Abstract
We have previously demonstrated that the antiapoptotic gene mcl-1 is activated by interleukin-3 (IL-3) in Ba/F3 pro-B cells through two promoter elements designated the CRE-2 and SIE motifs. While the CRE-2-binding complex contains the CREB protein and is activated by IL-3 through the phosphatidylinositol 3-kinase/Akt-dependent pathway, the identity and cytokine activation pathway of the SIE-binding complex remains unclear. In this report, we demonstrated that PU.1 is one component of the SIE-binding complex. A chromatin immunoprecipitation assay further confirmed that PU.1 binds to the mcl-1 promoter region containing the SIE motif in vivo. While IL-3 stimulation does not significantly alter the SIE-binding activity of PU.1, it markedly increases PU.1's transactivation activity. The latter effect coincides with the increased phosphorylation of PU.1 following IL-3 activation of a p38 mitogen-activated protein kinase (p38MAPK)-dependent pathway. A serine-to-alanine substitution at position 142 significantly weakens PU.1's ability to be phosphorylated by the p38MAPK immunocomplex. Furthermore, this S142A mutant is impaired in the ability to be further stimulated by IL-3 to transactivate the mcl-1 reporter through the SIE motif. Taken together, our results demonstrate that IL-3 stimulation of mcl-1 gene transcription through the SIE motif involves phosphorylation of PU.1 at serine 142 by a p38MAPK-dependent pathway.
The mcl-1 gene was originally identified as an early gene induced during differentiation of ML-1 myeloid leukemia cells (22). Its product contains some structural motifs that characterize it as a member of the Bcl-2 family of proteins. The wild-type Mcl-1 protein has antiapoptotic activity (5, 38, 55), whereas an alternatively spliced variant harboring only the BH3 domain is a proapoptotic molecule (1, 3). Mcl-1 expression is induced by a number of growth factors or cytokines, including interleukin-3 (IL-3), IL-5, IL-6, granulocyte-macrophage colony-stimulating factor (GM-CSF), vascular endothelial growth factor, alpha interferon, and epidermal growth factor (5, 13, 15, 24). However, the signaling pathway activated by the individual growth factor/cytokine receptor, which leads to increased expression of the Mcl-1 protein, is largely uncharacterized.
We have previously demonstrated that mcl-1 is an immediate-early gene activated by the GM-CSF and IL-3 signaling pathways and that the mcl-1 gene product is one component of the viability response of these two cytokines (5). Cytokine activation of the mcl-1 gene is regulated at the transcriptional level and requires the membrane-distal region between amino acids 573 and 755 of the common β chain of the GM-CSF and IL-3 receptors (5). Through cloning and extensive characterization of the mcl-1 promoter, we have found that the IL-3 inducibility of this gene in Ba/F3 pro-B cells is mediated mainly through two upstream DNA motifs located at positions −70 (the CRE-2 site) and −87 (the SIE site) (49). Interestingly, these two promoter elements can each confer IL-3 inducibility on a heterologous promoter but work additively in mediating IL-3 response via two different signaling pathways. While the CRE-2-binding complex (which contains the CREB protein) is induced and activated by IL-3 via activation of the phosphatidylinositol 3-kinase (PI3-K)/Akt-dependent pathway, the identity and the IL-3 activation pathway of the SIE-binding complex remain to be determined (49).
PU.1 is a member of the Ets family of transcription factors, and this family of proteins is characterized by the presence of a DNA-binding domain that recognizes a core DNA element containing the 5′-GGAA/T-3′ motif (16, 28, 31). The expression of PU.1 is restricted specifically to cells of the hematopoietic lineage. These include B cells, macrophages, mast cells, neutrophils, and early erythroblasts (6, 10, 12, 20, 32, 37). Knockout mouse studies have demonstrated that PU.1 deficiency results in the absence of morphologically normal B cells and macrophages, disrupted granulopoiesis, and aberrant T lymphopoiesis (29, 41). This phenotype suggests that PU.1 may directly or indirectly regulate some of the genes required for the development of either lymphoid or myeloid lineages. Consistent with this finding, many B-cell- and myeloid-specific genes, including those encoding immunoglobulins, receptors, and enzymes, have been reported to be directly regulated by PU.1 or have a potential PU.1-binding site in their promoters (7, 26, 53).
In this study, we explored the identity and the IL-3 activation pathway of the transcription factor that binds to the SIE element of the mcl-1 gene promoter. By expression library screening, oligonucleotide pulldown, gel shift, and chromatin immunoprecipitation assays, we found that the Ets family of transcription factor PU.1 is one component of the SIE-binding complex in IL-3-dependent Ba/F3 cells. While IL-3 treatment of cells does not significantly alter the SIE-binding activity of PU.1, it markedly stimulates the transactivation activity of PU.1. The latter effect involves phosphorylation of PU.1 at serine 142 following IL-3 stimulation through a p38 mitogen-activated protein kinase (p38MAPK)-dependent pathway.
MATERIALS AND METHODS
Plasmid construction.
The hemagglutinin (HA) epitope-tagged PU.1 expression vector (pcDNA3HA/PU.1) was constructed by reverse transcription-PCR amplification of the total RNA isolated from Ba/F3 cells with the following primers: sense, 5′-TGGAATTCTGTTACAGGCGTGCAAAATG-3′; antisense, 5′-ATGCTCGAGGATCAGTGGGGCGGGAGG-3′. The PCR product was then restricted with EcoRI and XhoI and cloned into a pcDNA3 derivative, pcDNA3-HA (H.-W. Peng and H.-F. Yang-Yen, unpublished results), that would direct the synthesis of an HA-tagged protein in vivo by transfection into mammalian cells or in vitro by the use of a transcription-translation-coupled reticulocyte lysate system (Promega). The HA/PU.1dlN133 expression vector (pcDNA3 HA/PU.1dlN133), which directs the synthesis of a mutant PU.1 protein without the N-terminal 133 amino acids, was constructed by PCR amplification of the wild-type template with primers 5′-CGGGATCCGATGAGGAGGAGGGTG-3′ and 5′-ATGCTCGAGGATCAGTGGGGCGGGAGG-3′. The resultant PCR products were then cloned into the pcDNA3-HA vector. pQE30/PU.1 and pQE30/PU.1dlN133 are expression vectors that would direct the synthesis of the histidine-tagged wild-type or N-terminally truncated PU.1 protein in Escherichia coli and were constructed by inserting an appropriate DNA fragment from the PU.1 cDNA into the pQE30 vector (Qiagen). All of the constructs generated via cloning steps involving PCR were sequenced to confirm their primary structures. pQE30/PU.1S142A, pQE30/PU.1S148A, and pQE30/PU.1SS142/148AA are identical to pQE30/PU.1, except that the nucleotide sequence encoding the serine residue at position 142, 148, or both was mutated to generate an alanine codon(s). These three constructs were generated by site-directed mutagenesis of each individual region, and the mutated nucleotides were confirmed by sequencing. pcDNA3HA/PU.1S142A, pcDNA3HA/PU.1S148A, and pcDNA3HA/PU.1SS142/148AA were derived by subcloning the cDNA inserts from pQE30/PU.1S142A, pQE30/PU.1S148A, and pQE30/PU.1SS142/148AA, respectively, into the pcDNA3-HA vector. The dominant negative mutant forms of p38α and p38β (i.e., p38α[AF] and p38β[AF], respectively) were gifts of Jiahuai Han (14, 50, 54).
