Abstract
HGK (hepatocyte progenitor kinase-like/germinal center kinase-like kinase) is a member of the human STE20/mitogen-activated protein kinase kinase kinase kinase family of serine/threonine kinases and is the ortholog of mouse NIK (Nck-interacting kinase). We have cloned a novel splice variant of HGK from a human tumor line and have further identified a complex family of HGK splice variants. We showed HGK to be highly expressed in most tumor cell lines relative to normal tissue. An active role for this kinase in transformation was suggested by an inhibition of H-RasV12-induced focus formation by expression of inactive, dominant-negative mutants of HGK in both fibroblast and epithelial cell lines. Expression of an inactive mutant of HGK also inhibited the anchorage-independent growth of cells yet had no effect on proliferation in monolayer culture. Expression of HGK mutants modulated integrin receptor expression and had a striking effect on hepatocyte growth factor-stimulated epithelial cell invasion. Together, these results suggest an important role for HGK in cell transformation and invasiveness.
The mammalian STE20/mitogen-activated protein kinase kinase kinase kinase (MAP4K) family consists of 28 serine/threonine kinases related in their catalytic domains (reviewed in reference 14). By analogy with the prototype STE20 kinase in Saccharomyces cerevisiae, mammalian MAP4K kinases are likely to regulate changes in transcription, cytoskeletal organization, and cell cycle progression in response to extracellular signals (17). Comparison of the overall domain structure places these kinases into two structural classes, the p21-activated protein kinases (PAKs) (1) and germinal center kinases (GCKs) (28). The GCK kinases lack the regulatory Cdc42/Rac-interacting domain found in the PAKs, having instead an N-terminal kinase domain and a C-terminal extension of variable length. Unlike PAKs, several GCKs appear to be activated in the absence of stimuli when overexpressed (3, 10, 38), suggesting that these kinases are regulated either by oligomerization or by binding of negative regulatory factors.
GCK kinases show little sequence homology outside of the kinase domain and are further broken down into nine subfamilies (14, 31a). HGK (hepatocyte progenitor kinase-like/GCK-like kinase) is one of four members of the GCK group IV (recently renamed the MSN subfamily) that also includes TNIK, MINK, and NRK/NESK (13, 22, 27, 35, 51). HGK, TNIK, and MINK are highly homologous in their kinase and C-terminal domains (about 92 and 87% amino acid identity, respectively), with variability in the intervening region that is less conserved (53% between HGK and TNIK). NRK/NESK is more divergent, with only 59% homology in the kinase domain and 37% homology in the C-terminal domain. The C-terminal domain is a citron homology (CNH) domain, named for citron rho-interacting kinase (CRIK), where it was first described (16, 31). CNH domains are found not only in the GCK group IV/MSN kinases but also in group I GCKs (KHS subfamily) (14, 31a), as well as in proteins other than kinases, including human Vam6p homologs, involved in membrane vesicle docking and fusion, and the S. cerevisiae Rho GDP-exchange factor Rom1p (9). The CNH domain has been shown to be important for protein-protein interactions. In CRIK itself, this domain mediates association with rho family GTPases (30, 31). In Vam6p, it mediates association with lysosomes (9). In GCK (a group I/KHS kinase), this domain is important for homodimerization, leading to the activation of mitogen-activated protein kinase/extracellular signal-regulated kinase MEKK1 (10). In mouse HGK, the CNH domain has been shown to interact with MEKK1 and also recently shown to mediate interaction of this kinase with the cytoplasmic domain of β1-integrin receptors (37, 43).
Orthologs of the HGK group are known in both Drosophila melanogaster and Caenorhabditis elegans, called misshapen (msn) and mig-15, respectively (46, 53). msn and mig-15 show striking sequence conservation with HGK in both the kinase and CNH domains, with more than 80% amino acid identity in both domains.
msn functions in multiple signaling pathways, including those downstream of frizzled (Wnt receptor) in determining epithelial polarity, in the regulation of dorsal closure, and in neuronal targeting. In the Wnt pathway, msn bifurcates from the β-catenin/cadherin signaling pathway at the upstream signaling protein coded by disheveled, signaling to a separate pathway leading to the activation of both JNK and p38 (36). In dorsal closure, msn also signals through a JNK module that mediates the spreading of epithelial sheets to close the embryo through induction of transforming growth factor β (44, 46). In photoreceptor neuronal targeting, msn functions downstream of dock, the Drosophila homolog of the Nck adapter protein, to guide axons to their target (40, 44). In nematodes, mig-15 functions in the processes of muscle arm targeting, where muscle cells extend cytoplasmic protrusions to synapse with motor neurons during development, as well as in neuroblast migration (53). The common factor to both model systems is the involvement of an HGK homolog in processes requiring cell migration or cell shape change.
The mouse ortholog of HGK, Nck-interacting kinase (NIK), was cloned in an interaction screen with the SH3 domain of the Nck adapter protein (43). NIK has been shown to be activated by stimulation of the ephrin receptor family (3). Ephrins are cell surface ligands shown to be involved in cell migration and tissue remodeling (52) and have been proposed to be involved in inside-out integrin regulation (15, 33). Consistent with a positive function for NIK in cell migration and morphogenesis, NIK−/− knockout mice showed an early embryonic-lethal phenotype, with defects in mesoderm differentiation and migration (50). This embryonic lethality indicates a lack of functional redundancy between the GCK group IV family members during development.
Human HGK was previously cloned from a macrophage cDNA library (51). Yao et al. demonstrated that HGK activated JNK in transient transfections and further suggested that this activation occurred through the mitogen-activated protein kinase kinase kinase transforming growth factor β-activated kinase (TAK1) rather than MEKK1.
We present the characterization of an alternatively spliced form of HGK identified from a human glioblastoma library and provide an analysis of a complex family of alternatively spliced HGK isoforms. We found HGK mRNA to be broadly overexpressed in tumor cell lines relative to normal adult tissue. Ectopic expression of various mutants of the tumor-derived HGK isoform suggested an active role for this protein in cell transformation and invasion as well as in reducing the adhesive properties of tissue culture cells. HGK appeared to function in these processes through modulation of growth factor and integrin receptor signaling.
MATERIALS AND METHODS
Cloning of HGK gene.
The complete HGK cDNA was constructed from two overlapping PCR clones isolated with an SNB19 human glioblastoma single-stranded cDNA library as a template. The original PCR product containing the catalytic domain was isolated with degenerate oligonucleotides corresponding to conserved motifs in kinase domains. Homology with the murine ortholog NIK and the C. elegans ZC504.4 contig were observed, and primers from the N terminus of NIK were used in combination with primers derived from the original PCR isolate to generate the N-terminal half of the gene. A Smith-Waterman search of the public expressed sequence tag (EST) database identified a human EST whose open reading frame was related to the C. elegans gene (mig-15) and terminated in an identical Trp residue. A primer was designed to the 3′ end and used in a PCR assay with a primer derived from the 5′ end of the HGK cDNA.
Bioinformatic analysis of HGK gene.
A Blast search with our human HGK cDNA sequence was run against the Celera (version R26) and the public (release of 20 June 2001) genomic sequence databases to identify long contigs that covered the gene. Those contigs were then compared with EST databases from the public domain, Incyte (LifeSeqGold), and in-house sequence databases and with the NCBI nonredundant nucleotide database. Sequences that were identified by the Blast search were aligned to both the Celera and Human Genome Project genomic sequences with the Sim4 program (18) and visualized with an in-house genome browser.
Manual inspection was used to identify ESTs and cDNA sequences whose mapping to the genome showed alternative exon usage, and each sequence was aligned with a consensus long sequence (containing all alternative spliced modules) in order to confirm the differences between alternative splice forms. When there were sequence polymorphisms between genomic, cDNA, and EST data sources, the polymorphic regions were compared with Blast against all EST and public sources used and against raw genomic reads from Celera to determine which nucleotide was seen most frequently at the point of dispute.
Cloning HGK cDNA fragments from human tumor cell lines.
Total RNA was prepared from A549, H1299, and 293T cells with an RNeasy kit (Qiagen). In addition, Universal Reference RNA (containing a mixture of RNAs purified from 10 tumor cell lines) was purchased from Clontech. Reverse transcription reactions were performed with a Taqman kit (ABI); 2 μg of the cDNA was used in PCRs with Taq-Platinum polymerase (Invitrogen) with two primer sets to generate products between 1 and 3 kb. Products were size selected and cloned into pCR-II with a TA cloning kit (Invitrogen). Between 13 and 17 clones were fully sequenced and analyzed from each of the four RNA sources.
Expression analysis.
Northern blots were performed with standard techniques. The cells and tissues were described previously (5). Northern blots were prepared by running 10 μg of total RNA isolated from 60 human tumor lines and 21 adult tissues and 1 fetal tissue on a denaturing 1.2% formaldehyde-agarose gel and transferred to a nylon membrane. Filters were hybridized with a random-primed [32P]dCTP-labeled probe derived from a 404-bp NaeI-SmaI fragment from a coding region of HGK conserved in all splice variants.
The multiple tissue blot from which the tumor versus normal data were extracted was prepared as follows. cDNA libraries derived from 489 tissue or cell line sources were immobilized onto nylon membranes. The amplified cDNA library was manually arrayed onto nylon membranes with a 384-pin replicator. DNA spotting on arrays was quantified by treating nondenatured arrays with SYBR Green (1:100,000 in 50 mM Tris, pH 8.0) for 2 min. After washing with 50 mM Tris, pH 8.0, the fluorescent emission was quantified with a Phosphorimager (Molecular Dynamics). The amount of the arrayed DNA was used to normalize the hybridization signal. The same 32P-labeled HGK probe used for the Northern blots was hybridized to the array blot. Blots were washed and quantified with a Phosphorimager (Molecular Dynamics). The data were standardized for statistical analysis across the different tissue types with range standardization to convert measurements to a common scale starting at 0 and ending at 1. All statistical analysis was carried out with SYSTAT 9.01 (SPSS, Inc.).
Mutagenesis method and location of point mutations.
Mutations in the full-length cDNA were generated to change threonine 187 to glutamate (T187E), threonine 191 to glutamate (T191E), and lysine 54 to arginine (K54R) with a QuickChange PCR mutagenesis kit (Stratagene) and oligonucleotides T187E (CTGTGGGGCGGAGAAATGAATTCATAGGCACTCCC, GGGAGTGCCTATGAATTCATTTCTCCGCCCCACAG), T191E (GAAATACGTTCATAGGCGAGCCCTACTGGATGGC, GCCATCCAGTAGGGCTCGCCTATGAACGTATTTC), and K54R (GTTGGCAGCCATCAGAGTTATGGATGTCACTGAGG, CCTCAGTGACATCCATAACTCTGATGGCTGCCAAC). Wild-type and mutant forms of HGK were subcloned into pcDNA3myc(−)B vectors (Invitrogen), removing all 3′ untranslated sequence and generating a C-terminal Myc epitope tag.
