intensity was in the geometric phase of amplification and averaged for triplicate samples. Values for BLBP were normalized to GAPDH values and used to calculate fold change in gene expression with the following equation described by K. Livak (PE-ABI; Sequence Detector User Bulletin 2): 2−ΔΔCT. CT represents the cycle number at the chosen amplification threshold; ΔCT = CT (BLBP) − CT (GAPDH); ΔΔCT = ΔCT (Nf1−/− TXF) − ΔCT (Nf1+/+).
Western analysis.
Mouse Schwann cells were grown to 90% confluency on 100-mm-diameter dishes and lysed in cold radioimmunoprecipitation assay buffer (150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl, pH 8.0) supplemented with a protease inhibitor cocktail (Sigma). Cell lysates were scraped into tubes on ice and incubated for 20 min, followed by cold centrifugation at 14,000 × g for 15 min. Supernatants (∼20 μg) were mixed with SDS electrophoresis buffer and boiled for 5 min before separation through an SDS-15% polyacrylamide gel, followed by transfer to a polyvinylidene difluoride membrane (Bio-Rad). The membrane was blocked with a 5% milk solution, probed with affinity-purified anti-BLBP antibodies diluted 1:50,000 (20), stripped, and reprobed with anti-Ras C10 diluted 1:1,000 (Upstate Biotechnology) as a protein loading control. Signals were detected with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz) in combination with the ECL Plus developing system (Amersham Pharmacia Biotech).
Nf1:p53 mutant tumor cells were grown until confluent and lysed (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM dithiothreitol). Cell lysates containing 50 μg of protein were subjected to analysis by SDS-14% polyacrylamide gel electrophoresis and immunoblotting with anti-BLBP antibodies, as described above. The membrane was stripped and reprobed with antitubulin antibodies (Sigma) for establishing relatively equal amounts of loaded protein.
Immunolabeling.
Cells were fixed with 4% paraformaldehyde, blocked with 10% goat serum, and permeabilized with 0.5% Triton X-100 (Bio-Rad). Mouse Schwann cells on LabTek slides were stained with anti-BLBP antibodies (20), anti-p75NGFR antibodies (Chemicon AB1554), or anti-S100 antibodies (Dako) diluted 1:500, followed by goat anti-rabbit fluorescent secondary antibodies (Jackson Immunoresearch). Cell nuclei were labeled with 5 μg of bis-benzimide (Sigma)/ml. AlexaFluor 488 phalloidin antibodies (Molecular Probes) recognized F-actin. In DRGN cocultures, neurofilament was visualized by incubation with anti-NF-15g1 antibodies (38) diluted 1:5, followed by donkey anti-mouse Cy3-labeled secondary antibodies (Jackson Immunoresearch).
For nerve injury experiments, 3- to 4-month-old wild-type mice were deeply anesthetized with Avertin (2,2,2-tribromoethanol; Aldrich) (0.5 to 0.8 ml, intraperitoneally). The skin overlying the sciatic nerve was cut, muscles were parted, and the sciatic nerve was exposed. For nerve crush, sciatic nerve 2 mm medial to the sciatic notch was crushed by mechanical pressure three times with a sharp pair of no. 5 Dumont tweezers. For nerve transection and deflection, the sciatic nerve was cut 2 mm medial to the sciatic notch, and the cut ends were deflected to opposite sides and secured under muscle masses to avoid regeneration. After surgeries, muscles were realigned and the skin was closed with surgical staples. Animals were evaluated after 4, 7, 15, and 30 days (n = 3 per time interval) postcrush and 21 days posttransection (n = 3). Nonlesioned nerves were used as controls. Mice were perfused with 4% paraformaldehyde, and distal ends of the sciatic nerve were dissected, postfixed, and processed for paraffin embedding. Distal segments of the sciatic nerve were sectioned at 6 μm. Sections were air dried, deparaffinized, hydrated, incubated in Digest-All 3 pepsin (Zymed) at 37°C for 10 min, rinsed in phosphate-buffered saline, and processed for immunolabeling. Sections were labeled with polyclonal rabbit antineurofilament (NF-178.3 diluted 1:150) (38), polyclonal rabbit anti-BLBP (diluted 1:500), and rabbit anti-myelin basic protein (a gift from D. Colman, diluted 1:500), followed by appropriate fluorescent secondary antibodies.
Transient transfection.
Wild-type mouse Schwann cells were plated onto poly-l-lysine-coated LabTek slides (Nalge-Nunc International) and grown to ∼75% confluency. Plasmid DNA (1 μg) was incubated for 20 min at room temperature with 1.5 μl of FuGENE 6 transfection reagent (Roche) that had been previously diluted in serum-free DMEM (Cellgro). The DNA-FuGENE complex was then added directly to the cells in culture. Cells were fixed in 4% paraformaldehyde 48 h later. Transfected DNA constructs included CNP-(HA)HRas12V, CNP-EGFR, CNP-Script, and pZIPRasH(17N) (39). Human EGFR (52) and hemagglutinin (HA)-tagged (HA)HRas12V (55) DNA fragments were subcloned downstream of the glial cell-specific 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP) promoter (12). CNP-Script consists of the CNP promoter alone in a Bluescript (Stratagene) background. Expression of exogenous protein was detected by immunolabeling with mouse anti-HA (Santa Cruz) followed by donkey anti-mouse Cy3-conjugated secondary antibodies (Jackson Immunoresearch) or goat anti-human EGFR antibodies (Santa Cruz) followed by donkey anti-goat tetramethyl rhodamine isocyanate-labeled secondary antibodies (Jackson Immunoresearch). BLBP expression was detected with rabbit anti-mouse BLBP antibodies (20) followed by donkey anti-rabbit Cy2-labeled secondary antibodies (Jackson Immunoresearch).
Cell survival assay.
Nf1−/− TXF cells were plated (50,000 cells/well) in 24-well plates and incubated overnight in DMEM plus 10% fetal bovine serum. The next day, serum-containing medium was removed and cells were cultured for 5 days in serum-free N2 medium alone or supplemented with one of the following components: EGF (50 ng/ml), rabbit IgG (100 μg/ml), anti-BLBP antibodies (1:300, 1:1,000 [2], and 1:5,000), FTI (1 μM FTI 745.832, a gift of Merck Research Laboratories), and dimethyl sulfoxide (0.5 μl). Supplemental factors were added to triplicate wells and replenished at day 2.5. To establish a baseline cell number, cells were trypsinized and counted from three wells in N2 alone prior to the addition of any factor (day 0). Final cell counts were taken at day 5.
Cell migration assay.
The migratory response of Nf1−/− TXF mouse Schwann cells to BLBP was measured by the Boyden chamber assay (9). Schwann cells were detached from the tissue culture plates with a cell scraper (Costar), resuspended at a density of 1.5 × 105/ml in N2 medium (DMEM-F-12 with N2 supplements and 0.1% bovine serum albumin; Gibco-BRL), and plated on the upper well of a 24-transwell plate (8-μm-pore-size polycarbonate membrane; Costar). Cells in the upper wells were coincubated with anti-BLBP antibodies (20) at a concentration of 80 μg/ml. For control wells, the same concentration of IgG was used as mock anti-BLBP. The lower well of the 24-well plate contained 800 μl of N2 medium with the same ingredients as those in the upper well. Both sides of the polycarbonate membrane of the transwell were precoated with 50 μl of poly-l-lysine (Fisher Scientific)/ml. Cells were incubated for 15.5 h at 37°C in a humidified atmosphere of air with 10% CO2. Cells that had not migrated were removed from the upper surface with cotton swabs. Thereafter, the membranes were fixed with cold alcohol-acetic acid (95:5), stained with bis-benzimide (1 μg/ml; Sigma), and mounted onto glass slides with Fluoromount (Electron Microscopy Sciences). Migration was quantified by counting cells on the entire membrane under a fluorescence microscope.
Electron microscopy.