Cell culture.
Ba/F3 cells (mouse bone marrow-derived, IL-3-dependent pro-B cells) were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 50 μM β-mercaptoethanol, 2 mM l-glutamine, 100 U of penicillin G per ml, 100 μg of streptomycin per ml, and 2% WEHI-3B conditioned medium as a source of IL-3. Ba/F3-HAPU.1 is a Ba/F3 derivative stably overexpressing the HA-tagged PU.1 protein. This Ba/F3 derivative was generated by electroporation with the HA-tagged PU.1 expression vector (pcDNA3HA/PU.1) and selected in growth medium supplemented with 500 μg of G418 per ml. To examine whether IL-3 stimulates phosphorylation of PU.1 in vivo, Ba/F3-HAPU.1 cells (2 × 107) were first deprived of IL-3 for 4 h and then cultured in phosphate-free medium (Dulbecco modified Eagle medium without phosphate; GIBCO-BRL) for 2 h more. Fifteen minutes prior to IL-3 stimulation, cells were fed with 1 mCi of [32P]orthophosphate (Perkin-Elmer Life Sciences). Eighty minutes after IL-3 treatment, cells were lysed in radioimmunoprecipitation assay buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 1% sodium deoxycholate) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg of aprotinin per ml, and 1 μg of leupeptin per ml and the cell lysates were immunoprecipitated with anti-HA antibody (Roche Applied Sciences). The immunoprecipitated complexes were then resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and the specific bands were revealed by autoradiography or subsequently detected by immunoblotting with anti-HA antibody. For experiments with chemical inhibitors, the following concentrations were used: SB203580, 20 μM; wortmannin, 0.1 μM; anisomycin, 50 ng/ml. All inhibitors (purchased from Calbiochem) were added 30 min prior to IL-3 treatment of cells.
Screening of SIE-binding proteins.
A 32P-labeled concatemer of the double-stranded SIE oligonucleotide probe (sense strand sequence, 5′CTTTTACGGGAAGTCC3′) was used to screen a mouse day 15 embryonic cDNA expression library (Clontech). Briefly, the nitrocellulose filters were immersed in 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG) before they were placed onto the phage plates. After 4 h of incubation at 37°C, these membranes were transferred into SIE binding buffer (10 mM HEPES [pH 7.9], 75 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 10 mM MgCl2, 10% glycerol) containing 6 M guanidine-HCl and the proteins on the membranes were denatured for 5 min. To allow the denatured proteins to be renatured, the membranes were subsequently transferred into a series of binding buffers containing decreasing concentrations of guanidine-HCl (5 min for each step). Following the renaturation steps, the membrane filters were allowed to bind the SIE probe for 18 h at 4°C. After a few washes in SIE binding buffer, positive clones were revealed by autoradiography. After the tertiary screening, all positive clones were purified and their inserts were PCR amplified and sequenced.
Oligonucleotide pulldown assay.
Ba/F3 cells with or without IL-3 stimulation were lysed in NP-40 lysis buffer (50 mM Tris-HCl [pH 8.0], 0.5% NP-40, 150 mM NaCl, 0.1 mM EDTA, 10 mM NaF, 1 mM PMSF, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 mM Na3VO4, 10% glycerol, 1 mM dithiothreitol [DTT]), and 200 μg of lysates from each group was incubated with 1 μg of biotinylated SIE oligonucleotide dimers in the presence of 2 μg of poly(dI-dC). After 1 h of incubation at 4°C, 40 μl of agarose-streptavidin was added to the reaction mixture and the incubation was continued for 1 h more. The SIE-binding complex was then precipitated by centrifugation and washed twice with NP-40 lysis buffer and twice with SIE binding buffer before it was resolved by SDS-PAGE and subsequently analyzed by immunoblotting with anti-PU.1 antibody (Santa Cruz Biotechnology).
Reporter plasmids and luciferase assay.
mcl-1 reporter plasmids p(−203/+10)mcl-luc, −203/+10dlC, and −203/+10mS have been previously described (49). The p(−203/+10)mcl-luc construct contains a luciferase reporter gene that is driven by the mcl-1 gene promoter element between −203 and +10. Plasmids −203/+10dlC and −203/+10mS are identical to p(−203/+10)mcl-luc, except that the CRE-2 site at position −70 and the SIE site at position −87 are mutated, respectively. Plasmids pGL-8xSIE and pGL-2xCRE-2 were derived by inserting eight or two copies of the oligonucleotide fragments containing the SIE and CRE-2 sites, respectively, into the SmaI site of the pGL2-promoter vector (Promega). In both cases, insertion of an SIE or CRE-2 element conferred IL-3 inducibility on the downstream simian virus 40 minimal promoter (49; see Fig. 5B). The sense strand sequences of these two oligonucleotide fragments are as follows: SIE, 5′CTTTTACGGGAAGTCC3′; CRE-2, 5′TCGCCTGCGTCAGCACG3′. To analyze the promoter activity of these reporter genes under various conditions, these plasmids, together with an appropriate expression vector, were transiently introduced into Ba/F3 cells by electroporation with a Bio-Rad Gene Pulser II RF Module system as previously described (5). Electroporated cells were seeded in growth medium with or without murine IL-3 (mIL-3) (R&D Systems). Twelve hours after reseeding, cells were harvested and assayed for luciferase activity. All luciferase activities were normalized to the amounts of proteins present in the cell lysates and, unless specified otherwise, plotted as activities relative to that obtained from cells cotransfected with a control vector and stimulated with IL-3. The latter luciferase activity was assigned a value of 100%. To analyze the effects of various dominant negative mutant forms in the reporter gene assays, electroporated cells were recovered in mIL-3-containing medium for 12 h prior to being deprived of mIL-3 for 6 h and restimulated with mIL-3 for 3 h more.