In vitro HGK kinase assay.
RIE-1 cells expressing Myc-tagged HGK were lysed with 50 mM HEPES (pH 7.0)-150 mM NaCl,-1.5 mM MgCl2-1.0 mM EGTA-10% glycerol-1% Triton-10 mM pyrophosphate-1 mM Na3VO5-1 mM dithiothreitol-0.1 mg of 4-(2-aminoethyl)benzenesulfonyl fluoride per ml-2 to 10 μg each of aprotinin, pepstatin, leupeptin, and E-64 per ml as protease inhibitors. The lysates were spun at 14,000 rpm (15,800 × g) in an Eppendorf microcentrifuge for 10 min at 4°C. The supernatants were immunoprecipitated with protein G-Sepharose beads (Fisher) with an anti-Myc monoclonal antibody (mouse ascites fluid made with the 9E10 hybridoma cell line). The beads were washed twice with the lysis buffer and twice with kinase assay buffer (20 mM Tris [pH 7.4], 200 mM NaCl, 0.5 mM dithiothreitol, 10 mM MgCl2). The washed beads were resuspended in kinase assay buffer with 100 μM ATP and 0.5 μCi of [γ-32P]ATP per μl and incubated for 20 min in an Eppendorf Thermomixer at 30°C at maximal mixing velocity. Then 4 μg of myelin basic protein (Sigma) was added, and the reaction was allowed to continue for 10 min. The reaction was stopped by boiling in Laemmli sodium dodecyl sulfate (SDS) sample buffer for 10 min.
Focus-forming assays.
Focus formation assays were performed according to standard procedures (12). NIH 3T3 cells, obtained from Tony Hunter, were used at a low passage number, and the RIE-1 cells were obtained from Channing Der. NIH 3T3 cells were transfected with a total of 6 μg of DNA and 18 μl of Lipofectamine per 10-cm dish, and 0.3 to 0.5 μg of SV40sport-H-RasV12 along with 1 to 10 μg of pcDNA3-HGK mutants was transfected along with plasmid pBSK+ as carrier DNA. Empty vector (pcDNA3 from Invitrogen) was included as a negative control. RIE-1 cells were transfected with a total of 15 μg of DNA and 50 μl of Lipofectamine per 10-cm dish with the same plasmid ratios as for NIH 3T3. Foci were stained with 0.1% crystal violet in 20% methanol and counted from duplicate plates 12 to 13 days after transfection.
Generation of stable cell lines with retroviral infection.
HGK full-length cDNAs including a Myc epitope tag on the C terminus were subcloned into the pLXSN or pBABE retroviral vector. pLXSN-HGK and pBABE-HGK constructs were cotransfected into 293T cells with an Ecotropic retroviral packaging vector (29) with Lipofectamine. Viral supernatants were used to infect RIE-1 (Ecotropic) cells. Three successive rounds of infection were done approximately every 8 to 12 h to maximize the rate of infection. Pools of cells resistant to G418 (pLXSN) or puromycin (pBABE) were generated after 1 week of selection. These pools were used in some experiments, and individual clones were grown out of the pools as selected by screening for higher expression of the HGK proteins.
Monolayer cell growth rates.
Stable pools or clones of RIE-1 cells expressing HGK mutants were plated at low cell density in 12-well trays in medium with 10%, 1%, or 0.1% fetal bovine serum. Cell numbers were counted from two wells per cell line per day on a Coulter particle counter (Beckman-Coulter).
Soft agar assays.
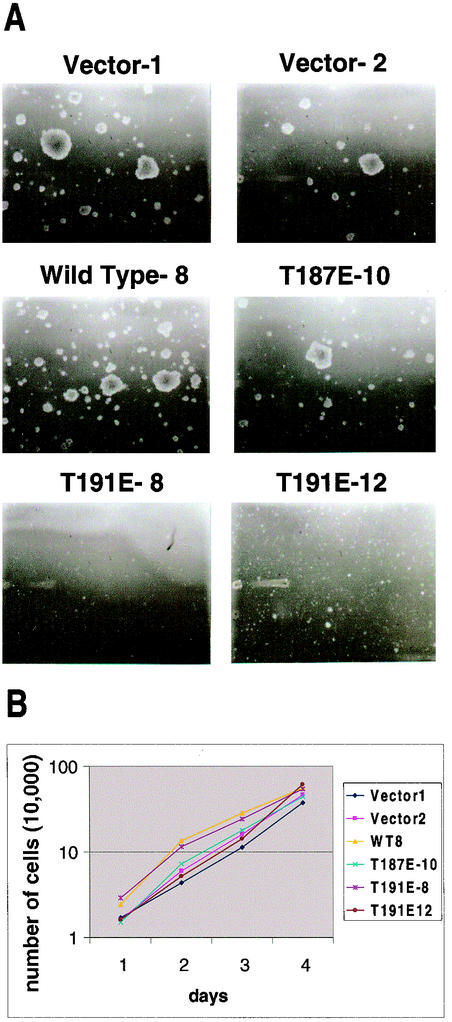
Stable RIE-1 cell lines were plated in 0.3% Bactoagar (Gibco-BRL)-Dulbecco's modified Eagle's medium (DMEM)-10% fetal calf serum (Gibco-BRL) on top of a 0.6% Bactoagar-DMEM base layer. For each clone, one six-well tray was plated with the following number of cells per well: 0, 1.0 × 103, 5.0 × 103, 1.0 × 104, 5.0 × 104, and 1.0 × 105. Assays were photographed after 4 weeks of growth. Photographs shown in Fig. 6 were of the wells with 1.0 × 104 cells/well.
FIG. 6.
HGK kinase activity is required for growth of RIE-1 cells in soft agar but not growth in monolayer. (A) Shown are photomicrographs (10× magnification) of colonies formed in soft agar from RIE-1 stable clones after 28 days of growth. (B) Shown is a semilogarithmic plot of growth curves generated for the same six RIE clones shown in A over a 4-day period, with duplicate wells counted every 24 h.
Cell spreading assay.
RIE-1 clones expressing wild-type HGK, activated mutant (T187E), inactive mutant (T191E), and empty vector as controls were removed from the dish with trypsin-EDTA and counted, and equal numbers of cells were replated on dishes coated with a 10-μg/ml solution of fibronectin (Chemicon FC014). Cells were then photographed at 10× magnification at various time points. The number of spread cells was quantified by counting the number of phase-dark cells.
Cell adhesion assay.
Standard-curve wells were prepared by adding 50 μl of polylysine (Sigma; 100 μg/ml) per well to wells, incubating at room temperature for 5 min, washing with phosphate-buffered saline (PBS), and air drying. For experimental wells, fibronectin (Chemicon, FC104) was diluted in PBS to the appropriate working concentration, added at 100 μl/well to 96-well trays, and incubated for 60 min. Fibronectin solution was removed, and the wells were blocked with bovine serum albumin (Sigma, tissue culture grade) for 30 min and washed with PBS. RIE-1 cells were removed from plates with trypsin-EDTA solution (Gibco-BRL), counted, and resuspended in 0.1% bovine serum albumin at 5 × 105 cells/ml. Then 50 μl of cells was added to each experimental well, and cells were allowed to adhere for 30 and 60 min. The medium was then removed from all wells, and nonadherent cells were gently washed away with PBS. Then 5% glutaraldehyde was added for 20 min at room temperature. Wells were washed with PBS. Crystal violet solution (0.1% crystal violet dye in 20% methanol) was added to each well and incubated for 60 min on a platform shaker. Stained cells were first photographed and then solubilized in 100 μl of 10% acetic acid for 1 to 2 h, and the absorbance at 550 nm was measured.
Fluorescence-activated cell sorting analysis.
Cells were detached from dishes with trypsin-EDTA, counted, and resuspended at 2 × 107 cells/ml in PBS-1% fetal bovine serum. Anti-integrin receptor antibodies (Pharmingen) 553350 (α5), 555003 (β1), 553715 (β1), 553343 (β3), 550344, and 553957 (isotype controls) were diluted to 20 μg/ml, mixed with cells in a 1:1 volume ratio, and incubated in the dark for 20 min at 4°C. Cells were washed twice with PBS-1% fetal bovine serum (wash buffer). Then 100 μl of secondary antibody (Pharmingen), fluorescein isothiocyanate-conjugated anti-rat immunoglobulin G2a 553896 or fluorescein isothiocyanate-conjugated anti-hamster immunoglobulin M 554036) at 10 μg/ml in PBS-1% fetal bovine serum was added to the cells and incubated for 20 min in the dark at 4°C. Cells were washed twice and resuspended in 500 μl of wash buffer. Cells were analyzed for relative FL1 intensity for cells stained with different primary antibodies and compared with a secondary-antibody staining only as well as with a primary antibody isotype control with a Becton-Dickinson fluorescence-activated cell sorter machine and the CellQuest program. The mean fluorescence intensity values shown in Fig. 8 represent the mean fluorescence intensity values measured for the specific integrin antibody minus the mean fluorescence intensity value for the corresponding isotype control antibody.
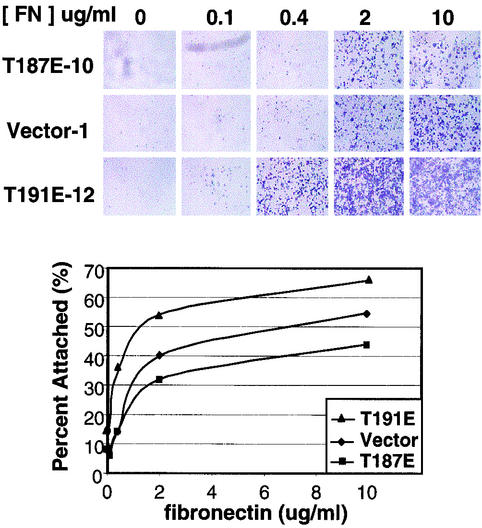
FIG. 8.