For electron microscopy, mice were perfusion fixed with glutaraldehyde. Sciatic nerves were dissected and postfixed for 1 h in 2% OsO4 plus 0.6% potassium ferrocyanide. After the osmicated nerves were dehydrated and embedded, thin sections (70 to 80 nm) were cut and stained with uranyl acetate and lead citrate.
RESULTS
BLBP mRNA expression is elevated in mouse Nf1−/− TXF cells.
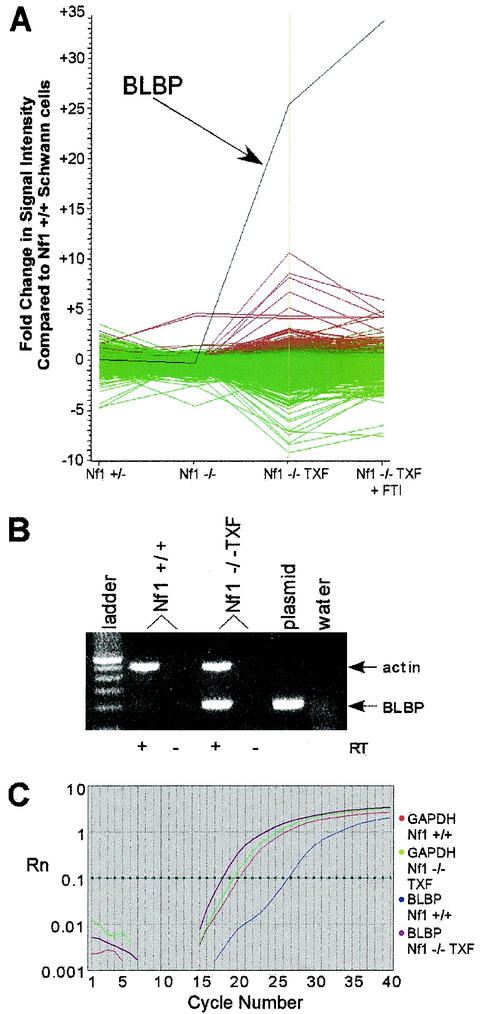
For each of three Nf1 mutant Schwann cell samples (Nf1+/−, Nf1−/−, and Nf1−/− TXF) and Nf1−/− TXF treated with FTI, mRNA was isolated and used as a template to synthesize Cy5-labeled cDNA. Cy3-labeled cDNA generated from reverse transcription of normal mouse Schwann cell mRNA was used as the control for each comparison. Cy3- and Cy5-labeled cDNA probes were hybridized simultaneously to a glass slide containing a microarray of nearly 9,000 cDNA targets (including 3,205 known gene sequences and 4,649 expressed sequence tags; Incyte Genomics MouseGEM 1.0). Relative intensities of Cy3 versus Cy5 fluorescent signals for each cDNA target sequence were obtained and analyzed with GeneSpring computer software (Silicon Genetics). Only 278 sequences had differential expression that was more than threefold higher or lower than that of the wild type in at least one comparison (Fig. 1A). Nf1−/− TXF cells had the greatest number of changes. This result was expected, because the Nf1−/− TXF cells have a more dramatic change in phenotype than do the Nf1−/− and Nf1+/− mutant cells (30). Although FTI treatment inhibited growth of the Nf1−/− TXF cells, expression of a few genes was normalized, including some known Ras targets. Expression of one target cDNA, BLBP, was dramatically altered in the Nf1−/− TXF cells, 26-fold above wild-type levels. BLBP expression was not significantly affected by FTI treatment, in which it was 35-fold upregulated.
FIG. 1.
mRNA expression analysis in Nf1 mutant mouse Schwann cells. (A) Microarray analysis was used to compare genome-wide expression levels between normal mouse Schwann cells and Nf1 mutant Schwann cells. The control for each comparison was Cy3-labeled cDNA generated from normal mouse Schwann cell mRNA. For each of four Nf1 mutant Schwann cell samples (Nf1+/−, Nf1−/−, Nf1−/− TXF, and Nf1−/− TXF treated with FTI), mRNA was used as a template to synthesize Cy5-labeled cDNA. Cy3- and Cy5-labeled cDNA probes were hybridized simultaneously to the Incyte Genomics MouseGEM 1.0 cDNA microarray. Relative intensities of Cy3 versus Cy5 fluorescent signals for each cDNA target sequence were analyzed with GeneSpring software. The most changes were observed in the Nf1−/− TXF cells (genes upregulated in Nf1−/− TXF are red; genes downregulated in Nf1−/− TXF are green). Expression of one target cDNA, BLBP (black line), was 26-fold above normal in the Nf1−/− TXF cells and not normalized by FTI treatment. (B) RT-PCR analysis confirmedthe microarray result of elevated BLBP expression in Nf1−/− TXF cells. Reverse transcriptase (RT) was omitted from duplicate samples to control for DNA contamination. Primers for BLBP (∼200-bp amplicon) and actin control primers (∼500-bp amplicon) were included in the mixture for each 40-cycle reaction. The plasmid positive control for BLBP amplification is the UniGEM clone (Incyte Genomics) containing the BLBP cDNA insert spotted on the microarray. (C) Quantitative real-time PCR of BLBP normalized to GAPDH resulted in a 145-fold change over expression in Nf1−/− TXF cells compared to wild-type mouse Schwann cells. Rn, fluorescent signal intensity; horizontal starred line, chosen threshold at geometric phase of amplification.
Further analyses were conducted to confirm the results of the microarray hybridization on the mRNA level by RT-PCR. After 40 cycles of amplification, BLBP was barely detectable in wild-type Schwann cell cDNA but abundant in Nf1−/− TXF cDNA (Fig. 1B). Quantitative real-time PCR of BLBP, normalized to GAPDH, also confirmed overexpression of BLBP in Nf1−/− TXF cells (Fig. 1C). The change in expression, approximately 145-fold, is higher than that reported for the microarray data and suggests that the fluorescent signal was saturated in the microarray method (40, 41, 44). The 145-fold change is likely to represent the actual difference in expression, because the real-time PCR data were obtained in triplicate with two independent mRNA samples.
BLBP protein is elevated in Nf1−/− TXF cells and localized to the cytoplasm and cell surface.
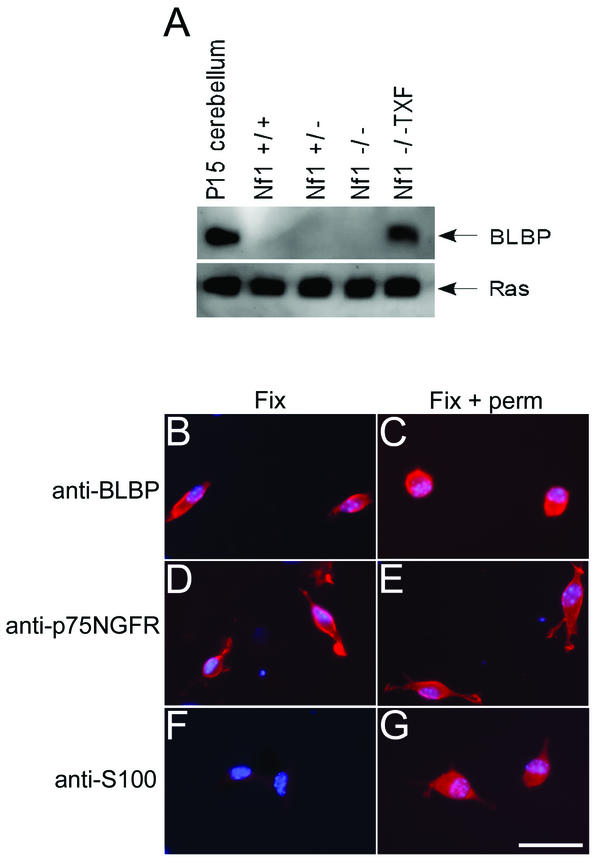
Changes in mRNA expression levels generally reflect changes in protein levels, although not in all cases (25, 27). Western analysis was used to confirm differential BLBP expression at the protein level. Lysates from wild-type, Nf1+/−, Nf1−/−, and Nf1−/− TXF Schwann cells were probed with an anti-BLBP antibody, the gift of N. Heinz (20). A 15-kDa protein corresponding to BLBP was detected in the Nf1−/− TXF cells only (Fig. 2A), coinciding with the cDNA microarray data. BLBP was not detected in conditioned medium from Nf1−/− TXF cells (data not shown), suggesting that BLBP is not secreted at appreciable levels.