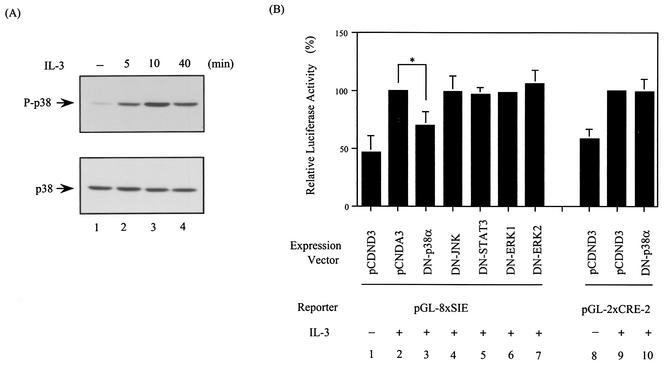
FIG. 5.
Dominant negative mutant form of p38α attenuates the IL-3 inducibility of the SIE but not the CRE-2 reporter. (A) p38MAPK is activated by IL-3. Cell lysates from Ba/F3 cells stimulated with IL-3 for various times were analyzed by Western blotting with an antibody recognizing the active form (P-p38) (top) or all forms (bottom) of p38MAPK. (B) Ba/F3 cells transfected with the indicated reporter plasmid along with a control vector (lanes 1, 2, 8, and 9) or a vector expressing the dominant negative mutant form of p38α (lanes 3 and 10) or other proteins, as indicated (lanes 4 to 7), were stimulated with or without IL-3 for 12 h before the cell lysates were prepared and analyzed for luciferase activity. The results shown are averages of three independent experiments done in duplicate and are plotted as described in the legend to Fig. 3. The difference between the results shown in lanes 2 and 3 is statistically significant (*, P < 0.001).
Gel shift assays.
Gel shift assays were carried out essentially as previously described (49). Briefly, the 32P-labeled oligonucleotide probe (∼0.2 to 0.5 ng) containing the SIE site (the same oligonucleotides as described above for reporter gene construction) was incubated with 8 μg of nuclear extracts or 1.5 μl of in vitro-translated PU.1 in SIE binding buffer containing 1.5 μg of poly(dI-dC). After 20 min of incubation at room temperature, the reaction mixtures were resolved in a 5% native polyacrylamide gel (acrylamide/bisacrylamide ratio, 80:1) at 4°C and the specific protein complexes were visualized by autoradiography. For antibody supershifting experiments, 1 μg of anti-PU.1 (Santa Cruz Biotechnology) or control rabbit immunoglobulin G (IgG) was included in the binding reaction mixture. For competition experiments, a 100-fold molar excess of unlabeled wild-type or mutant SIE (mSIE) oligonucleotides was included in the binding reaction mixture. The sense strand sequence of an mSIE oligonucleotide is as follows: 5′CTTTTAgGatccGTCC3′ (mutated nucleotides are in lowercase).
Western blotting.
Cells to be analyzed were lysed in a buffer containing 50 mM HEPES (pH 7.4), 100 mM NaCl, 1 mM EGTA, 20 mM NaF, 20 mM Na4P2O7, 1 mM Na3VO4, 1 mM PMSF, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, and 1% Triton X-100. Following lysis, the lysates were resolved on an SDS-containing 10% polyacrylamide gel, transferred to polyvinylidene difluoride nylon membrane (Millipore), and probed with antibodies specific to mouse Mcl-1 (raised in a rabbit with recombinant histidine-tagged mouse Mcl-1), α-tubulin (Amersham, Buckinghamshire, England), active p38 (phospho-p38/Thr180Tyr182), or all forms of p38 (both from Cell Signaling Technology). The specific bands were detected by horseradish peroxidase-conjugated goat antibody to rabbit IgG and revealed by an ECL (enhanced chemiluminescence) Western blot system (Amersham Pharmacia Biotech).
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was carried out essentially as described by Saccani et al. (40), with a minor modification. Briefly, Ba/F3 cells, with or without prior stimulation with IL-3, were treated with 1% formaldehyde for 15 min. The cross-linked chromatin was then prepared and sonicated to an average size of 300 to 400 bp prior to being immunoprecipitated with antibody specific to PU.1, CREB, p53 (a gift of Yang-Sun Lin, Institute of Biomedical Sciences, Academia Sinica), or control rabbit IgG at 4°C overnight. After reversal of cross-linking, the immunoprecipitated chromatin was PCR amplified with various sets of primers as indicated below. The amplified DNA product was resolved by agarose gel electrophoresis, subsequently transferred onto nylon membrane, and subjected to Southern blot analysis with a 32P-labeled mcl-1 promoter-specific probe spanning the region between −175 and +38. For PCR amplification of specific regions (A to D; see Fig. 2) of the mcl-1 genomic locus, the following sets of primers were used: region A (nucleotide [nt] −1329 to nt −1062), mMcl1-1329 (5′-CAGCTTGTGTCAAGTTGATATTAAGTCTAACC-3′) and mMcl1-1062 (5′-CCATTTATGGTGGCTAGGCCTATAGCTC-3′); region B (nt −519 to nt −351), mMcl1-519 (5′CCAGGGTTTAACTCCCAGCACCCACC-3′) and mMcl1-351 (5′-AACTCTGCATACCCCAGGCTGGTCC-3′); region C (nt −175 to nt +38), mMcl1-175 (5′-AAGCCGCGAGAGCGCTCCGGCCGGAAG-3′) and mMcl1+38 (5′-ACGCCGCAGGCTGAGGGGAAGGAGC-3′); region D (nt +268 to nt +502), mMcl1+268 (5′-AAAGGCGGCTGCATAAGTCGCCCGGC-3′) and mMcl1+502 (5′-TCCTCTTCCTCCTCGGGCGGCGGCGG-3′). The primers used to amplify a promoter region of a negative control gene (E4BP4) were E4BP4+1 (5′-CAGAAAGGACCTCCTCGTCCTACAGAC-3′) and E4BP4-360 (5′-TCTGCTGGACCACATAGTCCAAGGCAAAGA-3′).