HGK kinase activity interferes with cell attachment. RIE-1 stable clones expressing active and inactive alleles of HGK as well as vector controls were detached with trypsin and plated on various concentrations of fibronectin (FN). After 60 min, unattached cells were washed away, and the remaining cells were stained with crystal violet. (A) Photomicrographs (2× magnification) show the presence of remaining cells at different concentrations of fibronectin. (B) By comparison with a standard curve of cells plated on polylysine, the relative amount of cells was quantified by measuring the amount of solubilized dye by optical density. The graph shows the relative optical density at 550 nm as a function of fibronectin concentration for the different RIE-1 clones.
Three-dimensional invasion-tubulogenesis assay.
Matrigel (Matrigel basement membrane matrix, catalog no. 40234, Becton Dickinson Labware) was thawed on ice. Then 50 μl of Matrigel was added per well in a 96-well plate and incubated for 20 min at 37°C to induce gel formation. RIE-1 cells were suspended at 2 × 104 cell/ml and mixed 1:1 with Matrigel on ice. Then 0.1 ml of the mixture was added per well. The next day, 0.1 ml of DMEM with and without 80 ng/ml of hepatocyte growth factor (HGF) (R&D 294-hg-005) was added on top of each Matrigel cell plug. Fresh medium with and without HGF was added every 3 days. Photographs were taken after 6 days in culture.
Boyden chamber (directional) invasion assay.
The Boyden chamber (directional) invasion assay was done with Biocoat Matrigel Invasion 24-well chambers (Becton Dickinson). Inserts were first rehydrated at room temperature. With trypsin-EDTA, cell suspensions were prepared in DMEM-1% bovine serum albumin at 105 cells/ml. HGF was added to the bottom chamber at 50 to 100 ng/ml. Then 0.5 ml of cell suspension was added to the inserts, and the chambers were incubated for 48 h at 37°C in a 5% CO2 atmosphere. After incubation, the noninvading cells were removed from the upper surface of the membrane with a moistened Q-tip swab. The inserts were transferred to crystal violet (0.2% crystal violet in 10% buffered Formalin) stain solution (1 min) and then washed with water. Inserts were allowed to air dry before being photographed.
Cell stimulation with HGF and analysis of cellular phosphoproteins.
RIE cells were grown until confluent and then serum starved overnight. After stimulation with HGF (50 ng/ml; 294-HG, R&D Systems), cells were lysed with HNTG (50 mM HEPES [pH 7.4], 150 mM NaCl, 10% glycerol, 0.5% Triton X-100,1 mM NaVO4, and protease inhibitors [1.4 μM E-64, 10 μM bestatin, 1 μM leupeptin, 0.3 μM aprotinin, 1 μM pepstatin, and 1 mM phenylmethylsulfonyl fluoride]). For determination of HGF-stimulated phosphorylation of Met, the Met protein was isolated by immunoprecipitation with anti-Met antibody (SC-162; Santa Cruz Biotechnology) and analyzed by Western blotting with antiphosphotyrosine antibody (SC-7020B; Santa Cruz Biotechnology). For analysis of phospho-STAT3, aliquots of whole-cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting with anti-phospho-STAT3 antibodies (anti-phospho-STAT3-Y705, 06658, Upstate Biotechnology; anti-phospho-STAT3-S727, SC-8001, anti-phospho-ERK, SC-7383, Santa Cruz Biotechnology; anti-phospho-AKT, 9271, Cell Signaling).
To ensure equal protein loading, membranes were stripped and reprobed with antibodies against STAT3 (anti-STAT3, 7179, anti-ERK1, SC93, and anti-ERK2, SC-154, Santa Cruz Biotechnology) or, in one case (AKT), a parallel gel was run followed by Western blotting with antibodies against the respective proteins (anti-AKT, 9272; Cell Signaling). For coimmunoprecipitation of HGK and STAT3, confluent 293T cells transfected with HGK or RIE stable clones expressing wild-type HGK with or without stimulation with HGF were lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.2], 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS, 1 mM NaVO4, and protease inhibitors [1.4 μM E-64, 10 μM bestatin, 1 μM leupeptin, 0.3 μM aprotinin, 1 μM pepstatin, and 1 mM phenylmethylsulfonyl fluoride]). Soluble proteins were subjected to immunoprecipitation with anti-STAT3 antibody 9132 (Cell Signaling) for 2 h at 4°C, and the immunoprecipitates were analyzed by SDS-PAGE followed by Western blotting with anti-Myc tag antibodies 9E10 and anti-STAT-3.
Nucleotide sequence accession number.
The GenBank accession number for the novel HGK splice variant is AY212247.
RESULTS
Cloning of HGK gene and identification of a family of splice variants.
We have identified a novel HGK (HPK-like/GCK-like kinase) gene product from a human SNB19 glioblastoma cell line library. The cDNA isolated was 3,798 bp long, encoding a protein of 1,239 amino acids. No polyadenylation signal was found in the original clone. This HGK gene product contained multiple splice variations compared with published human HGK forms (26, 51) and the mouse ortholog, NIK (43).
In view of these differences, we performed a full bioinformatic analysis of the HGK gene. A family of HGK transcripts were identified by comparison of the sequence of our cloned HGK cDNA with the genomic sequence as well as with public and Incyte ESTs and public cDNAs mapping to that locus. The HGK gene is located on chromosome 2 at 2q11.2 and contains 33 exons, including nine alternatively spliced modules (exons or partial exons created by use of alternative splice sites; Fig. 1 and Table 1) within the coding region. All modules encode a whole number of codons, allowing any combination of modules to be used while maintaining the open reading frame. In addition, examination of HGK ESTs revealed a long 3′ untranslated region of 3,442 additional base pairs. Within this region are three alternative polyadenylation sites that would give transcripts of ≈5.0, 6.3, and 8.1 kb.
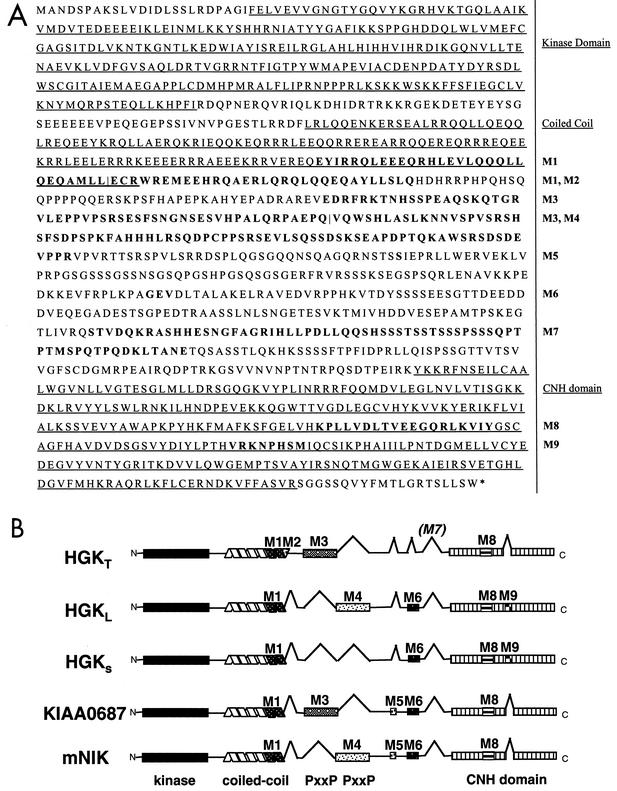
FIG. 1.
(A) HGK amino acid sequence: predicted maximal HGK protein containing all possible alternatively spliced modules. The kinase domain, the coiled-coil domain, and the CNH domain are underlined. Alternatively spliced modules M1 to M9 are shown in bold and labeled to the right of the sequence. For adjacent modules M1/M2 and M3/M4, a vertical line defines their boundaries. (B) Schematic illustration of the domain structure of known HGK splice variants compared with mouse NIK. Alternatively spliced modules are indicated with inverted V's when absent and alternative patterning where present. The HGK gene product that we cloned from a tumor cell line (HGKT) contains alternative modules M1, M2, M3, and M8. The two HGK cDNAs from a human macrophage library contain M1, M6, M8, and M9 (HGKS; short version) and M1, M4, M6, M8, and M9 (HGKL; long version). KIAA0687 isolated from brain has modules M1, M3, M5, M6, and M8. The mouse NIK clone contains M1, M4, M5, M6, and M8.
TABLE 1.
Alternative sequence modules in HGK
| Mod- ule | Frame | Length (nucleotides/ amino acids) | Position (amino acid no. in max form) | Origin | No. of EST database sequences with/without this module | No. of tumor cell line cDNAs with/without this module | Comments |
|---|---|---|---|---|---|---|---|
| M1 | 1 | 87/29 | 428 | Exon | 41/1 | 27/14 | Encodes C-terminal extension of coiled-coil domain; similar module found in TNIK; adjacent to M2 |
| M2 | 1 | 93/31 | 495 | Alternative splice acceptor | 14/28 | 12/29 | Adjacent to MI |
| M3 | 1 | 162/54 | 570 | Exon | 14/33 | 7/50 | Similar module found in TNIK; contains 2 PxxP motifs; adjacent to M4 |
| M4 | 1 | 231/77 | 623 | Exon | 12/35 | 1/56 | Contains 2 PxxP motifs; adjacent to M3 |
| M5 | 1 | 3/1 | 736 | Alternative splice acceptor | 23/9 | 39/18 | Adds a serine residue |
| M6 | 1 | 9/3 | 819 | Exon | 20/20 | 10/46 | Adds 3 residues |
| M7 | 1 | 192/64 | 921 | Exon | 21/16 | 9/47 | Contains a serine-rich sequence |
| M8 | 1 | 57/19 | 1215 | Intron readthrough | 34/1 | 24/0 | Encodes part of CNH domain; similar sequence seen in other human GCK type IV kinases |
| M9 | 1 | 24/8 | 1258 | Alternative splice donor | 11/17 | 3/20 | Encodes part of CNH domain; similar sequence not seen in other CNH domains |
Figure 1A contains the predicted maximal HGK amino acid sequence from all 33 exons. The alternatively spliced modules, M1 to M9, are shown in bold. EST evidence indicated that each alternative module was present in multiple but not all transcripts (Table 1). All HGK splice forms contained the N-terminal kinase domain, a coiled-coil domain, and a C-terminal citron homology (CNH) domain. The close HGK family member TNIK also has three alternatively spliced exons, two of which are equivalent to modules M1 and M3 (22).