FIG. 2.
BLBP protein analysis in mouse Schwann cells. (A) Western analysis confirmed BLBP microarray data at the protein level. Cell lysates from wild-type, Nf1+/−, Nf1−/−, and Nf1−/− TXF Schwann cells were probed for BLBP expression. A 15-kDa protein corresponding to BLBP was detected in the Nf1−/− TXF cells only. Lysate from postnatal day 15 (P15) mouse cerebellum was used as a positive control. Blots were stripped and reprobed with anti-Ras antibody as a loading control. (B to G) Cells were fixed and immunolabeled with anti-BLBP, anti-p75NGFR, or anti-S100 antibodies, with (Fix + perm) or without (Fix) permeabilization of cell membranes. Labeling was detected with a rhodaminated secondary antibody (red). Cell nuclei were labeled with bis-benzimide (blue). BLBP is detected on the cell surface (B) and after permeabilization (C) in Nf1−/− TXF cells. The membrane protein p75NGFR is detected with (E) or without (D) permeabilization. The cytoplasmic S100 protein is detected only after permeabilization (F and G). The scale bar in panel G equals 50 μm and also applies to panels B to F.
Previous studies showed that anti-BLBP antibodies were capable of preventing extension of radial glial cell processes and subsequent neuronal migration in mixed primary cerebellar cultures (2, 20), suggesting that BLBP resided at the cell surface. To localize BLBP in Nf1−/− TXF cells immunolabeling was conducted. The plasma membrane of fixed Nf1−/− TXF cells was labeled by anti-BLBP with (Fig. 2C) or without (Fig. 2B) addition of detergent to permeabilize cell membranes. Control experiments confirmed that fixed, nonpermeabilized Nf1−/− TXF cells could be labeled with an antibody recognizing the extracellular domain of a membrane protein (p75NGFR, Fig. 2D) but not a cytosolic protein (S100, Fig. 2F). Detection of protein without permeabilization of cell membranes indicates that BLBP is associated, at least in part, with the cell surface. BLBP was not localized to the nuclei of Nf1−/− TXF cells (Fig. 2C).
BLBP expression is elevated in mouse and human neural crest-derived tumor cell lines.
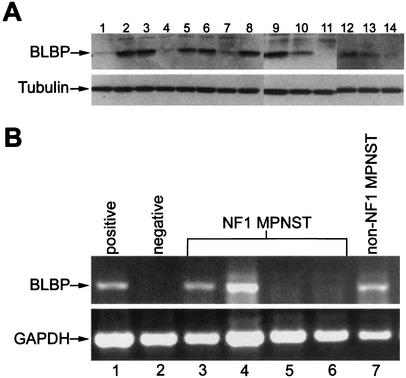
While Nf1 heterozygous mice do not form neurofibromas (10, 28), mice heterozygous for both Nf1 and p53 mutations develop neural crest-derived malignancies associated with NF1 patients, including MPNST and triton tumors, malignant peripheral sarcomas containing cells expressing muscle markers (14, 53). Cell lines derived from the Nf1:p53 mutant tumors (53) were analyzed for BLBP expression by Western blotting. BLBP was detected in 12 of the 14 cell lines examined (Fig. 3A), including malignant triton tumors and an MPNST. This result establishes a correlation between BLBP expression and Nf1-mediated tumorigenesis.
FIG. 3.
BLBP expression analysis in neural crest-derived tumor cell lines. (A) BLBP protein is detected by Western blotting in 12 of 14 neural crest-derived tumor cell lines from Nf1:p53 double mutant mice. Lanes 1 to 3, 5, 7 to 11, and 13 contain lysates from malignant triton tumor cell lines; lane 14 contains an MPNST lysate; lysates in lanes 4, 6, and 12 were obtained from unclassified tumors. Cell lines are as follows: lane 1, 38-1-3-7; lane 2, 40-1-7; lane 3, 67C-4; lane 4, 32-5-24; lane 5, 33-2-20; lane 6, 38-2-17-8; lane 7, 67A-4; lane 8, 39-2-11; lane 9, 61E-4; lane 10, 37-3-8-17; lane 11, 32-7-33; lane 12, 41-2-9; lane 13, 61C-4; lane 14, 32-5-30-2. Tubulin represents a control for loading equal amounts of protein in each lane. (B) BLBP is expressed in three of five MPNST cell lines. BLBP was amplified in a single-step RT-PCR from DNase-treated total RNA samples. GAPDH was amplified in a separate reaction to confirm the integrity of each sample. Lane 1, positive control, human embryonic kidney 293 cells; lane 2, negative control, HeLa cells; lane 3, NF1 patient MPNST cell line 190-8; lane 4, NF1 patient MPNST cell line 188-3; lane 5, NF1 patient MPNST cell line ST88-14; lane 6, NF1 patient MPNST cell line T265; lane 7, non-NF1 patient MPNST cell line S26T.
BLBP expression was then examined in human MPNST cell lines. BLBP mRNA was analyzed by RT-PCR, because antibodies capable of detecting human BLBP were not available. Four of the five cell lines were derived from NF1 patients. Three of the five cell lines tested were positive for BLBP (Fig. 3B); one of these was from the patient who did not have NF1. Therefore, half of the NF1-associated cell lines were BLBP positive.
BLBP expression can be induced by EGFR expression in wild-type mouse Schwann cells.
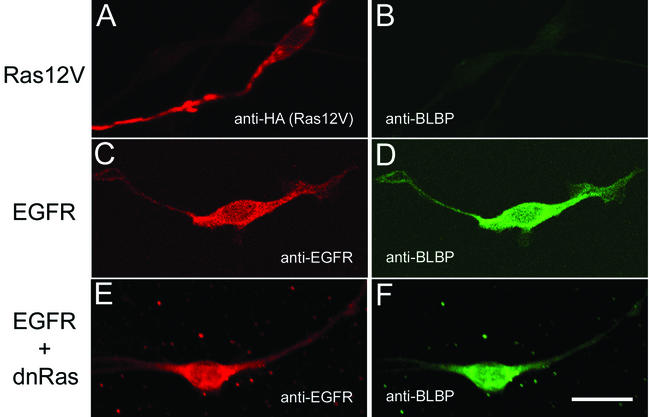
Candidate signaling pathways were tested for their ability to regulate BLBP expression in Schwann cells. Nf1−/− TXF mouse Schwann cells have elevated levels of activated Ras (30, 31; N. Ratner, unpublished data) and express EGFR (17). Human MPNST cell lines also have high Ras-GTP and EGFR expression (17, 18). Wild-type mouse Schwann cells do not express EGFR or BLBP. Therefore, to test the involvement of Ras and EGFR signaling pathways in BLBP expression, wild-type mouse Schwann cells were transfected with a constitutively active Ras mutant, CNP-(HA)HRas12V, or with a human EGFR, CNP-EGFR, and assayed for BLBP expression by immunolabeling. Schwann cells expressing (HA)HRas12V (Fig. 4A) have elevated levels of Ras-GTP (data not shown) and yet do not express BLBP (Fig. 4B). EGFR induced BLBP expression (Fig. 4C and D), with or without EGF in the culture medium. Schwann cells do produce EGFR ligands (S. J. Miller and N. Ratner, submitted for publication) that could stimulate EGFR signaling in an autocrine or paracrine fashion. EGFR induction of BLBP places BLBP expression downstream of EGFR.
FIG. 4.