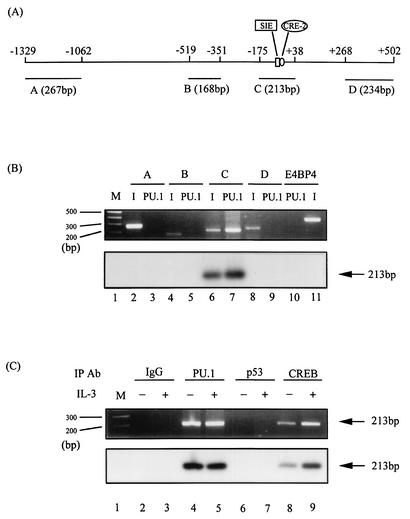
FIG. 2.
PU.1 binds to the mcl-1 gene promoter in vivo. (A) Schematic representation of the mcl-1 genomic locus spanning the promoter region. Fragments A to D (sizes are indicated in base pairs) are those predicted to be generated by a specified pair of primers as described in Materials and Methods. (B) ChIP analysis of PU.1 binding to the mcl-1 gene locus. Formaldehyde-cross-linked chromatin was immunoprecipitated with PU.1 antibody and processed for PCR amplification as described in Materials and Methods, by using primer sets that would specifically amplify fragments A to D, as indicated in panel A. As a positive control, PCR amplification was also carried out with input DNA from chromatin prior to the immunoprecipitation (I) step (lanes 2, 4, 6, 8, and 11). Serving as a negative control, a primer set that would specifically amplify the promoter region of the E4BP4 gene was included in the assay mixture (lanes 10 and 11). At the bottom is the result of a Southern blot analysis of the same gel with a probe spanning the mcl-1 promoter region between −175 and +38. All of the chromatin used in this assay was isolated from Ba/F3 cells stimulated with IL-3. (C) ChIP analysis similar to that described for lane 7 of panel B, except that chromatin was isolated from cells with (lanes 3, 5, 7, and 9) or without (even-numbered lanes) IL-3 treatments and the immunoprecipitation (IP) step was carried out with various antibodies (Ab), as indicated. At the bottom is the result of a Southern blot analysis of the same gel with the same probe as described for panel B. M stands for DNA size markers (100-bp ladders).
In vitro kinase assay.
IL-3-stimulated Ba/F3 cells were lysed in p38 lysis buffer (10 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 2 mM EGTA, 10 mM NaF, 0.2% Triton X-100, 50 mM β-glycerophosphate, 2 mM Na3VO4, 1 mM DTT, 1 mM PMSF, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml), and the cell lysates were immunoprecipitated with a p38MAPK antibody (kindly provided by Jiahuai Han). After three washes with p38 lysis buffer and two with kinase buffer (20 mM HEPES [pH 7.5], 20 mM MnCl2, 20 mM MgCl2, 2 mM DTT, 0.1 mM Na3VO4, 10 mM NaF, 25 mM β-glycerophosphate), the p38 immunocomplex was resuspended in 40 μl of kinase buffer containing 10 μCi of [γ-32P]ATP and 5 μg of wild-type or mutant PU.1 purified from bacteria (XL-1-blue) transformed with pQE30/PU.1, pQE30/PU.1dlN133, pQE30/PU.1S142A, pQE30/PU.1S148A, or pQE30/PU.1SS142/148AA. After 30 min of incubation at 30°C, the reaction products were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Specific bands were first revealed by autoradiography and later by staining of the membrane with Coomassie brilliant blue.
RESULTS
PU.1 binds to the SIE element of the mcl-1 promoter in vitro.
To identify the transcription factor that is involved in IL-3 stimulation of murine mcl-1 gene transcription through the SIE motif, a mouse day 15 embryonic cDNA expression library was screened with a 32P-labeled concatemer of double-stranded SIE oligonucleotide (see Materials and Methods). Of 5 × 105 clones screened, 23 were positive and sequence analysis revealed that they represented clones derived from 11 distinct cDNAs. Among these positive clones, the one encoding the Ets family of transcription factor PU.1 was first characterized, as PU.1 cDNA was pulled out four times (all with different sizes) in our library screen and Western analysis (data not shown) revealed that PU.1 was expressed in our model cell line, Ba/F3, for studying mcl-1 gene regulation by the IL-3 signaling pathway (49).
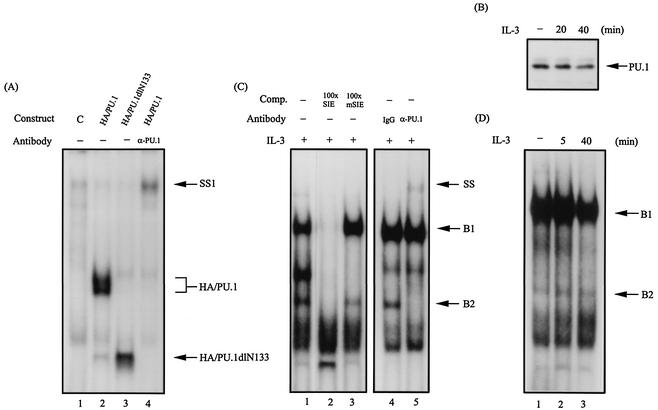
To investigate whether PU.1 indeed binds to the SIE element of the mcl-1 gene promoter, the following experiments were carried out. We first examined whether PU.1 prepared from the in vitro translation system would recognize the SIE oligonucleotide probe. As shown in Fig. 1A, the rabbit reticulocyte lysate programmed with a vector encoding either the HA-tagged full-length PU.1 (HA/PU.1) protein or a mutant protein retaining the DNA-binding domain (HA/PU.1dlN133) gave rise to a specific protein-DNA complex that was not observed with the reticulocyte lysate programmed with an empty control vector (compare lanes 2 and 3 to lane 1). The identity of the protein-DNA complex formed with HA/PU.1 in this assay was further confirmed either by its ability to be supershifted by a PU.1 antibody (lane 4) or by its ability to be competed out by a 100-fold molar excess of unlabeled wild-type SIE but not mSIE oligonucleotide (data not shown). We next examined whether the SIE-binding complex previously identified with the Ba/F3 cell lysates (49) contained the PU.1 protein. To address this issue, two different approaches were taken. First, the oligonucleotide pulldown assay (see Materials and Methods) was employed. In this experiment, the PU.1 protein was found to be present in the protein complex coprecipitated with the biotinylated SIE oligonucleotide as revealed by immunoblotting analysis of this complex with antibody specific to PU.1 (Fig. 1B). The pulldown of PU.1 in this experiment was specific to the SIE element, as it was specifically inhibited by oligonucleotides containing the wild-type SIE site but not by oligonucleotides containing the mSIE site (data not shown). It is worth noting that the formation of the SIE-PU.1-binding complex was independent of stimulation by IL-3 (compare lanes 1, 2, and 3 in Fig. 1B). The second approach was to employ the antibody supershift assay with the SIE oligonucleotide probe. Figure 1C shows that, in this approach, in addition to the major complex (designated B1) identified in our previous report (49), a minor, lower-molecular-weight protein-DNA complex (designated B2) was detected in an autoradiogram developed after a longer exposure time. Like the B1 complex, the B2 complex bound specifically to the SIE site, as it was competed out by the wild-type SIE-containing oligonucleotide but not by the mSIE-containing oligonucleotide (Fig. 1C, lanes 1 to 3). Unexpectedly, it was the B2 but not the B1 complex that was specifically supershifted by the PU.1 antibody (compare lanes 4 and 5 in Fig. 1C). Similar to the results shown in Fig. 1B, IL-3 did not significantly affect the formation of the B2 complex (compare lanes 1 to 3 in Fig. 1D).