The cDNAs encoding HGK isoforms isolated from different tissue sources each contained a distinct subset of modules. A schematic comparison of the domain structures of the four cloned human HGK isoforms as well as the mouse ortholog NIK is shown in Fig. 1B. The human HGK isoforms include our tumor-derived HGK isoform (HGKT), the original HGK clones isolated from macrophages (HGKS, short version; HGKL, long version) (51), as well as another splice variant cloned from brain, published with the reference number KIAA0687 (26).
There was no alternative splicing in the kinase domain, but the coiled-coil region can be alternatively spliced into short and long forms. The HGKT form contains both M1 and M2 and therefore has a longer coiled coil than the other forms. The M3 and M4 modules show sequence similarity to each other and contain PxxP motifs that can interact with SH3 domain-containing proteins. Our tumor-derived HGK (HGKT) contains M3, while the short form (HGKS) isolated from human macrophage lacks M3/M4, and the long form (HGKL) and NIK contain M4. M5 and M6 are small exons encoding one and three amino acids, respectively. M7 is very rich in serine and is not present in any of the isolated full-length HGK clones. The CNH domain contains two alternatively spliced modules, of which the M8 but not the M9 module is conserved in CNH domains found in other protein families.
We extended our study to survey which isoforms of HGK are expressed in tumor cells. With reverse transcription-PCR, we cloned multiple cDNA fragments of HGK from RNA isolated from several tumor cell lines, including A549 and H1299 lung carcinoma cells, 293T transformed kidney cells, as well as a mixed pool of RNA from 10 tumor cell line sources (Clontech's Universal Reference RNA). Table 1 shows the prevalence of each module in cDNAs isolated from these tumor cell lines; the results from the four tumor cell line sources were pooled and are shown together, as the distribution of modules from each was similar. We identified at least five different splice isoforms from each tumor cell line source, indicating that the tumor cells possessed multiple forms of HGK. The most striking difference observed when the tumor cell line data was compared with EST data was that the tumor cell clones largely lacked the PxxP-containing module M4. Only 2% of the tumor-derived clones contained M4. Similarly, the M3 PxxP motif was absent in most clones. In addition, the tumor cell lines also generally lacked the serine-rich M7 module and the M9 module in the CNH domain.
HGK expression analysis.
A ∼400-bp fragment from a conserved region of HGK present in all splice variants was used to probe Northern blots containing RNA purified from various normal human tissues as well as from 60 cell lines from the National Cancer Institute (NCI) tumor panel (Fig. 2A). Transcripts of ∼4.7 and ∼6.5 kb were detected, comparable in size to two of the transcripts predicted from the analysis of ESTs. In normal tissue, HGK mRNA was expressed at readily detectable levels only in tissue samples from brain and testis. In contrast, HGK mRNA was expressed at high levels in 40 of the 60 tumor cell lines derived from a variety of tissues (Fig. 2A).
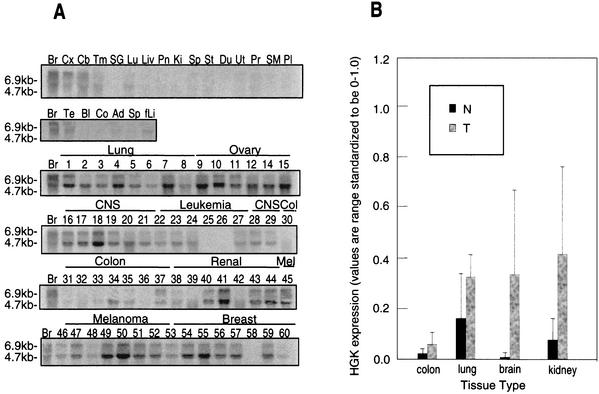
FIG. 2.
(A) HGK mRNA is overexpressed in human tumor cell lines. Total RNA was purified from various human tissues (top two rows) as well as from human tumor cell lines from the standard tumor panel from the National Cancer Institute. Abbreviations for normal tissues: (first row) Br, brain; Cx, cortex; Cb, cerebellum; Tm, thymus; SG, salivary gland; Lu, lung; Liv, liver; Pn, pancreas; Ki, kidney; Sp, spleen; St, stomach; Du, duodenum; Ut, uterus; Pr, prostate; SM, skeletal muscle; Pl, placenta; (second row) Br, brain; Te, testis; Bl, bladder; Co, colon; Ad, adipose tissue; fLi, fetal liver. Numbering for the NCI tumor panel: 1, HOP-92; 2, EKVXl 3, NCI-H23; 4, NCI-H226; 5, NCI-H322; 6, NCI-H460; 7, NCI-H522; 8, A549; 9, HOP-62; 10, OVCAR-3; 11, OVCAR-4; 12, OVCAR-5; 14, IGROV1; 15, SK-OV-3; 16, SNB-19; 17, SNB-75; 18, U251; 19, SF-288; 20, SF-295; 21, SF-539; 22, CCRF-CEM; 23, K-562; 24, MOLT-4; 25, HL-60; 26, RPMI-8226; 27, SR; 28, DU-145; 29, PC-3; 30, HT-29; 31, HCC-2998l; 32, HCT116; 33, SW620; 34, COLO-205; 35, HCT-15; 36, M-12; 37, UO-31; 38, SN12C; 39, A498; 40, Caki-1; 41, RXF-393; 42, ACHN; 43, 786-0; 44, TK-10; 45, LOXIMVI; 46, Malme-3 M; 47, SK-MEL-2; 48, SK-MEL-5; 49, SK-MEL-28; 50, UACC-62; 51, UACC-257; 52, M14; 53, MCF-7; 54, MCF-7/ADR; 55, Hs578T; 56, MDA-MB-231; 57, MDA-MB-431; 58, MDA-N; 59, BT-549, and 60, T-47D. (B) Quantification of HGK in tumor versus normal cells from selected individual tissues. The bar graph shows the relative signal from PhosphorImager quantification of dot blot arrays from tumor cell lines and normal tissues. From left to right: colon tumor-normal, 2.7-fold, P = 0.210; lung tumor-normal, 2.0, P = 0.034; brain tumor-normal, 46.3, P = 0.007; kidney tumor-normal, 5.5, P = 0.076. P values were calculated with the Mann-Whitney test.
The same probe was also hybridized to an array of cDNAs amplified from a broader array of tissue and tumor cell line sources. Quantification of the probed array by phosphorimaging revealed that across all tumor-derived sources (cell lines and tissues) where a normal tissue comparison was available, HGK was increased in expression by an average of 7.7-fold in tumor versus normal tissues. This difference was statistically significant at P = 0.000001 (Mann-Whitney test). Quantification of the same data broken down by individual tumor type is shown in Fig. 2B. We observed the highest upregulation in glioblastoma samples (46.3-fold relative to expression in normal brain).
Analysis of HGK mutants: effects on growth and transformation.
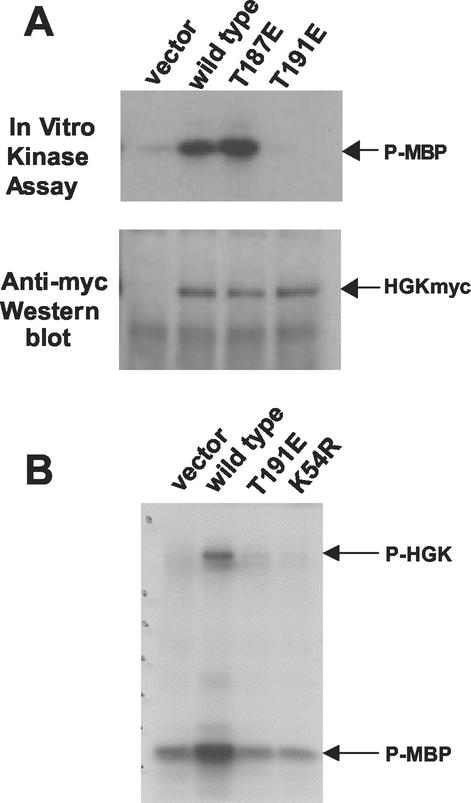
Overexpression of HGK mRNA in a variety of tumor cells suggests that HGK may play a role in tumorigenesis. In order to test for a causative role for this kinase in cell transformation, we constructed mutant forms of HGKT in mammalian expression vectors to test in tissue culture cells. Based on homology with other related kinases, threonine 187 and threonine 191 in the HGK activation loop were predicted to be potential regulatory phosphorylation sites. We mutated each of the two residues to glutamic acid to create activating alleles of HGK. We also mutated the catalytic lysine to an arginine (K54R). In vitro kinase assays performed on extracts from rat intestinal epithelial cells (RIE-1) infected with retroviruses expressing each of the HGK mutants used in subsequent biological experiments are shown in Fig. 3. HGK wild-type kinase showed strong activity, while mutation of K54 to R inactivated the kinase, as expected. Comparison of the kinase activity of wild-type HGK and T187E showed that the latter had a slight increase in in vitro catalytic activity. Mutating T191 to E, in contrast, abolished the kinase activity. This T191E mutant was used in addition to K54R as a dominant-negative allele in subsequent experiments.
FIG. 3.
Kinase activity of HGK mutants in RIE-1 cells. HGK wild-type and mutant proteins were immunoprecipitated from stable pools of RIE-1 cells infected with HGK retroviruses. (A) Comparison of the T187E and T191E activation loop mutations with wild-type kinase. The top panel shows an autoradiogram of samples analyzed by SDS-PAGE, showing the relative amount of 32P incorporated into myelin basic protein (MBP) after incubation of the immunoprecipitate and [γ-32P]ATP for 20 min at 30οC. The bottom panel shows the relative amount of HGK proteins in the immunoprecipitate. (B) Comparison of the T191E and K54R catalytically inactive HGK mutants with wild-type HGK. An autoradiogram shows the relative amount of 32P incorporation both into HGK protein (autophosphorylation) and into added MBP in the immunoprecipitate.