BLBP expression analysis following transfection of H-Ras12V or EGFR into wild-type mouse Schwann cells. Wild-type mouse Schwann cells were transiently transfected with CNP-(HA)HRas12V or CNP-EGFR and assayed for induction of BLBP expression. (A) Detection of HA-tagged H-Ras12V protein with anti-HA antibodies. (B) Absence of BLBP in H-Ras12V-expressing cell (A). (C and E) Detection of EGFR protein with anti-EGFR antibodies. (D) Detection of BLBP with anti-BLBP antibodies in the wild-type mouse Schwann cells expressing EGFR (C). (F) BLBP expression is maintained in Schwann cells expressing EGFR and dominant-negative Ras (dnRas) (E). The scale bar in panel F equals 20 μm and also applies to panels A to E. Single cells are representative of all cells positive for the transfected construct indicated. Transfection efficiency was ∼6%.
Ras activation is also downstream of EGFR signaling (58). Although BLBP was not detected in Schwann cells expressing constitutively active H-Ras12V, signaling through Ras may still be required to induce BLBP expression. To determine if EGFR-induced expression of BLBP is Ras independent, a dominant-negative inhibitor of Ras, pZIPRasH(17N) (39), was cotransfected with EGFR into wild-type Schwann cells (Fig. 4E and F). The dominant-negative Ras inhibits downstream extracellularly regulated kinase phosphorylation (data not shown) but not BLBP expression (Fig. 4F). Control experiments with empty vector alone or in cotransfection did not affect BLBP expression (data not shown).
Previous studies identified multiple transcriptional regulatory elements in the BLBP gene (21). One of these sequences is specific to radial glial cells and dependent on neuregulin signaling (21), inducing BLBP expression with GGF (2). GGF failed to upregulate BLBP expression in wild-type mouse Schwann cells, as did contact of wild-type Schwann cells with axons (data not shown).
Inhibition of BLBP alters Nf1−/− TXF cell morphology but does not affect cell growth or migration.
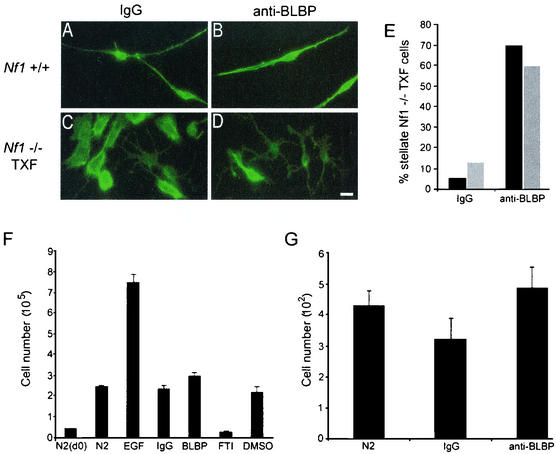
The majority of Nf1−/− TXF cells exhibit a process-free precursor-like shape, whereas approximately 10% are stellate. To assess the significance of BLBP expression in morphological transformation, BLBP-blocking antibodies were added to serum-starved Nf1−/− mouse Schwann cell cultures. Anti-BLBP antibodies, previously shown to block radial glial cell process outgrowth (20), did not prevent the formation of Nf1−/− TXF colonies (data not shown). Furthermore, inhibition of BLBP did not reverse Nf1−/− TXF cells to a wild-type Schwann cell shape, with characteristic slender cell body and two or three long processes. Approximately 60% of the cells, however, were converted to stellate morphology (Fig. 5A to E). This effect was not observed with control IgG, and no differences in cell death were observed between cells treated with IgG and those treated with anti-BLBP antibodies. Results were not significantly different between cells plated on poly-l-lysine alone and those plated on poly-l-lysine plus laminin-coated glass. Wild-type mouse Schwann cells were unaffected by anti-BLBP antibodies.
FIG. 5.
In vitro BLBP-blocking antibody experiments. (A to E) Anti-BLBP antibodies promote stellate Nf1−/− TXF cell morphology without affecting wild-type Schwann cell morphology. Wild-type (A and B) or Nf1−/− TXF (C and D) mouse Schwann cells were preincubated with rabbit IgG (A and C) or anti-BLBP antibodies (B and D) and plated onto glass coated with poly-l-lysine. After a 48-h incubation, cells were fixed and immunolabeled with AlexaFluor 488 phalloidin (green) recognizing F-actin. The scale bar in panel D equals 15 μm and also applies to panels A to C. (E) Approximately 70% of Nf1−/− TXF cells are stellate upon blocking BLBP, compared to ∼10% of those treated with IgG. All cells in each treatment group described above (∼200) were counted and assigned stellate or rounded morphology. Results are shown as percentages of stellate cells. No significant differences were observed between cells plated on poly-l-lysine alone (black bars) and cells plated on poly-l-lysine plus laminin (gray bars). (F) Anti-BLBP has no effect on Nf1−/− TXF cell number. Cells were plated in 24-well dishes (50,000 cells/well) and exposed to serum-free medium (N2), or serum-free medium supplemented with the indicated factors, in triplicate (EGF, mitogen; IgG, antibody control; anti-BLBP antibodies [1:300]; FTI, Ras-inhibitory drug shown to reduce Nf1−/− TXF cell number; dimethyl sulfoxide [DMSO], drug carrier control). Cells were counted at day 0, before the addition of any factor, to establish a baseline cell number. Factors were added at day 0 and day 2.5. Final cell counts were taken at day 5. Error bars reflect standard deviations in a two-tailed Student t test. (G) Anti-BLBP has no effect on Nf1−/− TXF cell migration. Cells were incubated with anti-BLBP antibodies or rabbit IgG and assayed for their ability to migrate through transwell filters. Cell nuclei were stained with bis-benzimide 15.5 h after plating and counted. Error bars reflect standard deviations in a two-tailed Student t test.
The anti-BLBP antibodies that induced morphological changes in Nf1−/− TXF cells did not affect cell growth or migration. Nf1−/− TXF cells proliferate in the absence of serum or added growth factors, and yet anti-BLBP failed to diminish their growth (Fig. 5F). Thus, BLBP is not directly responsible for Nf1−/− TXF-enhanced cell proliferation but represents a marker of the phenotype. Nf1-deficient mouse Schwann cells are also invasive (30) and migratory (Y. Huang and N. Ratner, unpublished data). Nf1−/− TXF cells exhibit enhanced migration, which was not inhibited by anti-BLBP antibodies (Fig. 5G). Therefore, blocking BLBP is not sufficient to prevent Nf1−/− TXF cell migration.
Inhibition of BLBP reverses the neuron-glial cell interaction defect in Nf1−/− TXF cells.
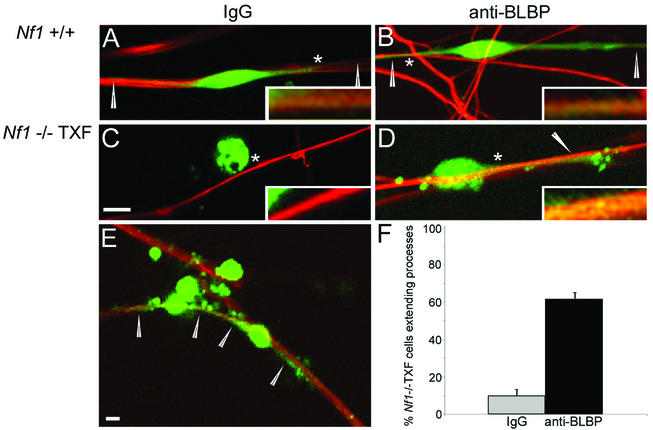
Wild-type and Nf1−/− Schwann cells maintain close contact with axons. A striking characteristic of the Nf1−/− TXF cell phenotype is their colonization away from neurites (30). Studies reporting the ability of anti-BLBP antibodies to inhibit neuronal migration along radial glial cells (2, 20) suggested that BLBP might function in neuron-Schwann cell interaction. Wild-type mouse DRG neurons, stripped of endogenous Schwann cells, were cultured with dye-labeled wild-type or Nf1−/− TXF Schwann cells in the presence or absence of anti-BLBP antibodies. Wild-type Schwann cells adhered to axons and showed extension of processes along axons as expected, regardless of IgG (Fig. 6A) or anti-BLBP antibodies (Fig. 6B). Nf1−/− TXF cells occasionally adhered to neurites but rarely extended processes (Fig. 6C). In contrast, when anti-BLBP antibodies were present (Fig. 6D and E), 62% of Nf1−/− TXF cells extended processes along axons (in comparison to 10% of those preincubated with control IgG). While blocking BLBP allowed Nf1−/− TXF cells to extend processes along neurites, the extent of process outgrowth never reached that of wild-type cells.