FIG. 1.
PU.1 is one component of the SIE-binding complex. (A) Gel shift assay with the 32P-labeled SIE probe using in vitro-translated HA/PU.1 (lane 2) or HA/PU.1dlN133 (lane 3). Lane 4, same as lane 2 except that the anti-PU.1 antibody was included in the assay. Lane 1 is the result of a control experiment using the reticulocyte lysates programmed with an empty expression vector. SS1, the HA/PU.1-DNA complex supershifted by the PU.1 antibody. (B) PU.1 is present in the complex pulled down by the SIE-containing oligonucleotide. The oligonucleotide pulldown assay using lysates of Ba/F3 cells with or without prior stimulation with IL-3 was carried out as described in Materials and Methods. The protein complex pulled down by this assay was analyzed by Western blotting with an antibody specific for PU.1. (C) Lane 1, same as lane 2 of panel A, except that nuclear extracts from IL-3-stimulated Ba/F3 cells were used in the gel shift assay. Lanes 2 and 3 are results of the same experiment with the inclusion of a 100-fold molar excess of unlabeled wild-type SIE- and mSIE (see Materials and Methods for the bases changed in the nucleotide sequence)-containing oligonucleotides, respectively, as a cold competitor (Comp.). SS denotes the B2 complex supershifted by the PU.1 antibody. (D) Same as lane 1 of panel C, except that the assay was carried out with nuclear extracts from Ba/F3 cells with or without IL-3 treatments.
PU.1 binds to the mcl-1 promoter in vivo.
We next examined whether PU.1 bound to the mcl-1 gene promoter in vivo. To address this issue, the ChIP assay was performed. For this experiment, we designed five sets of primers (see Materials and Methods) that would specifically amplify either a promoter region of a negative control gene (−360 to +1 of the E4BP4 gene) or some designated regions (A to D) of the mcl-1 gene locus, as illustrated in Fig. 2A. The results shown in Fig. 2B indicated that, while all of the primer sets generated a PCR fragment of the predicted size, as indicated in Fig. 2A, from the chromatin DNA prior to the immunoprecipitation step (lanes 2, 4, 6, 8, and 11, top), only region C, containing the SIE motif, was specifically precipitated by the PU.1 antibody and subsequently PCR amplified (lane 7, top). The identity of the amplified C fragment was further confirmed by Southern blotting analysis of the same gel shown in the upper part of Fig. 2B with a genomic DNA probe spanning the region between −175 and +38 (Fig. 2B, bottom, lanes 6 and 7). Next, we examined whether coprecipitation of the C fragment was indeed due to specific binding of PU.1 to this region. As shown in Fig. 2C, neither an isotype-matched rabbit IgG (lanes 2 and 3) nor an antibody recognizing an irrelevant transcription factor (p53, lanes 6 and 7) precipitated this C fragment. On the other hand, an antibody recognizing the active form of the CREB protein (phospho-CREB) specifically coprecipitated the same C fragment in an IL-3-inducible manner (Fig. 2C, lanes 8 and 9). The latter result was consistent with our previous finding that CREB binds to the CRE-2 element right next to the SIE motif of the mcl-1 gene promoter and its binding activity is stimulated by IL-3 treatment of cells (49). Similar to the results shown in Fig. 1 B and D, binding of PU.1 to the C fragment was constitutive, i.e., independent of stimulation by IL-3 (compare lanes 4 and 5 of Fig. 2C). Taken together, these results indicate that PU.1 binds to the mcl-1 gene promoter in vivo.
PU.1 is involved in IL-3 stimulation of mcl-1 gene transcription through binding to the SIE motif.
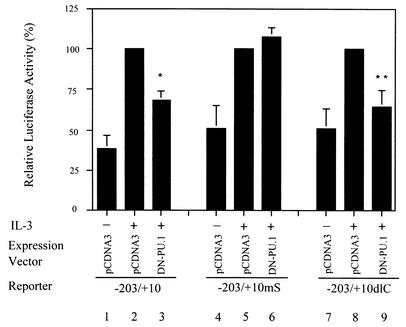
Next, we examined whether PU.1 plays a role in IL-3 stimulation of mcl-1 gene transcription. To address this issue, the reporter gene assay using the Ba/F3 cell system as previously described (49) was performed. In this experiment, cells were cotransfected with various mcl-1 reporter genes together with a control vector or a vector expressing mutant PU.1 (HA/PU.1dlN133) that was previously shown to have a dominant negative effect (43). As previously noted (49), IL-3 stimulated (about three- to fourfold) the luciferase activity of cells transfected with a luciferase reporter driven by the mcl-1 promoter region between −203 and +10, i.e., the p(−203/+10)mcl-luc construct (see reference 49, and compare lanes 1 and 2 in Fig. 3). Mutation of the CRE-2 or SIE motif resulted in a reporter that still responded to IL-3 stimulation but to a lesser extent (about twofold) (see reference 49, and compare lanes 4 and 5 and lanes 7 and 8 in Fig. 3). Interestingly, coexpression of the dominant negative mutant form of PU.1 (HA/PU.1dlN133) significantly attenuated the IL-3 inducibility of the mcl-1 gene reporters containing the SIE motif, i.e., both the p(−203/+10)mcl-luc and −203/+10dlC reporters (compare lanes 2 and 3 and lanes 8 and 9 in Fig. 3). In contrast, the IL-3 inducibility of the −203/+10mS reporter with the mSIE site was not affected by the dominant negative mutant form of PU.1 (compare lanes 5 and 6 in Fig. 3). Taken together, these results suggest that PU.1 is involved in IL-3 stimulation of mcl-1 gene transcription and that its effect is mediated through binding to the SIE motif.
FIG. 3.