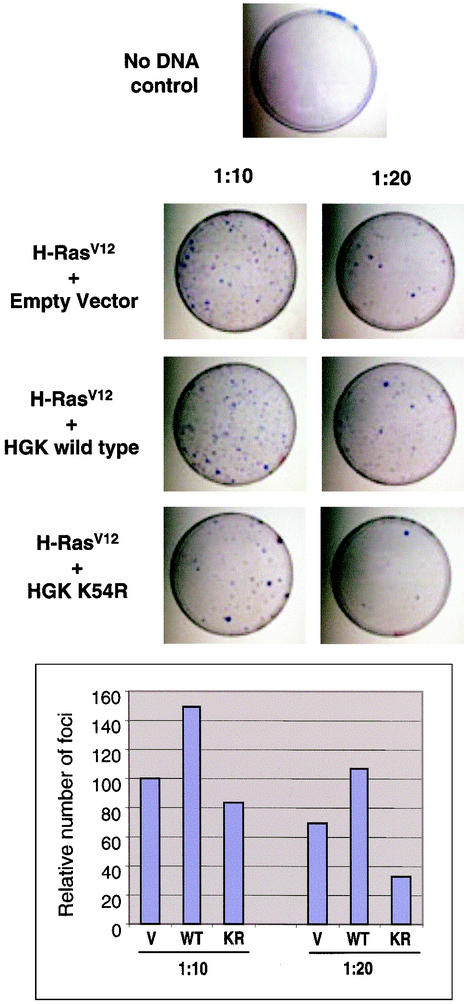
To test for a possible role for HGK in focus formation, we coexpressed different alleles of HGK in NIH 3T3 cells together with an activated allele of H-Ras (valine 12). While expression of wild-type and T187E mutant HGKs did not induce focus formation by itself (data not shown), coexpression of the inactive HGK K54R mutant with H-RasV12 reduced the frequency of Ras-induced foci in a dose-dependent fashion compared with coexpression of wild-type HGK (Fig. 4). When the number of foci was tabulated, the K54R mutant was found to reduce focus formation rates by approximately 50% at a DNA ratio of 1:20 with respect to H-RasV12. In addition, coexpression of wild-type HGK reproducibly increased the number of foci generated by H-RasV12, indicating some level of cooperation. A similar rate of inhibition of focus formation was observed in RIE-1 cells cotransfected with H-RasV12 and either inactive form of HGK (K54R or T191E; data not shown). These data indicate that HGK kinase activity is important for transformation by Ras in this assay.
FIG. 4.
Dominant negative mutant HGK can suppress Ras-induced focus formation in NIH 3T3 cells. (Top) Images of tissue culture plates stained with crystal violet to visualize the foci are shown from a representative experiment with NIH 3T3 cells. The relative number of foci per plate generated by H-RasV12 in the presence of a 10- or 20-fold molar excess of plasmid encoding HGK wild-type, HGK K54R inactive mutant kinase, or vector control is shown. The panel on the top shows background levels of foci generated by transfection of a vector control (no H-RasV12). (Bottom) The bar graph shows the relative number of foci per plate (setting the number of foci in the Ras/vector control 1:10 at 100%). The data represent an average of two experiments.
To further elucidate the role of HGK in cell growth and transformation, we compared both monolayer and anchorage-independent growth of stable clones of RIE-1 cells expressing various alleles of HGK isolated from the retrovirally infected pools. Relative expression levels of Myc-tagged HGK in different clones are shown in Fig. 5. RIE-1 stable clones were suspended in soft agar in the presence of 10% serum. Clones expressing wild-type HGK and the kinase-active mutant (T187E) formed small colonies similar to vector control clones, indicating that HGK activity alone was not sufficient for full transformation of this cell line. However, we noticed that expression of kinase-inactive HGK (T191E) completely abrogated the ability of these cells to grow in soft agar (Fig. 6A, bottom two panels).
FIG. 5.
HGK expression levels in stable clones of RIE-1 cells. (A)Western blot of SDS-6% PAGE on which HGK-Myc protein immunoprecipitated with anti-Myc antiserum was separated is shown. Vector clones contain an empty pLXSN virus. Active clones contain the T187E HGK mutant virus, and inactive clones contain the T191E HGK virus.
To test whether HGK expression affected cell growth in monolayer culture, the relative cell doubling time for RIE-1 cells expressing mutant forms of HGK was assessed (Fig. 6B). All clones showed similar growth rates in monolayer cultures in both 1% serum (shown) and 10% serum (data not shown). These data suggest that HGK kinase activity is not directly required for mitogenic signaling from serum growth factors when cells are attached yet functions in anchorage-independent growth.
HGK mutants: effects on cell adhesion and integrin receptor levels.
The ability of inactive HGK to suppress the growth of RIE-1 cells in soft agar suggested that this kinase may function in pathways that regulate the anchorage dependence of normal cell growth. The anchorage dependence of growth in adherent cells is mediated by essential signal transduction pathways downstream of activated integrin receptors (19, 20). We therefore examined the effects of HGK on integrin receptor function.
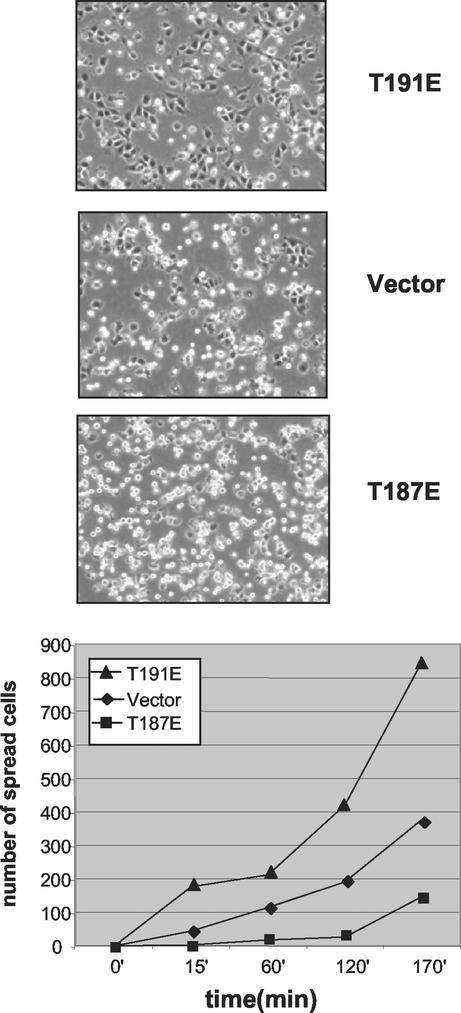
RIE-1 cells expressing wild-type, active, or inactive HGK were tested for the ability to attach and spread on the integrin ligand fibronectin. The rate of cell spreading was measured as a function of time. Expression of kinase-active (T187E) HGK decreased the rate at which cells spread compared with vector control clones (Fig. 7). Conversely, expression of the inactive (T191E) HGK significantly increased the rate of cell spreading. This spreading phenotype was observed with multiple clones expressing each mutant form of HGK.
FIG. 7.
Role for HGK in cell spreading. RIE-1 stable clones expressing active and inactive alleles of HGK as well as vector controls were detached with trypsin and plated on fibronectin. Photomicrographs (10× magnification) were taken at various time points thereafter. The graph shows the relative number of spread (phase-dark) cells counted in three separate fields for each clone as a function of time. Representative photographs from the 170-min time point are shown on top.
Cell spreading is a function of both integrin receptor engagement and changes in cytoskeleton. To assess whether HGK has effects on integrin receptor-ligand interaction, we tested these stable cell lines in cell attachment assays. RIE-1 cells were plated on different concentrations of fibronectin, and percent attachment as a function of fibronectin levels was calculated based on a polylysine standard curve to eliminate background due to differences in cell number and plating efficiency. Compared with the vector control, the inactive-kinase-expressing clones demonstrated increased rates of adhesion, while the active-kinase-expressing clones (T187E) showed a reduced rate of adhesion (Fig. 8). Similar results were obtained with multiple clones. Therefore, HGK appears to negatively regulate RIE-1 cell adhesion to fibronectin.
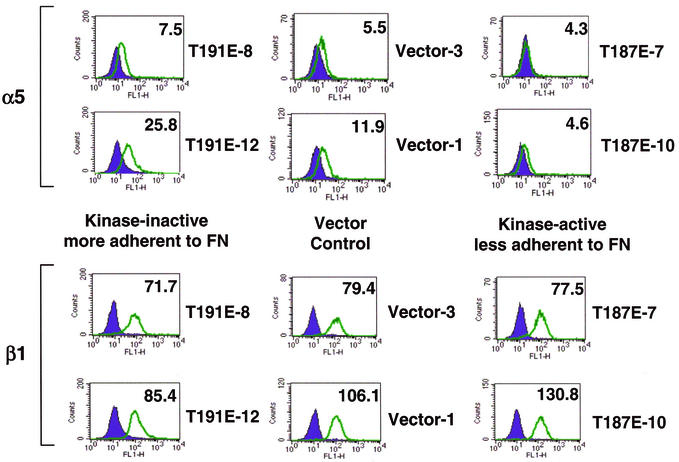
Major fibronectin receptors in epithelial cells include α5β1 and αvβ3. Function-blocking integrin antibodies were used to show that RIE-1 cell adhesion to fibronectin is mediated, at least in part, by α5β1 receptors. Adhesion of these cells to fibronectin was completely blocked by treatment of cells with anti-β1 antibodies and partially blocked by treatment with anti-α5 antibodies, while anti-β3 antibodies had no effect (data not shown). The level of α5 and β1 integrin receptor subunits on the cell surface of HGK-expressing RIE-1 clones was measured by fluorescence-activated cell sorting. While there was no effect on β1 levels, the α5 levels were somewhat higher in the inactive-kinase-expressing clones and lower in the active-kinase-expressing clones relative to vector controls (Fig. 9).
FIG. 9.
HGK mutant expression affects cell surface α5 integrin subunit levels. By fluorescence-activated cell sorting analysis, the level of cell surface integrin receptors was compared for different RIE-1 stable clones. Histograms show the relative fluorescence intensity of cells stained with fluorescein isothiocyanate-labeled integrin receptor antibodies. The level of specific integrin receptor (α5 subunit on top and β1 subunit on the bottom) are shown as open green peaks. Isotype controls are shown as solid purple peaks. The relative shift of the green peak to the right relative to the isotype control indicates the level of that particular receptor. Numbers in the upper right corner of each histogram are the mean fluorescence intensity (normalized to the relevant isotype control).
HGK effect on cell invasion and morphogenesis.