FIG. 6.
Mouse neuron-Schwann cell coculture. Anti-BLBP promotes extension of Nf1−/− TXF cell processes along axons. Wild-type (A and B) or Nf1−/− TXF (C and D) mouse Schwann cells labeled with Cell Tracker green were preincubated with rabbit IgG (A and C) or anti-BLBP antibodies (B and D) and seeded onto DRGN cultures stripped of endogenous Schwann cells. Two days after seeding, cocultures were fixed and stained with antineurofilament antibodies followed by Cy3 (red)-conjugated secondary antibodies. Confocal images obtained with Zeiss LSM Image Browser software are shown. Single cells are representative of the majority observed with each treatment. Arrowheads indicate Schwann cell processes. The asterisk indicates the region which is magnified fivefold in the inset. The scale bar in panel C equals 5 μm and also applies to panels A, B, and D. (E) Lower magnification (scale bar, 5 μm) of Nf1−/− TXF on DRGN cultures in the presence of anti-BLBP antibodies. Arrowheads indicate processes from two cells extending along neurites; other cells lack processes. (F) Extension of Nf1−/− TXF cell processes in the presence of anti-BLBP antibodies is statistically significant. The percentages of Nf1−/− TXF cells extending processes along axons in the presence of control IgG (gray bar) or anti-BLBP antibodies (black bar) are graphed. Error bars reflect standard deviations in a Student t test (P = 0.003).
BLBP expression in vivo is upregulated 2 to 4 weeks after nerve injury.
If BLBP expression were involved in neuron-glial interactions, it seemed possible that it would mark a specific stage of neuron-Schwann cell interaction. To test the hypothesis that neuronal interaction regulates BLBP expression by Schwann cells in vivo, nerve injury experiments were conducted. The reinnervation of Schwann cells by axons following a nerve crush recapitulates neuron-glial interactions in development.
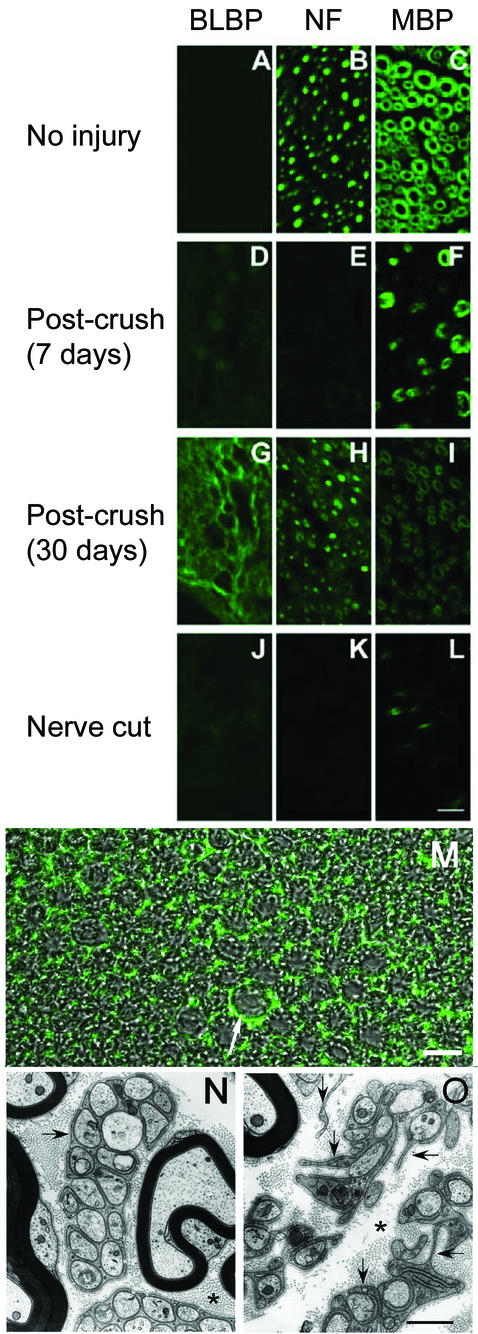
Sciatic nerves of wild-type adult mice were crushed, and distal nerve segments were isolated 4 to 30 days after injury. Tissue sections were labeled with highly specific BLBP antibodies (20). BLBP was not detected in uninjured sciatic nerve (Fig. 7A), as expected (34). BLBP was also not present 4 to 7 days postinjury (nonspecific labeling appeared to be associated with macrophages [Fig. 7D]). This time point represents a period of axon degeneration and temporary loss of Schwann cell contact (3). In contrast, BLBP expression was robust 14 to 30 days after crush injury (Fig. 7G), when many thinly myelinated axons are present. To further confirm a dependence of BLBP expression on axon interaction, adult mouse sciatic nerve was cut, and the distal stump was isolated 21 days later, when axons are absent. No labeling was detected with BLBP antibodies (Fig. 7J).
FIG. 7.
BLBP expression in adult mouse nerve following lesion. BLBP expression is upregulated in adult mouse Schwann cells 2 to 4 weeks following nerve crush injury. Sciatic nerves of anesthetized wild-type adult mice were isolated without injury (A to C), at 7 days after nerve crush (D to F), at 30 days after nerve crush (G to I), or at 21 days after nerve cut (J to L). Nerve cross sections were labeled with anti-BLBP antibodies (BLBP) (A, D, G, and J), antineurofilament (NF) (B, E, H, and K) to label axons, or anti-myelin basic protein (MBP) (C, F, I, and L) to stain myelin; staining was detected by immunofluorescence. For each antibody, confocal images were obtained at the same settings to match relative intensities. The scale bar in panel L equals 10 μm and also applies to panels A to K. (M) Confocal differential interference contrast and fluorescent image of sciatic nerve 30 days postcrush stained with anti-BLBP antibodies. The arrow indicates BLBP detected outside of the myelin sheath. Scale bar, 10 μm. (N) Electron micrograph of wild-type adult mouse nerve. (O) Electron micrograph of nerve 28 days after crush injury. Arrows indicate multiple Schwann cell processes. Asterisks mark collagen-rich matrix. The scale bar equals 500 nm and also applies to panel N.
The pattern of BLBP immunolabeling in the distal stump at 14 to 30 days after nerve crush (Fig. 7G) did not overlap that of myelin sheaths (Fig. 7I) or axons (Fig. 7H). Rather, BLBP appeared to be between myelinated axons (Fig. 7M). To define elements present in this location, we carried out electron microscopy on distal nerve segments 26 days after a crush injury. The matrix surrounding myelinated fibers is filled with abundant non-myelin-forming Schwann cell processes (Fig. 7O). Some Schwann cell processes are not associated with axons, but most enclose one to four unmyelinated axons. This appearance is in striking contrast to that of the uncrushed nerve, in which non-myelin-forming Schwann cells are much more limited and patchy in their distribution and each contains large groups of unmyelinated fibers in delineated packets (Fig. 7N). Membrane-delineated Schwann cell processes unassociated with axons never were observed in the uncrushed nerve. In the crushed nerve, in addition to Schwann cells, there were a limited number of fibroblasts, lacking a basal lamina, and occasional phagocytic cells that each contained ∼20 degenerating nerve fibers. The only cell type seen by electron microscopy of the regenerating nerve that had a pervasive distribution like that seen by BLBP immunocytochemistry was the Schwann cell. This correlation of BLBP expression with Schwann cell process outgrowth and/or retraction is consistent with a role for BLBP in process remodeling.