Dominant negative mutant form of PU.1 attenuates the IL-3 inducibility of mcl-1 reporters with an intact SIE motif. Ba/F3 cells were transiently transfected with reporter genes as indicated along with a control or the HA/PU.1dl133 (DN-PU.1) expression vector. After transfection, cells were cultivated in medium with or without IL-3 for 12 h before cell lysates were prepared and analyzed for luciferase activity. The results shown are averages of three independent transfection experiments done in duplicate. For experiments with each indicated reporter, the luciferase activity of that particular reporter in cells cotransfected with a control vector and with IL-3 stimulation was assigned a value of 100% and all other results obtained with cells transfected with the same reporter were normalized to this activity. The differences between the results shown in lanes 2 and 3 and those shown in lanes 8 and 9 are statistically significant (*, P < 0.001; **, P = 0.002).
SIE-mediated IL-3 stimulation of mcl-1 transcription involves activation of p38MAPK.
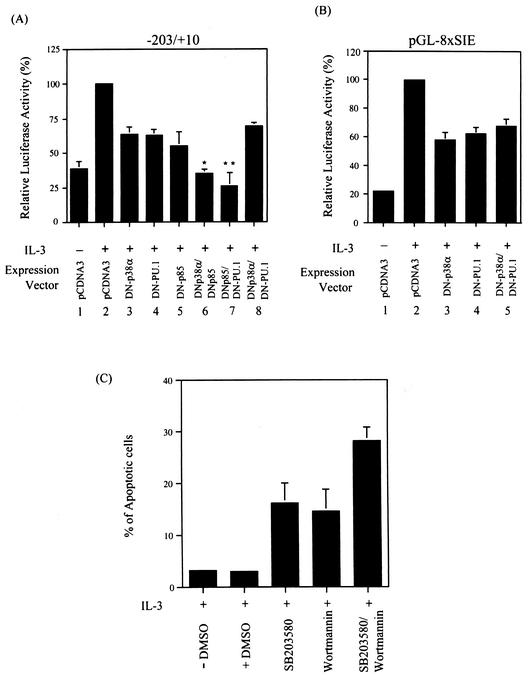
We have previously demonstrated that IL-3 activation of mcl-1 gene transcription through the SIE motif occurs via an unknown but PI3-K/Akt-independent pathway (49). To determine which signaling pathway is involved in this regulatory process, we first tested whether any known kinase inhibitors would inhibit the IL-3 inducibility of an SIE reporter configured in a heterologous promoter (pGL-8xSIE). Of the various chemical inhibitors tested, SB203580, which specifically inhibits the α and β isoforms of p38MAPK, manifested the best inhibitory effect (data not shown). We next examined whether SB203580 would inhibit IL-3 stimulation of mcl-1 expression in vivo. To address this issue, Northern blotting analysis was carried out. The results shown in Fig. 4A indicated that, like the PI3-K inhibitor wortmannin (49), SB203580 also inhibited IL-3 stimulation of mcl-1 mRNA expression (compare lanes 3 and 4 to lane 2). While either inhibitor had an approximately 50% inhibitory effect, both inhibitors, when added together to cells, nearly completely blocked IL-3 stimulation of mcl-1 expression. A similar inhibitory effect was also observed at the protein level (Fig. 4B). As p38MAPK was indeed activated by IL-3 in Ba/F3 cells, as was evident from the appearance of a phosphorylated form (at positions Thr-180 and Tyr-182) of p38 following IL-3 treatment (Fig. 5A), we next examined whether the IL-3 inducibility of the SIE reporter (pGL-8XSIE) could be blocked by a dominant negative mutant form of p38MAPK. In Ba/F3 cells, the α isoform of p38MAPK (p38α) is expressed in a much larger quantity than the β isoform (p38β) (α/β ratio, >20:1; data not shown). We therefore first tested the blocking ability of the dominant negative mutant form of p38α (DN-p38α). The results shown in Fig. 5B indicated that coexpression of DN-p38α, but not the dominant negative mutant forms of other irrelevant molecules, as indicated in the figure, attenuated IL-3's stimulatory effect on SIE reporter activity (compare lanes 2 and 3). This inhibitory effect was specifically mediated through the SIE motif, as under the same conditions, the IL-3 inducibility of the CRE-2 reporter (pGL-2XCRE-2) was not affected by DN-p38α (compare lanes 9 and 10 in Fig. 5B).
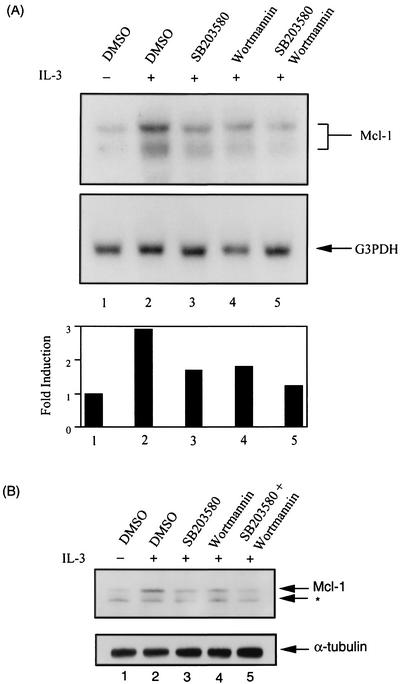
FIG. 4.
p38MAPK inhibitor (SB203580) attenuates IL-3 stimulation of mcl-1 mRNA and protein expression. (A) Ba/F3 cells deprived of IL-3 were pretreated with various inhibitors, as indicated, for 30 min prior to stimulation with IL-3 for 1 h. After IL-3 stimulation, total RNA was isolated from these cells and analyzed by Northern blotting with a probe specific for detection of the murine mcl-1 mRNA. The same blot was later stripped and hybridized with a glyceraldehyde-3-phosphophate dehydrogenase (G3PDH)-specific probe. At the bottom is a quantitative analysis of the mRNA levels shown at the top. The amount of mcl-1 mRNA in each group was normalized to that of G3PDH mRNA and plotted as a ratio relative to results obtained with cells deprived of IL-3 (lane 1). (B) Same conditions as in panel A, except that cells were lysed and 100 μg of protein lysates was analyzed by immunoblotting with antibodies specific to mouse Mcl-1 and α-tubulin, respectively. The asterisk points to an unknown protein that was also recognized by the mouse Mcl-1 antibody. DMSO, dimethyl sulfoxide.