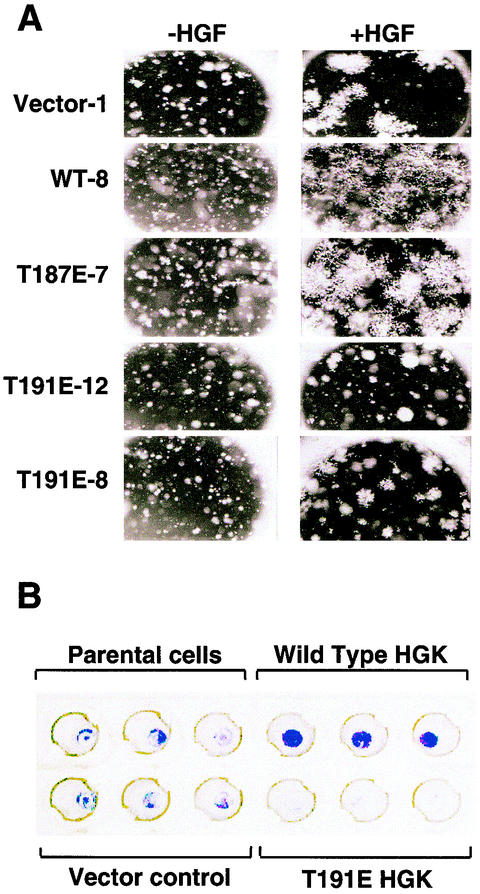
RIE-1 cells were used to test the effects of HGK mutant expression on cell growth, morphogenesis, and invasion in Matrigel. In a three-dimensional invasion or tubulogenesis assay (2, 34), cells were suspended in Matrigel and cultured in the presence and absence of hepatocyte growth factor (HGF). As was seen previously with kidney epithelial cells, HGF was both mitogenic and morphogenic for RIE-1 cells in this assay, as demonstrated by inducing them to grow and invade the surrounding Matrigel, forming large lattice-like networks (Fig. 10A, compare vector control with and without HGF). In the absence of HGF, RIE-1 cells survived in the Matrigel but showed a slower growth rate. Clones expressing either the wild-type or the T187E mutant of HGK gave larger networks of invading cells than the vector controls within the same time frame. However, cells expressing the inactive T191E form of HGK were almost fully blocked in their invasive response, forming spheroid colonies. In contrast to their inability to invade, the T191E mutant cells appeared to be able to respond mitogenically to HGF and proliferate (Fig. 10A, compare T191E clones with and without HGF). Similar effects were observed when cells expressing the mutant allele K54R were tested (data not shown). These data suggest that blocking HGK kinase-dependent signaling specifically blocked the morphogenic response of the cells to HGF.
FIG. 10.
(A) Role for HGK in RIE cell morphogenesis. HGK kinase expression affects RIE-1 cells in a tubulogenesis assay. Shown are photomicrographs (3.2× magnification) of colonies formed in Matrigel after 6 days of growth. Each clone is shown with and without added HGF. Vector control is an RIE-1 clone with pLXSN alone. WT-8 is a clone expressing wild-type HGK. T187E-7 is a clone expressing the active HGK mutant; T191E-12 and T191E-8 are two clones expressing the inactive HGK mutant. (B) Role for HGK in RIE cell invasion. Boyden chambers in which the cells of the lower membrane are fixed and stained with crystal violet are shown. The amount of staining is proportional to the number of cells that successfully invaded through the Matrigel plug to colonize the lower membrane after stimulation with 50 ng of human HGF per ml for 4 days. Triplicate wells are shown from an experiment with RIE-1 stable pools expressing vector alone, wild-type, or inactive (T191E) HGK.
Next we asked whether HGK plays a role in directional invasion (chemotaxis toward HGF). To address this question, RIE-1 cells were seeded in a Boyden chamber coated with Matrigel, and the rate of invasion was measured. RIE-1 stable pools overexpressing wild-type HGK showed significantly increased migration through the Matrigel towards HGF compared with either the vector control or parental cells (Fig. 10B). In contrast, the inactive-HGK-expressing pools were significantly retarded in invasive response.
Effects of HGK mutants on HGF intracellular signaling.
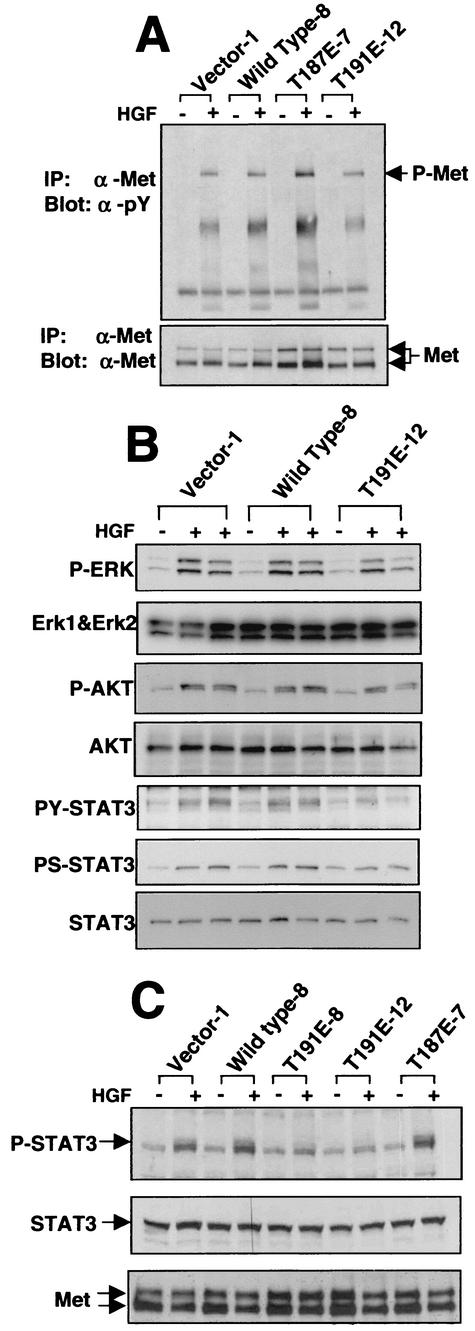
In view of the ability of the HGK mutants to interfere with HGF-induced morphogenic and invasive responses, effects on immediate downstream signaling from the HGF receptor, Met, were examined in RIE-1 clones expressing HGK mutants. The level of Met receptor expression as well as the level of receptor phosphorylation in response to HGF stimulation was not significantly affected (Fig. 11A and C). Next we examined three of the major signaling pathways that have been reported to function in HGF-induced morphogenesis, the mitogen-activated protein kinase pathway, the phosphatidylinositol 3-kinase/AKT pathway, and STAT3 activation (11, 41, 42).
FIG. 11.
HGK mutant expression affects HGF-induced STAT3 phosphorylation. Western blots of cell lysates prepared from starved RIE-1 cells with and without stimulation with 100 ng of HGF per ml are shown. (A) Western blots of anti-Met receptor immunoprecipitates (IP) probed with antiphosphotyrosine (α-pY) antibody in the top panel and anti-Met receptor antibody in the bottom panel are shown. (B) A panel of Western blots prepared from total cell lysates stimulated for 0, 30, and 60 min with HGF are shown. From top to bottom: anti-phospho-mitogen-activated protein kinase, anti-ERK1 plus anti-ERK2, anti-phospho-AKT, anti-AKT anti-phospho-STAT3 (Y705), anti-phospho-STAT3 (S727), and anti-STAT3. (C) Total cell lysates from unstimulated cells and cells 30 min after HGF stimulation. The top panel was probed with an anti-phospho-Y705 STAT3-specific antibody; the middle panel was probed with an anti-STAT3 antibody; the bottom panel shows anti-Met receptor immunoprecipitations from the same lysates, probed with an anti-Met antibody. Data from RIE-1 clones expressing empty vector (vector), wild-type HGK, T187E HGK (active mutant), and T191E HGK (inactive mutant) are shown.
As shown in Fig. 11B, we observed little or no effect on both ERK1/ERK2 and AKT activation with HGF stimulation. In contrast, we noted an attenuation of STAT3 phosphorylation on both of the regulatory residues, tyrosine 705 and serine 727, in the cells expressing the kinase-inactive HGK. To rule out the possibility that this effect was due to clonal variation, we examined HGF stimulation of STAT3 phosphorylation in two independent clones expressing inactive HGK (T191E-8 and T191E-12). The panel shown in Fig. 11C illustrates that STAT3 tyrosine phosphorylation was inhibited in both of these clones. In addition, a small increase in STAT3 Y705 phosphorylation was observed in the clones expressing the active forms (wild type or T187E) of HGK in some experiments (Fig. 11C). We also observed STAT3 serine 727 phosphorylation to be inhibited in both kinase-inactive clones (data not shown). We confirmed the protein band observed as STAT3 by immunoprecipitation with a STAT3 antibody, followed by probing with the phospho-STAT3 antibodies (data not shown). These data suggest that HGK functions in the activation of STAT3 downstream of the stimulated Met receptor. No increase in basal STAT3 phosphorylation was observed in the RIE clones expressing active HGK forms or in 293T cells transiently transfected with wild-type HGK, indicating that this kinase cannot activate STAT3 on its own but rather modulated the stimulation of STAT3 phosphorylation with HGF stimulation (Fig. 11 and data not shown).
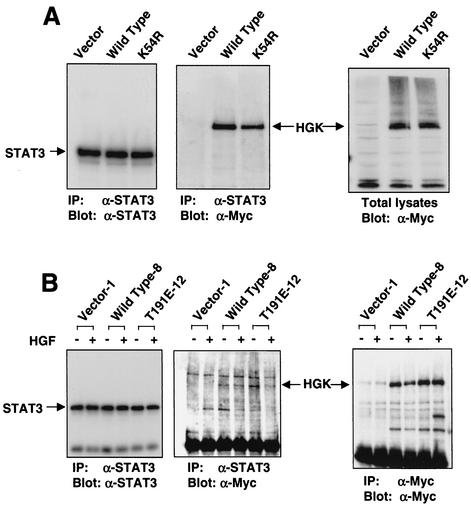
To further investigate the involvement of HGK in STAT3 activation, we designed experiments to test whether HGK and STAT3 were associated in cells. As shown in Fig. 12A, we transfected 293T cells with wild-type and kinase-inactive HGK, followed by immunoprecipitation of proteins with anti-STAT3. We observed that the transfected HGK coimmunoprecipitated with endogenous STAT3. The association between STAT3 and HGK was not dependent on HGK kinase activity, as the wild-type and kinase-inactive proteins coimmunoprecipitated with similar efficiency. We next preformed a similar coimmunoprecipitation from the RIE stable clones with and without HGF stimulation (Fig. 12B). In this cell system as well, we observed that HGK was retained in the STAT3 immunoprecipitation. These data suggest that HGK and STAT3 are part of the same signaling complex and suggest a direct involvement of HGK in modulation of STAT3 phosphorylation in response to HGF.
FIG. 12.