DISCUSSION
Tumorigenesis is a multistep process, involving inactivation of tumor suppressor genes and activation of oncogenes, that results in uncontrolled cell growth and behavior. Apart from mutations in NF1 and p53, molecular changes contributing to the progression of neurofibroma-MPNST have not been identified (13). Here we utilized cDNA microarray technology to identify a gene, BLBP, that is dramatically upregulated in Nf1−/− TXF mouse Schwann cells that have characteristics of tumor cells including rapid proliferation and loss of interaction with axons. BLBP belongs to a family of fatty acid binding proteins that have been implicated previously in the pathogenesis of several malignancies (11, 15, 16, 42). BLBP binds fatty acids in vitro and may function as a lipid transporter, but the in vivo BLBP ligand has not been identified (4, 43, 56). It is of interest that fatty acids inhibit the GAP activity of neurofibromin and of p120GAP (8, 23, 26, 47). Our data support the utility of the Nf1 mutant mouse tissue culture system and microarray methodology for identification of genes relevant to peripheral nerve tumors.
BLBP and EGFR are both abnormally expressed in tumor models of NF1. Nf1−/− TXF mouse Schwann cells express EGFR (17) and, as we have shown here, BLBP. We have as yet been unable to analyze BLBP expression in neurofibromas or MPNST tissue sections due to the lack of anti-human BLBP antibodies. All human MPNST cell lines tested express EGFR (17), as do all of the Nf1:p53 mouse peripheral nerve tumor-derived cell lines (36, 53). A subset of the tested MPNST cell lines and Nf1:p53 cell lines that express EGFR have increased BLBP, just as BLBP is expressed in a subset of human malignant glioma cell lines (22). Although no obvious phenotypic differences were observed between BLBP-positive and BLBP-negative MPNST cell lines, BLBP expression could signify a specific stage in tumor formation. Molecular changes that accumulated in some MPNST cell lines could diminish EGFR-induced BLBP expression. Alternatively, cell lines with low BLBP expression, while expressing EGFR, could have low activation of EGFR signaling.
Transfection of wild-type mouse Schwann cells with EGFR induced BLBP expression. In vivo, BLBP is expressed in Schwann cell precursors (34) and following nerve injury. In neither case is EGFR expressed. Alternatively, Schwann cells express the ErbB2 and ErbB3 neuregulin receptors that are closely related to EGFR (1); indeed, EGFR and ErbB2 form heterodimers and activate similar signaling cascades (58). It appears that EGFR but not neuregulin stimulation of ErbB2-ErbB3 signals increases BLBP expression in Schwann cells. Omitting or increasing GGF, a neuregulin isoform, in culture media did not affect BLBP expression in wild-type or Nf1−/− TXF mouse Schwann cells (data not shown). GGF is included in routine culture of all Schwann cells, and yet only the Nf1−/− TXF cells express BLBP. It remains possible that in vivo neuregulin signaling affects BLBP expression at restricted times during development, as neuregulin signaling in radial glia can induce BLBP expression (2, 21). It appears likely that in Schwann cells high signaling strength caused by increased receptor levels and/or activation of specific downstream signaling pathways is required for BLBP expression.
One of the major conclusions of this study is that EGFR expression, but not Ras-GTP, is sufficient to drive BLBP expression in Schwann cells. Based upon Drosophila melanogaster mutants, a model including receptor tyrosine kinase activation of a mitogen-activated protein kinase signaling cascade was proposed elsewhere to explain BLBP expression (21). Mitogen-activated protein kinase signaling downstream of H-Ras activation does not appear to explain our results. Ras-mediated signaling is hyperactive in Schwann cells lacking Nf1 (18, 24, 31, 49), and yet Nf1−/− mouse Schwann cells do not express BLBP. In addition, transfection of a constitutively active H-Ras mutant into wild-type mouse Schwann cells failed to induce BLBP expression, and a dominant-negative H-Ras was unable to block EGFR-mediated expression of BLBP. Furthermore, exposure to FTI, an inhibitor of H-Ras function in Schwann cells and of proliferation in Nf1−/− TXF cells (30), did not alter BLBP expression. This demonstrates that farnesylation-sensitive Ras proteins, such as H-Ras (46), are not necessary for BLBP expression. These lines of evidence strongly support the conclusion that Ras-GTP is not sufficient to drive BLBP expression in Schwann cells. Other signaling pathways downstream of EGFR activation (58) must therefore contribute to BLBP induction. Identification of BLBP as an EGFR target should allow us to define relevant pathways. Regulation of BLBP expression is partially understood; the BLBP promoter is a complex regulatory element (21). POU transcription factors (29) and nuclear factor I (7) are implicated in BLBP gene expression. Possibly non-Ras signaling downstream of EGFR will link to nuclear factor I activation and thus to BLBP expression.
Our in vitro assays suggest a possible functional role for BLBP in the pathogenesis of neurofibromas and/or MPNST, but not in initiation of cell transformation. Anti-BLBP antibodies failed to influence formation of Nf1−/− TXF colonies. The antibodies also did not affect cell proliferation or migration. However, blocking BLBP did facilitate process outgrowth from Nf1−/− TXF cells and normalized association of Nf1−/− TXF cells with axons, suggesting that BLBP inhibits stable interaction of Schwann cells with axons. This is important because neuron-glial disruption is characteristic of Schwann cells in human neurofibromas and MPNST. In our studies anti-BLBP antibodies stimulated process formation by Nf1−/− TXF cells, and yet the same antibodies inhibited process outgrowth from normal radial glial cells in vitro (2, 20). These opposite effects on process outgrowth may be due to aberrant signaling in the Nf1 mutant cells or different functions of BLBP in the central versus peripheral nervous systems.
Anti-BLBP antibodies affect glial cell process outgrowth when applied to cultured radial glial cells (2, 20) and Nf1−/− TXF Schwann cells, suggesting that BLBP is present at least in part at the cell surface. In radial glial cells, immunoelectron microscopy showed BLBP in the cytoplasm and nucleus, but membrane localization was not evaluated (20). In our immunolabeling studies, BLBP was detected in the cytoplasm and at the plasma membrane of cultured Nf1−/− TXF mouse Schwann cells. As the structure of BLBP does not suggest a transmembrane protein (4), BLBP may function in association with an as-yet-unidentified cell surface receptor. Surface localization may reflect a role for BLBP in fatty acid uptake and/or cell-cell interaction.
BLBP is expressed early in peripheral nerve development (34) when Schwann cells are segregating progressively smaller groups of axons (19, 54). Once Schwann cell-axon interactions are stabilized in adult nerve, BLBP expression is extinguished (34) (Fig. 7). BLBP expression was induced in 2 to 4 weeks after nerve crush injury, when L1 and N-CAM adhesion molecules, associated with nonmyelinating Schwann cells (37), are also upregulated. Using electron microscopy we showed that, 2 to 4 weeks after crush injury, nonmyelinating Schwann cell-axon bundles are maturing by dynamic Schwann cell process outgrowth. Our in vivo BLBP expression data thus support a role for BLBP as a marker of specific phases of neuron-glial cell interactions in the peripheral nervous system. When Schwann cell process formation is exuberant, BLBP expression is high. We do not yet know whether BLBP is required for process outgrowth and/or for the withdrawal of processes as axon-Schwann cell bundles mature. However, our studies of Nf1 mutant Schwann cells support the idea that BLBP can cause process withdrawal in certain cellular contexts.
In conclusion, global gene expression analysis of an Nf1 mutant mouse Schwann cell culture system resulted in the identification of a potential marker for Schwann cell transformation, BLBP. BLBP is expressed in malignant Schwann cell tumor cell lines. EGFR, also implicated in Schwann cell tumorigenesis (17), induced BLBP expression in wild-type Schwann cells. Although Ras activity is upregulated in Nf1 mutant Schwann cells, BLBP expression is not affected by Ras signaling. Our in vitro and in vivo data support a role for BLBP in neuron-Schwann cell interactions. Expression of BLBP inhibits stable association of Schwann cells with axons. The results of these experiments suggest a novel molecular pathway in peripheral nerve tumorigenesis, in which NF1 mutation leads to aberrant EGFR expression that results in elevated levels of BLBP and loss of axonal contact.