Given the fact that both PU.1 and p38α play a role in IL-3 activation of SIE-containing reporters, we next examined whether these two molecules function in the same pathway to activate transcription of the mcl-1 gene. To address this issue, a transfection experiment identical to that shown in Fig. 3, using the p(−203/+10)mcl-luc reporter but with various combinations of dominant negative mutant forms, was carried out. The p(−203/+10)mcl-luc reporter was first tested in this assay because it contains both the CRE-2 and SIE motifs and the IL-3 inducibility through these two elements is mediated via PI3-K (49)- and PU.1 (Fig. 3)-dependent pathways, respectively. Figure 6A shows that IL-3-activated p(−203/+10)mcl-luc reporter activity was partially inhibited by coexpression of DN-p38α, DN-PU.1, or a dominant negative mutant form of PI3-K (DN-p85) alone (lanes 2 to 5). While DN-p38α and DN-PU.1 each synergized with DN-p85 to inhibit IL-3 stimulation of this reporter activity (compare lanes 5 to 7), no synergism was observed when DN-p38α and DN-PU.1 were coexpressed with the reporter gene (compare lanes 3, 4, and 8). The lack of a synergistic effect between DN-p38α and DN-PU.1 was also evident in an experiment that was identical except for the use of the pGL-8XSIE reporter (Fig. 6B, lanes 3 to 5). Taken together, these results suggested that both p38α and PU.1 function in the same pathway to mediate IL-3 activation of mcl-1 gene transcription through the SIE motif. Next, we examined whether activation of the p38MAPK pathway is important to the cell survival activity of IL-3. As shown in Fig. 6C, like the case observed with the PI3-K inhibitor wortmannin (49), blocking of the p38 pathway by treatment of cells with SB203580 also induced apoptosis of cells cultivated in IL-3-containing medium. Furthermore, SB203580 worked additively with wortmannin to inhibit the survival activity of IL-3. This result, together with that illustrated in Fig. 4, suggests that the p38MAPK and PI3-K pathways both contribute to the regulation of Mcl-1 expression, as well as to the survival activity of IL-3.
FIG. 6.
(A and B) p38α and PU.1 work in the same pathway to regulate the IL-3 inducibility of SIE-containing reporters. (A) Ba/F3 cells transfected with the p(−203/+10)mcl-luc reporter along with a control vector or vectors expressing DN-PU.1, DN-p38α, and DN-p85 or a combination of these expression vectors were left untreated or stimulated with IL-3 for 12 h before cell lysates were prepared and analyzed for luciferase activity. The data shown here are representative results of three to five independent experiments performed in duplicate. Luciferase activity was plotted as described in legend to Fig. 3. The differences between the results shown in lanes 3 and 6 and between those shown in lanes 4 and 7 are statistically significant. (*, P = 0.0001; **, P = 0.003). (B) Same conditions as in lanes 1 to 4 and 8 of panel A, except that the pGL-8XSIE reporter was used in the assay. (C) The p38MAPK pathway is involved in the viability response of IL-3 in Ba/F3 cells. Ba/F3 cells cultivated in growth medium containing IL-3 were treated with various chemical inhibitors as indicated. Fifteen hours after each treatment, cells were harvested and the percentage of apoptotic cells in each group was quantified by flow cytometric analysis of cells with a sub-G1 DNA content. The data shown are representative results of two independent experiments performed in duplicate. DMSO, dimethyl sulfoxide.
IL-3-induced phosphorylation of PU.1 is mediated via a p38MAPK-dependent pathway.
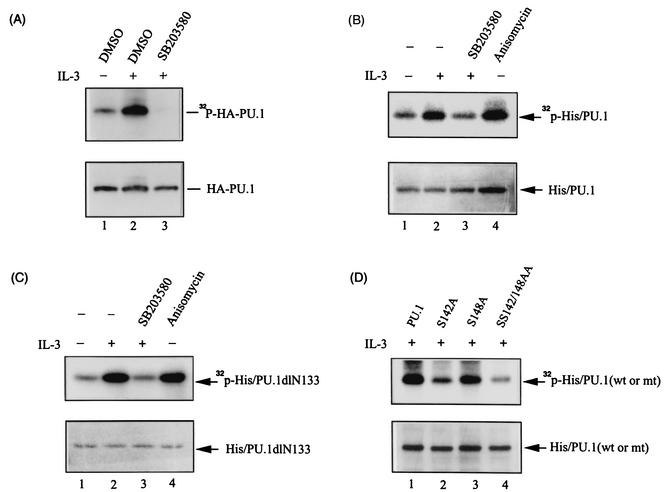
PU.1 is a phosphoprotein, and phosphorylation at serine 148 is required for its interaction with a B-cell-restricted transcription factor, PIP (NF-EM5), and activates the immunoglobulin κ 3′ enhancer (35). In the context of IL-3-regulated mcl-1 gene transcription, PU.1 plays a role in this process, yet its DNA-binding activity is not significantly influenced following treatment of cells with IL-3 (Fig. 1 and 2). These results suggest that PU.1 may undergo an IL-3-dependent posttranslational modification, e.g., phosphorylation, that then directly or indirectly activates mcl-1 gene transcription (see Discussion). To address this possibility, we next examined whether IL-3 induces phosphorylation of PU.1 in vivo. For this experiment, Ba/F3 cells stably overexpressing HA-tagged PU.1 were stimulated with IL-3 in the presence of [32P]orthophosphate and the cell lysates were analyzed as described in Materials and Methods. Figure 7A shows that following IL-3 treatment, HA-tagged PU.1 was indeed phosphorylated, as revealed by the detection of a 32P-labeled band with an approximate size of 38 kDa in the anti-HA immunoprecipitate (lane 2, top) that could be further recognized by an anti-HA antibody in the subsequent immunoblotting analysis (bottom). Furthermore, IL-3-induced phosphorylation of PU.1 was inhibited by pretreatment of cells with SB203580 (compare lanes 2 and 3 in Fig. 7A), suggesting that this phosphorylation event is mediated through a p38MAPK-dependent pathway. Next, we examined whether PU.1 could be phosphorylated in vitro by p38MAPK immunoprecipitated from cells treated with IL-3 or anisomycin (a known p38MAPK activator). Figure 7B shows that this was indeed the case. The p38MAPK immunocomplex purified in both cases phosphorylated the bacterially produced His-tagged PU.1 (His/PU.1) protein (lanes 2 and 4). Furthermore, the in vitro phosphorylation activity of the p38MAPK immunocomplex was indeed p38MAPK dependent, as the immunocomplex isolated from cells pretreated with SB203580 was devoid of this activity (lane 3). These results, together with those shown in Fig. 7A, suggest that IL-3 stimulates PU.1 phosphorylation through a p38MAPK-dependent pathway.