HGK kinase coimmunoprecipitated with STAT3. (A) 293T cells were transfected with Myc-tagged wild-type K54R (inactive) HGK and the vector control. Lysates were immunoprecipitated with anti-STAT3, followed by analysis on Western blots probed with anti-STAT3 (left) and anti-Myc (middle). Total lysates were analyzed with anti-Myc in parallel. (B) RIE-1 clones expressing wild-type and kinase-inactive (T191E-12) HGK were stimulated with HGF for 0 or 30 min Lysates were immunoprecipitated with both anti-Myc and anti-STAT3. The middle panel shows the STAT3 immunoprecipitate probed with anti-Myc. The left panel shows the same immunoprecipitate probed with anti-STAT3, and the right panel is an anti-Myc immunoprecipitate probed with anti-Myc for HGK. The HGK and STAT3 protein bands are indicated with arrows.
DISCUSSION
We identified a novel form of HGK in a human glioblastoma cell line cDNA library. Compared with other forms of HGK isolated from macrophage and brain, and with the mouse ortholog NIK, this form showed multiple different splice variations. We analyzed the overall genomic structure of the HGK gene and found it to include 33 exons, with nine regions (modules) of alternative splicing. EST data suggest that all of these alternative modules are present in expressed proteins, resulting in a complex family of HGK isoforms. While many genes are alternatively spliced, the HGK gene has an unusually large amount of alternative splicing. At least five different isoforms were identified from each of four tumor cell line sources, indicating that multiple isoforms are present in the same cell. Alternative splicing modulated the length of the coiled-coil domain, the composition of the CNH domain, and an array of PxxP motifs.
Given the variations in these protein-protein interaction domains, different splice forms are likely to be differentially localized and/or regulated. For example, the HGK clone isolated from glioblastoma cells has the PXXP-containing M3 module, while an alternative PXXP-containing module, M4, replaces M3 in the clones isolated from macrophage (51) and mouse adipocytes (43). The consensus sequences of the PXXP motifs in M3 and M4 differ so that HGK isoforms containing one or the other may interact with different SH3-containing proteins. Two PXXP motifs in M4 from NIK were shown to bind to the adapter protein Nck (43). The binding partners for the PXXP motifs in M3 are unknown and under investigation. The interactions mediated by these PXXP motifs may not be important for the function of HGK in tumor cells, as M3 and M4 are absent from the majority (88% and 98%, respectively) of the isoforms identified in tumor cell lines.
With a probe that detects all HGK splice forms, we found this kinase to be broadly overexpressed at the RNA level in a large number of tumor cell lines relative to most normal tissues. HGK does not map to a known tumor amplicon, yet the overabundance of HGK RNA in tumor cell lines versus normal tissue provides correlative evidence for a role for this kinase in tumorigenesis. An active role for HGK kinase activity in cell transformation is suggested by the fact that ectopic expression of inactive forms blocked oncogenically relevant processes in tissue culture cell models. It is important to note that both the NIH 3T3 and RIE-1 cells used in these studies express detectable levels of HGK, suggesting that the inactive mutant acts in a dominant negative manner (data not shown).
Expression of the inactive dominant negative mutant blocked focus formation and anchorage-independent growth, increased cell adhesion and spreading rates, and blocked invasion and morphogenesis in response to HGF. In contrast, overexpression of wild-type or active alleles slightly increased focus formation, had no effect on anchorage-independent growth, decreased cell adhesion and spreading, and potentiated the cells' invasive and morphogenic responses to HGF. Together these results suggest that HGK catalytic activity is required for anchorage-independent growth, causes the reduction of cell adhesion, and increases invasive properties.
The ability of dominant negative HGK to block the growth of RIE-1 cells in soft agar suggests that this kinase may function in pathways that regulate the anchorage dependence of normal cell growth, implicating HGK in the so-called outside-in signaling from integrin receptors (24). Focal adhesion kinase (FAK) has been shown to function in such pathways (21, 25, 49). There was no measurable difference in autophosphorylation of FAK or phosphorylation of the FAK substrate p130CAS during cell attachment in the stable cell lines expressing the various HGK mutants (data not shown), indicating that HGK may function either downstream or independently of FAK.
The effects of HGK mutants on integrin function and on integrin receptor expression indicate that this kinase does appear to induce changes in cell adhesion or inside-out signaling. Integrin mediated inside-out and outside-in signaling are often described as independent pathways, but there is evidence that effects on one impinge on the other (reviewed in references 4 and 23). We observed that cells with higher HGK kinase activity were less adherent to fibronectin. This effect may be explained, at least in part, by the reduced surface expression of integrin receptor α5 subunit in cells expressing active HGK. While a comprehensive analysis of all integrin subunits levels could not be done with available reagents for rat cells, we did note that HGK also had similar effects on α2 subunit levels but not on α1 subunit levels. Interestingly, NIK−/− mice show a phenotype similar to those of fibronectin−/− and α5 integrin receptor−/− mice, suggesting that NIK may also be involved in the α5 integrin receptor regulation during embryogenesis (50).
As the changes in α5 subunit cell surface expression were smaller than might be expected from the phenotypic changes observed, HGK may also affect integrin signaling by an additional mechanism. The recent report showing physical association, as well as colocalization in attaching cells of β1-integrin receptor with NIK (mouse HGK) strongly suggests that HGK plays a direct role in integrin receptor signaling in addition to affecting α-integrin receptor levels, as we have shown (37). Taken together, these observations point to both direct and indirect involvement of HGK in the regulation of integrin function and signaling.
This role for HGK in negative regulation of cell adhesion may be conserved in other HGK family members, since TNIK also appears to interfere with cell spreading in a kinase activity-dependent manner in transient-transfection assays (22). A reduction in cell spreading rates was also observed previously with other STE20 kinases, including STE20-like kinase (SLK) and the p21-activated kinases αPAK and PAK4 (32, 39, 48). In these three reports, the effects on cell spreading were attributed to effects on the cell cytoskeleton. Whether or not these other STE20 kinases modulated integrin-ligand binding in addition to effects on the cytoskeleton was not investigated. STE20 kinases show significant homology within the kinase domain and may share some cellular substrates, yet the lack of homology in the regulatory domains of these kinases suggest that they are not localized and/or regulated by the same mechanism. For example, the PAK kinases are directly regulated by rho family G-proteins, while HGK is not regulated by Rac or CDC42 and furthermore has been implicated to function upstream of Rac in the activation of JNK (45).
Increased HGK kinase activity through overexpression caused a striking increase in the rate of cell invasion and morphogenesis, while expression of a kinase-inactive mutant blocked invasion and morphogenesis. Ectopic expression of HGK was not sufficient to induce invasion on its own and instead potentiated the invasive response of cells to HGF. A similar potentiation role in HGF-induced invasion was shown for the cytoplasmic domain of α6β4 integrin receptors (47). Perhaps HGK also participates in the signaling complex with the Met receptor and the α6β4 integrin receptor. In examination of signaling downstream of the Met receptor, we observed a decrease in STAT3 phosphorylation on both Y705 and S727 with dominant negative HGK mutant expression. Both of these phosphorylation events have been shown to increase the activity of STAT3 as a transcription factor (reviewed in reference 8).
STAT3 does not appear to be a direct substrate of HGK in that STAT3 protein isolated from cells was not phosphorylated when active, recombinant glutathione S-transferase-HGK kinase domain was added in the presence of ATP (data not shown). However, HGK protein was found to copurify with STAT3 in immunoprecipitations of STAT3 from cells, suggesting that they are part of the same protein complex. HGK could modulate STAT3 activation by affecting its localization in the cell or by regulating the kinases and phosphatases that directly regulate STAT3. As STAT3 plays a role in HGF-induced invasion, blocking STAT3 activation may be one mechanism by which the inactive HGK mutants blocked RIE-1 cell invasion and morphogenesis (6). STAT3 has also been implicated in tumorigenesis in a number of different cell systems (7) and thus could also be important for the dominant negative effects of inactive HGK on focus formation and anchorage-independent growth. A more detailed investigation of the function of HGK in STAT3 activation will be the subject of a future study.
In conclusion, we present an analysis of the HGK gene with its complex family of isoforms. We present evidence that this human homolog of the msn and mig-15 kinases from Drosophila melanogaster and Caenorhabditis elegans functions in morphogenic pathways not only during embryogenesis (50) but also during tumorigenesis and/or invasion. Based on our observations and those of others, we suggest that this kinase may function in the integration of extracellular signals from soluble growth factors and from the extracellular matrix.
Acknowledgments
We thank Rhea Pugliese, Greg Plowman, and Ricardo Martinez for identification and isolation of the first HGK gene and Joseph Zachwieja for preparation of the ascites 9E10 antibodies. We thank Kong Chow and Jennifer Goldstein for sequencing a large number of HGK clones. We thank Tony Hunter (by way of Martin Broome) for the low-passage-number NIH 3T3 cells, Channing Der (by way of Sara Courtneidge) for the RIE-1 cells, and Paul Hughes for advice on and protocols for the integrin experiments.
This work was supported entirely by Sugen, Inc., a subsidiary of Pharmacia Corporation.