Acknowledgments
We are especially grateful for the generous donations of anti-BLBP antibodies from Nathaniel Heintz, FTI (L-745,832) from Jay Gibbs (Merck Research Laboratories), and recombinant human GGF2 from Mark Marchionni (Cambridge Neuroscience). We appreciate the assistance of Bruce Aronow and Sarah Williams at the University of Cincinnati Bioinformatics Core in analyzing microarray data. We acknowledge Frank Sharp for the use of his ABI Prism 7700 sequence detection system. We also thank Doug Lowy and Larry Sherman for critically reviewing the manuscript.
This work was supported by grants NIH NS28840 and DOD 17-01-1-07 to N.R. S.R.M. was the recipient of a National Multiple Sclerosis Society Advanced Postdoctoral Fellowship.
REFERENCES
- 1.Adlkofer, K., and C. Lai. 2000. Role of neuregulins in glial cell development. Glia 29:104-111. [DOI] [PubMed] [Google Scholar]
- 2.Anton, E. S., M. A. Marchionni, K.-F. Lee, and P. Rakic. 1997. Role of GGF/neuregulin signaling interactions between migrating neurons and radial glia in the developing cerebral cortex. Development 124:3501-3510. [DOI] [PubMed] [Google Scholar]
- 3.Arroyo, E. J., J. R. Bermingham, M. G. Rosenfeld, and S. S. Scherer. 1998. Promyelinating Schwann cells express Tst-1/SCIP/Oct-6. J. Neurosci. 18:7891-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balendiran, G. K., F. Schnutgen, G. Scapin, T. Borchers, N. Xhong, K. Lim, R. Godbout, F. Spener, and J. C. Sacchettini. 2000. Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J. Biol. Chem. 275:27045-27054. [DOI] [PubMed] [Google Scholar]
- 5.Basu, T. N., D. H. Gutmann, J. A. Fletcher, T. W. Glover, F. S. Collins, and J. Downward. 1992. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature 356:663-664. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, E., K. L. Stenvers, P. K. Lund, and B. Popko. 1994. Cloning and characterization of a cDNA encoding a novel fatty acid binding protein from rat brain. J. Neurochem. 63:1616-1624. [DOI] [PubMed] [Google Scholar]
- 7.Bisgrove, D. A., E. A. Monckton, M. Packer, and R. Godbout. 2000. Regulation of brain fatty acid-binding protein expression by differential phosphorylation of nuclear factor I in malignant glioma cell lines. J. Biol. Chem. 275:30668-30676. [DOI] [PubMed] [Google Scholar]
- 8.Bollag, G., and F. McCormick. 1991. Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature 351:576-579. [DOI] [PubMed] [Google Scholar]
- 9.Boyden, S. 1962. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J. Exp. Med. 115:453-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brannan, C. I., A. S. Perkins, K. S. Vogel, N. Ratner, M. L. Nordlund, S. W. Reid, A. M. Buchberg, N. A. Jenkins, L. F. Parada, and N. G. Copleland. 1994. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 8:1019-1029. [DOI] [PubMed] [Google Scholar]
- 11.Celis, J. E., M. Ostergaard, B. Basse, A. Celis, J. B. Lauredsin, G. P. Ratz, I. Anderson, B. Hein, H. Wolf, T. Orntoft, and H. H. Rasmussen. 1996. Loss of adipocyte-type fatty acid binding protein and other biomarkers is associated with progression of human bladder transitional cell carcinomas. Cancer Res. 56:4782-4790. [PubMed] [Google Scholar]
- 12.Chandross, K. J., R. I. Cohen, P. Paras, M. Gravel, P. E. Braun, and L. D. Hudson. 1999. Identification and characterization of early glial progenitors using a transgenic selection strategy. J. Neurosci. 19:759-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cichowski, K., and T. Jacks. 2001. NF1 tumor suppressor function: narrowing the GAP. Cell 104:593-604. [DOI] [PubMed] [Google Scholar]
- 14.Cichowski, K., T. S. Shih, E. Schmitt, S. Santiago, K. Reilly, M. E. McLaughlin, R. T. Bronson, and T. Jacks. 1999. Mouse models of tumor development in neurofibromatosis type 1. Science 286:2172-2176. [DOI] [PubMed] [Google Scholar]
- 15.Custer, R. P., and S. Sorof. 1984. Target polypeptide of a carcinogen is associated with normal mitosis and carcinogen-induced hyperplasia in adult hepatocytes. Proc. Natl. Acad. Sci. USA 81:7638-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, R., R. Hammamieh, R. Neill, M. Melhem, and M. Jett. 2001. Expression pattern of fatty acid-binding proteins in human normal and cancer prostate cells and tissues. Clin. Cancer Res. 7:1706-1715. [PubMed] [Google Scholar]
- 17.DeClue, J. E., S. Heffelfinger, G. Benvenuto, B. Ling, S. Li, W. Rui, W. C. Vass, D. Viskochil, and N. Ratner. 2000. Epidermal growth factor receptor expression in neurofibromatosis type-1 related tumors and NF1 animal models. J. Clin. Investig. 105:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeClue, J. E., A. G. Papageorge, J. A. Fletcher, S. R. Diehl, N. Ratner, W. C. Vass, and D. R. Lowy. 1992. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell 69:265-273. [DOI] [PubMed] [Google Scholar]
- 19.Dong, Z., A. Sinanan, D. Parkinson, E. Parmantier, R. Mirsky, and K. R. Jessen. 1999. Schwann cell development in embryonic mouse nerves. J. Neurosci. Res. 56:334-348. [DOI] [PubMed] [Google Scholar]
- 20.Feng, L., M. E. Hatten, and N. Heintz. 1994. Brain lipid binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron 12:895-908. [DOI] [PubMed] [Google Scholar]
- 21.Feng, L., and N. Heintz. 1995. Differentiating neurons activate transcription of the brain lipid binding protein gene in radial glia through a novel regulatory element. Development 121:1719-1730. [DOI] [PubMed] [Google Scholar]
- 22.Godbout, R., D. A. Bisgrove, D. Shkolny, and R. S. Day. 1998. Correlation of B-FABP and GFAP expression in malignant glioma. Oncogene 16:1955-1962. [DOI] [PubMed] [Google Scholar]
- 23.Golubic, M., J. A. Harwalkar, S. S. Bryant, V. Sundaram, R. Jove, and J. H. Lee. 1998. Differential regulation of neurofibromin and p120 GTPase-activating protein by nutritionally relevant fatty acids. Nutr. Cancer 30:97-107. [DOI] [PubMed] [Google Scholar]
- 24.Guha, A., N. Lau, I. Huvar, D. Gutman, J. Provias, T. Pawson, and G. Boss. 1996. Ras-GTP levels are elevated in human NF1 peripheral nerve tumors. Oncogene 12:507-513. [PubMed] [Google Scholar]
- 25.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, J. W., F. McCormick, and I. G. Macara. 1991. Regulation of Ras-GAP and the neurofibromatosis-1 gene product by eicosanoids. Science 252:576-579. [DOI] [PubMed] [Google Scholar]
- 27.Ideker, T., V. Thorsson, J. A. Ranish, R. Christmas, J. Buhler, J. K. Eng, R. Bumgarner, D. R. Goodlett, R. Aebersold, and L. Hood. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929-934. [DOI] [PubMed] [Google Scholar]
- 28.Jacks, T., T. S. Shih, E. M. Schmitt, R. T. Bronson, A. Bernards, and R. A. Weinberg. 1994. Tumor predisposition in mice heterozygous for a targeted mutation in NF1. Nat. Genet. 7:353-361. [DOI] [PubMed] [Google Scholar]
- 29.Josephson, R., T. Muller, J. Pickel, S. Okabe, K. Reynolds, P. A. Turner, A. Zimmer, and R. D. McKay. 1998. POU transcription factors control expression of CNS stem cell-specific genes. Development 125:3087-3100. [DOI] [PubMed] [Google Scholar]
- 30.Kim, H. A., B. Ling, and N. Ratner. 1997. Nf1-deficient mouse Schwann cells are angiogenic and invasive and can be induced to hyperproliferate: reversion of some phenotypes by an inhibitor of farnesyl protein transferase. Mol. Cell. Biol. 17:862-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, H. A., T. Rosenbaum, M. A. Marchionni, N. Ratner, and J. E. DeClue. 1995. Schwann cells from neurofibromin deficient mice exhibit activation of p21ras, inhibition of cell proliferation and morphological changes. Oncogene 11:325-335. [PubMed] [Google Scholar]
- 32.Kluwe, L., R. Friedrich, and V. Mautner. 1999. Loss of NF1 allele in Schwann cells but not fibroblasts derived from an NF1-associated neurofibroma. Genes Chromosomes Cancer 24:283-285. [DOI] [PubMed] [Google Scholar]
- 33.Kohl, N. E., S. D. Mosser, J. deSolms, E. A. Giuliani, D. L. Pompliano, S. L. Graham, R. L. Smith, E. M. Scolnick, A. Oliff, and J. B. Gibbs. 1995. Inhibition of farnesyltransferase induces regression of mammary and salivary carcinomas in ras transgenic mice. Nat. Med. 1:792-797. [DOI] [PubMed] [Google Scholar]
- 34.Kurtz, A., A. Zimmer, F. Schnutgen, G. Bruning, F. Spener, and T. Muller. 1994. The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development 120:2637-2649. [DOI] [PubMed] [Google Scholar]
- 35.Legius, E., D. A. Marchuk, F. S. Collins, and T. W. Glover. 1993. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumor suppressor gene hypothesis. Nat. Genet. 3:122-126. [DOI] [PubMed] [Google Scholar]
- 36.Li, H., S. Velasco-Miguel, W. C. Vass, L. F. Parada, and J. E. DeClue. 2002. Epidermal growth factor receptor signaling pathways are associated with tumorigenesis in the Nf1:p53 mouse tumor model. Cancer Res. 62:4507-4513. [PubMed] [Google Scholar]
- 37.Nieke, J., and M. Schachner. 1985. Expression of the neural cell adhesion molecules L1 and N-CAM and their common carbohydrate epitope L2/HNK-1 during development and after transection of the mouse sciatic nerve. Differentiation 30:141-151. [DOI] [PubMed] [Google Scholar]
- 38.Parysek, L. M., and R. D. Goldman. 1987. Characterization of intermediate filaments in PC12 cells. J. Neurosci. 7:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quilliam, L. A., K. Kato, K. M. Rabun, M. M. Hisaka, S. Y. Huff, S. Campbell-Burk, and C. J. Der. 1994. Identification of residues critical for Ras(17N) growth-inhibitory phenotype and for Ras interactions with guanine nucleotide exchange factors. Mol. Cell. Biol. 14:1113-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajeevan, M. S., D. G. Ranamukhaarachchi, S. D. Vernon, and E. R. Unger. 2001. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods 25:443-451. [DOI] [PubMed] [Google Scholar]
- 41.Ramdas, L., K. R. Coombes, K. Baggerly, L. Abruzzo, W. E. Highsmith, T. Krogman, S. R. Hamilton, and W. Zhang. 2001. Sources of nonlinearity in cDNA microarray expression measurements. Genome Biol. 2:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasmussen, H. H., T. F. Orntoft, H. Wolf, and J. E. Celis. 1996. Towards a comprehensive database of proteins from the urine of patients with bladder cancer. J. Urol. 155:2113-2119. [PubMed] [Google Scholar]
- 43.Richieri, G. V., R. T. Ogata, A. W. Zimmerman, J. H. Veerkamp, and A. M. Kleinfeld. 2000. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry 39:7197-7204. [DOI] [PubMed] [Google Scholar]
- 44.Rickman, D. S., M. P. Bobek, D. E. Misek, R. Kuick, M. Blaivas, D. M. Kurnit, J. Taylor, and S. Hanash. 2001. Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 61:6885-6891. [PubMed] [Google Scholar]
- 45.Rizvi, T. A., Y. Huang, A. Sidani, R. Atit, D. A. Largaespada, R. E. Boissy, and N. Ratner. 2002. A novel cytokine pathway suppresses glial cell melanogenesis after injury to adult nerve. J. Neurosci. 22:9831-9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sebti, S., and A. D. Hamilton. 1997. Inhibitors of prenyl transferases. Curr. Opin. Oncol. 9:557-561. [DOI] [PubMed] [Google Scholar]
- 47.Sermon, B. A., J. F. Eccleston, R. H. Skinner, and P. N. Lowe. 1996. Mechanism of inhibition by arachidonic acid of the catalytic activity of Ras GTPase-activating proteins. J. Biol. Chem. 271:1566-1572. [DOI] [PubMed] [Google Scholar]
- 48.Serra, E., T. Rosenbaum, U. Winner, R. Aledo, E. Ars, X. Estivill, H.-G. Lenard, and C. Lazaro. 2000. Schwann cells harbor the somatic NF1 mutation in neurofibromas: evidence of two different Schwann cell populations. Hum. Mol. Genet. 9:3055-3064. [DOI] [PubMed] [Google Scholar]
- 49.Sherman, L. S., R. Atit, T. Rosenbaum, A. D. Cox, and N. Ratner. 2000. Single cell Ras-GTP analysis reveals altered Ras activity in a subpopulation of neurofibroma Schwann cells but not fibroblasts. J. Biol. Chem. 275:30740-30745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu, F., T. K. Watanabe, H. Shinomiya, Y. Nakamura, and T. Fujiwara. 1997. Isolation and expression of a cDNA for human brain fatty acid-binding protein (B-FABP). Biochim. Biophys. Acta 1354:24-28. [DOI] [PubMed] [Google Scholar]
- 51.Tamanoi, F. 1993. Inhibitors of Ras: farnesyltransferases. Trends Biochem. Sci. 18:349-353. [DOI] [PubMed] [Google Scholar]
- 52.Velu, T. J., L. Beguinot, W. C. Vass, M. C. Willingham, G. T. Merlino, I. Pastan, and D. R. Lowy. 1987. Epidermal growth factor-dependent transformation by a human EGF receptor proto-oncogene. Science 238:1408-1410. [DOI] [PubMed] [Google Scholar]
- 53.Vogel, K. S., L. J. Klesse, S. Velasco-Miguel, K. Meyers, E. J. Rushing, and L. F. Parada. 1999. Mouse tumor model for neurofibromatosis type 1. Science 286:2176-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webster, H. D. 1993. Development of peripheral nerve fibers, p. 243-266. In P. J. Dyck, P. K. Thomas, P. A. Low, and J. F. Poduslo (ed.), Peripheral neuropathy. The W. B. Saunders Co., Philadelphia, Pa.
- 55.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80:533-541. [DOI] [PubMed] [Google Scholar]
- 56.Xu, L. Z., R. Sanchez, A. Sali, and N. Heintz. 1996. Ligand specificity of brain lipid-binding protein. J. Biol. Chem. 271:24711-24719. [DOI] [PubMed] [Google Scholar]
- 57.Yan, N., C. Ricca, J. Fletcher, T. Glover, B. R. Seizinger, and V. Manne. 1995. Farnesyltransferase inhibitors block the neurofibromatosis type 1 (NF1) malignant phenotype. Cancer Res. 55:3569-3575. [PubMed] [Google Scholar]
- 58.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. 2:127-137. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, Y., P. Ghosh, P. Charnay, D. K. Burns, and L. F. Parada. 2002. NF1 associated neurofibromas initiate in Schwann cells and require a haploinsufficient environment. Science 296:920-922. [DOI] [PMC free article] [PubMed] [Google Scholar]