REFERENCES
- 1.Bagrodia, S., and R. A. Cerione. 1999. Pak to the future. Trends Cell Biol. 9:350-355. [DOI] [PubMed] [Google Scholar]
- 2.Bao, Q., and R. C. Hughes. 1995. Galectin-3 expression and effects on cyst enlargement and tubulogenesis in kidney epithelial MDCK cells cultured in three-dimensional matrices in vitro. J. Cell Sci. 108:2791-2800. [DOI] [PubMed] [Google Scholar]
- 3.Becker, E., U. Huynh-Do, S. Holland, T. Pawson, T. O. Daniel, and E. Y. Skolnik. 2000. Nck-interacting Ste20 kinase couples Eph receptors to c-Jun N-terminal kinase and integrin activation. Mol. Cell. Biol. 20:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman, A. E., and N. I. Kozlova. 2000. Integrins: structure and functions. Membrane Cell Biol. 13:207-244. [PubMed] [Google Scholar]
- 5.Bischoff, J. R., L. Anderson, Y. Zhu, K. Mossie, L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, C. S. Chan, M. Novotny, D. J. Slamon, and G. D. Plowman. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boccaccio, C., M. Ando, L. Tamagnone, A. Bardelli, P. Michieli, C. Battistini, and P. M. Comoglio. 1998. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature 391:285-288. [DOI] [PubMed] [Google Scholar]
- 7.Bowman, T., R. Garcia, J. Turkson, and R. Jove. 2000. STATs in oncogenesis. Oncogene 19:2474-2488. [DOI] [PubMed] [Google Scholar]
- 8.Bromberg, J., and J. E. Darnell, Jr. 2000. The Role of STATs in transcriptional control and their impact on cellular function. Oncogene 19:2468-2473. [DOI] [PubMed] [Google Scholar]
- 9.Caplan, S., L. M. Hartnell, R. C. Aguilar, N. Naslavsky, and J. S. Bonifacino. 2001. Human Vam6p promotes lysosome clustering and fusion in vivo. J. Cell Biol. 154:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chadee, D. N., T. Yuasa, and J. M. Kyriakis. 2002. Direct activation of mitogen-activated protein kinase kinase kinase MEKK1 by the Ste20p homologue GCK and the adapter protein TRAF2. Mol. Cell. Biol. 22:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comoglio, P. M. 2001. Pathway specificity for Met signaling. Nat. Cell Biol. 3:E161-E162. [DOI] [PubMed] [Google Scholar]
- 12.Cox, A. D., and C. J. Der. 1994. Biological assays for cellular transformation. Methods Enzymol. 238:271-276. [DOI] [PubMed] [Google Scholar]
- 13.Dan, I., N. M. Watanabe, T. Kobayashi, K. Yamashita-Suzuki, Y. Fukagaya, E. Kajikawa, W. K. Kimura, T. M. Nakashima, K. Matsumoto, J. Ninomiya-Tsuji, and A. Kusumi. 2000. Molecular cloning of MINK, a novel member of mammalian GCK family kinases, which is up-regulated during postnatal mouse cerebral development. FEBS Lett. 469:19-23. [DOI] [PubMed] [Google Scholar]
- 14.Dan, I., N. M. Watanabe, and A. Kusumi. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220-230. [DOI] [PubMed] [Google Scholar]
- 15.Davy, A., and S. M. Robbins. 2000. Ephrin-A5 modulates cell adhesion and morphology in an integrin-dependent manner. EMBO J. 19:5396-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Cunto, F., E. Calautti, J. Hsiao, L. Ong, G. Topley, E. Turco, and G. P. Dotto. 1998. Citron rho-interacting kinase, a novel tissue-specific ser/thr kinase encompassing the Rho-Rac-binding protein Citron. J. Biol. Chem. 273:29706-29711. [DOI] [PubMed] [Google Scholar]
- 17.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3:573-581. [DOI] [PubMed] [Google Scholar]
- 18.Florea, L., G. Hartzell, Z. Zhang, G. M. Rubin, and W. Miller. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8:967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frisch, S. M., and H. Francis. 1994. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frisch, S. M., and E. Ruoslahti. 1997. Integrins and anoikis. Curr. Opin. Cell Biol. 9:701-706. [DOI] [PubMed] [Google Scholar]
- 21.Frisch, S. M., K. Vuori, E. Ruoslahti, and P. Y. Chan-Hui. 1996. Control of adhesion-dependent cell survival by focal adhesion kinase. J. Cell Biol. 134:793-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu, C. A., M. Shen, B. C. Huang, J. Lasaga, D. G. Payan, and Y. Luo. 1999. TNIK, a novel member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and regulates the cytoskeleton. J. Biol. Chem. 274:30729-30737. [DOI] [PubMed] [Google Scholar]
- 23.Giancotti, F. G., and E. Ruoslahti. 1990. Elevated levels of the alpha 5 beta 1 fibronectin receptor suppress the transformed phenotype of Chinese hamster ovary cells. Cell 60:849-859. [DOI] [PubMed] [Google Scholar]
- 24.Giancotti, F. G., and E. Ruoslahti. 1999. Integrin signaling. Science 285:1028-1032. [DOI] [PubMed] [Google Scholar]
- 25.Hungerford, J. E., M. T. Compton, M. L. Matter, B. G. Hoffstrom, and C. A. Otey. 1996. Inhibition of pp125FAK in cultured fibroblasts results in apoptosis. J. Cell Biol. 135:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa, K., T. Nagase, M. Suyama, N. Miyajima, A. Tanaka, H. Kotani, N. Nomura, and O. Ohara. 1998. Prediction of the coding sequence of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 5:169-176. [DOI] [PubMed] [Google Scholar]
- 27.Kanai-Azuma, M., Y. Kanai, M. Okamoto, Y. Hayashi, H. Yonekawa, and K. Yazaki. 1999. Nrk: a murine X-linked NIK (Nck-interacting kinase)-related kinase gene expressed in skeletal muscle. Mech. Dev. 89:155-159. [DOI] [PubMed] [Google Scholar]
- 28.Kyriakis, J. M., and J. Avruch. 1996. Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays 18:567-577. [DOI] [PubMed] [Google Scholar]
- 29.Landau, N. R., and D. R. Littman. 1992. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J. Virol. 66:5110-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madaule, P., M. Eda, N. Watanabe, K. Fujisawa, T. Matsuoka, H. Bito, T. Ishizaki, and S. Narumiya. 1998. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature 394:491-494. [DOI] [PubMed] [Google Scholar]
- 31.Madaule, P., T. Furuyashiki, T. Reid, T. Ishizaki, G. Watanabe, N. Morii, and S. Narumiya. 1995. A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 377:243-248. [DOI] [PubMed] [Google Scholar]
- 31a.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sandarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed]
- 32.Manser, E., H.-Y. Huang, T.-H. Loo, X.-Q. Chen, J.-M. Dong, T. Leung, and L. Lim. 1997. Expression of constitutive active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol. 17:1129-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao, H., E. Burnett, M. Kinch, E. Simon, and B. Wang. 2000. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat. Cell Biol. 2:62-69. [DOI] [PubMed] [Google Scholar]
- 34.Montesano, R., K. Matsumoto, T. Nakamura, and L. Orci. 1991. Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell 67:901-908. [DOI] [PubMed] [Google Scholar]
- 35.Nakano, K., J. Yamauchi, K. Nakagawa, H. Itoh, and N. Kitamura. 2000. NESK, a member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and is expressed during the late stages of embryogenesis. J. Biol. Chem. 275:20533-20539. [DOI] [PubMed] [Google Scholar]
- 36.Paricio, N., F. Feiguin, M. Boutros, S. Eaton, and M. Mlodzik. 1999. The Drosophila STE20-like kinase misshapen is required downstream of the Frizzled receptor in planar polarity signaling. EMBO J. 18:4669-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poinat, P., A. De Arcangelis, S. Sookhareea, X. Zhu, E. M. Hedgecock, M. Labouesse, and E. Georges-Labouesse. 2002. A conserved interaction between beta1 integrin/PAT-3 and Nck-interacting kinase/MIG-15 that mediates commissural axon navigation in C. elegans. Curr. Biol. 12:622-631. [DOI] [PubMed] [Google Scholar]
- 38.Pombo, C. M., J. H. Kehrl, I. Sanchez, P. Katz, J. Avruch, L. I. Zon, J. R. Woodgett, T. Force, and J. M. Kyriakis. 1995. Activation of the SAPK pathway by the human STE20 homologue germinal centre kinase. Nature 377:750-754. [DOI] [PubMed] [Google Scholar]
- 39.Qu, J., M. S. Cammarano, Q. Shi, K. C. Ha, P. de Lanerolle, and A. Minden. 2001. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol. Cell. Biol. 21:3523-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruan, W., P. Pang, and Y. Rao. 1999. The SH2/SH3 adaptor protein dock interacts with the Ste20-like kinase misshapen in controlling growth cone motility. Neuron 24:595-605. [DOI] [PubMed] [Google Scholar]
- 41.Stella, M. C., and P. M. Comoglio. 1999. HGF: a multipfunctional growth factor controlling cell scattering. Int. J. Biochem. Cell Biol. 31:1357-1362. [DOI] [PubMed] [Google Scholar]
- 42.Stuart, K. A., S. M. Riordan, S. Lidder, L. Crostella, R. Williams, and G. G. Skouteris. 2000. Hepatocyte growth factor/scatter factor-induced intracellular signalling. Int. J. Exp. Pathol. 81:17-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su, Y. C., J. Han, S. Xu, M. Cobb, and E. Y. Skolnik. 1997. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 16:1279-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su, Y. C., C. Maurel-Zaffran, J. E. Treisman, and E. Y. Skolnik. 2000. The Ste20 kinase misshapen regulates both photoreceptor axon targeting and dorsal closure, acting downstream of distinct signals. Mol. Cell. Biol. 20:4736-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su, Y. C., J. E. Treisman, and E. Y. Skolnik. 1998. The Drosophila Ste20-related kinase misshapen is required for embryonic dorsal closure and acts through a JNK mitogen-activated protein kinase module on an evolutionarily conserved signaling pathway. Genes Dev. 12:2371-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treisman, J. E., N. Ito, and G. M. Rubin. 1997. misshapen encodes a protein kinase involved in cell shape control in Drosophila. Gene 186:119-125. [DOI] [PubMed] [Google Scholar]
- 47.Trusolino, L., A. Bertotti, and P. M. Comoglio. 2001. A signaling adapter function for a6b4 integrin in the control of HGF-dependent invasive growth. Cell 107:643-654. [DOI] [PubMed] [Google Scholar]
- 48.Wagner, S., T. A. Flood, P. O'Reilly, K. Hume, and L. A. Sabourin. 2002. Association of the Ste20-like kinase (SLK) with the microtubule. J. Biol. Chem. 277:37685-37692. [DOI] [PubMed] [Google Scholar]
- 49.Xue, H., X. Chen, L. Benade, J. Owens, E. Shanbrom, W. Drohan, and L. H. Xu. 1995. Attenuation of the expression of the focal adhesion kinase induces apoptosis in tumor cells. Blood 86:791-796. [PubMed] [Google Scholar]
- 50.Xue, Y., X. Wang, Z. Li, N. Gotoh, D. Chapman, and E. Y. Skolnik. 2001. Mesodermal patterning defect in mice lacking the Ste20 NCK interacting kinase (NIK). Development 128:1559-1572. [DOI] [PubMed] [Google Scholar]
- 51.Yao, Z., G. Zhou, X. S. Wang, A. Brown, K. Diener, H. Gan, and T. H. Tan. 1999. A novel human STE20-related protein kinase, HGK, that specifically activates the c-Jun N-terminal kinase signaling pathway. J. Biol. Chem. 274:2118-2125. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, R. 1998. The Eph family of receptors and ligands. Pharmacol. Ther. 77:151-181. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, X., and E. M. Hedgecock. 1996. mig-15 encodes a novel Ser/Thr protein kinase of the Ste-20/p65PAK family. Worm Breeder's Gazette 14:76. [Google Scholar]