FIG. 7.
IL-3-induced phosphorylation of PU.1 occurs via a p38MAPK-dependent pathway. (A) In vivo phosphorylation of PU.1. Ba/F3-HAPU.1 cells with (lane 3) or without (lane 2) prior treatment with SB203580 were stimulated with IL-3 in the presence of [32P]orthophosphate as described in Materials and Methods. After labeling, the HA-PU.1 protein was immunoprecipitated from cell lysates by using the anti-HA antibody, resolved by SDS-PAGE, and subsequently revealed by autoradiography (top) or Western blotting with an anti-HA antibody (bottom). DMSO, dimethyl sulfoxide. (B) In vitro phosphorylation of PU.1 by the p38MAPK immunocomplex. The bacterium-produced His/PU.1 protein was phosphorylated in vitro by the p38MAPK immunocomplex as described in Materials and Methods. Lanes 1 and 2 are results of experiments with p38MAPK immunocomplexes isolated from cells without and with IL-3 treatment, respectively. Lane 3, same as lane 2, except that the p38MAPK complex was prepared from cells pretreated with SB203580 prior to IL-3 stimulation. Lane 4, same as lane 1, except that the p38MAPK complex was isolated from cells pretreated with anisomycin. (C) Same conditions as described for panel B, except that bacterium-produced His/PU.1dlN133 was used as the substrate. (D) Same conditions as described for lane 2 of panel B, except that various bacterium-produced PU.1 mutant forms, as indicated, were used as the substrate.
We next examined whether phosphorylation would affect the ability of PU.1 to mediate the IL-3 inducibility of the mcl-1 gene. The p38MAPK-dependent phosphorylation site of PU.1 was first mapped. We noticed that the PU.1 mutant without the N-terminal 133 amino acids (His/PU.1dlN133) was still phosphorylated in vitro by the p38MAPK immunocomplex (Fig. 7C). We therefore searched the remaining C-terminal region for motifs like Ser-Pro, Thr-Pro, and Ser-Asp that many p38MAPK downstream targets (direct or indirect) display (17, 45, 48, 50a, 54). Two candidate sites, one at serine 142 and the other at serine 148, were identified, and each (or both) was mutated to an alanine residue by site-directed mutagenesis. Figure 7D shows that a serine-to-alanine mutation at position 148 (S148A) did not significantly affect PU.1's ability to be phosphorylated by the p38MAPK immunocomplex (lane 3), whereas a mutation at position 142 (S142A) markedly attenuated PU.1 phosphorylation in the same in vitro kinase assay (lanes 2 and 4). These results suggest that serine 142 is one site to be phosphorylated in vitro by the p38MAPK immunocomplex.
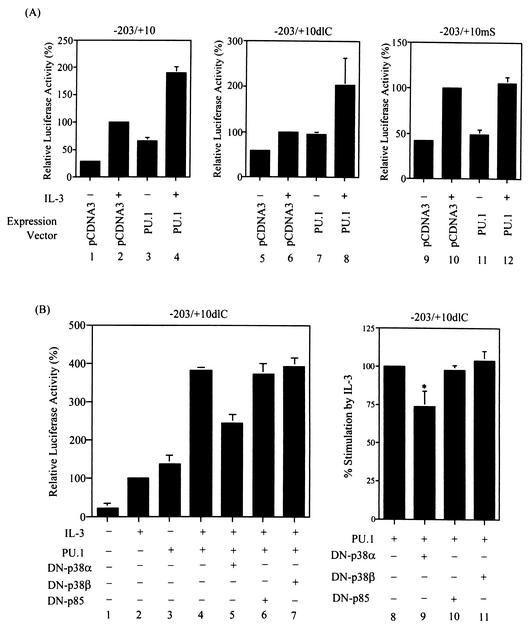
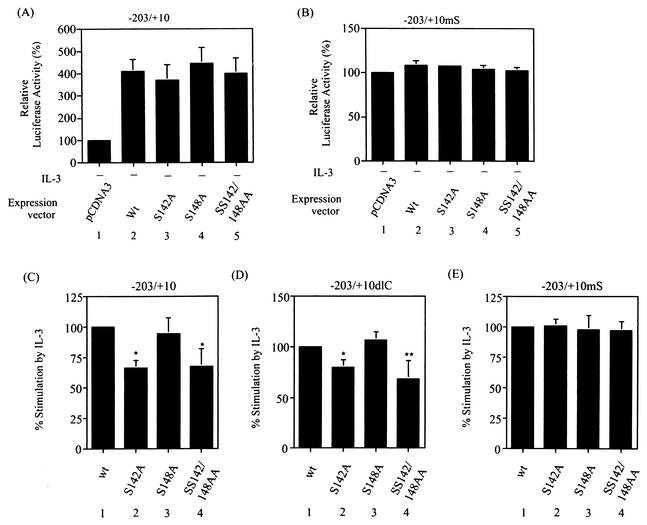
To examine whether p38MAPK-dependent phosphorylation at serine 142 is important for PU.1 to mediate the IL-3 response of the SIE motif, an assay that could specifically address this issue was first used. In the transient transfection experiments shown in Fig. 8A, we noticed that without IL-3 stimulation, overexpression of PU.1 could slightly transactivate both p(−203/+10)mcl-luc and −203/+10dlC (compare lanes 1 and 3 and lanes 5 and 7) but not the −203/+10mS reporter (compare lanes 9 and 11). With IL-3 stimulation, although the reporter activities of all three constructs were enhanced as previously noted (reference 49; compare lanes 1 and 2, lanes 5 and 6, and lanes 9 and 10 in Fig. 8A), a further increase in reporter activity due to overexpression of PU.1 was observed again only in cells transfected with p(−203/+10)mcl-luc or −203/+10dlC (compare lanes 2 and 4 and lanes 6 and 8) but not in cells transfected with the −203/+10mS reporter (compare lanes 10 and 12). These results suggest that IL-3-enhanced transactivation activity of PU.1 on mcl-1 reporters requires the presence of the SIE motif. We next examined whether IL-3-enhanced transactivation activity of PU.1 is dependent on activation of p38MAPK. To address this issue, the same transfection experiment as described in the middle part of Fig. 8A, except with the inclusion of a DN-p38 expression vector, was carried out. In this experiment, the −203/+10dlC reporter was used because it could eliminate the stimulation effect of IL-3 through the CRE-2 site via the PI3-K/Akt-dependent pathway (49). Figure 8B shows that coexpression of DN-p38α but not DN-p38β or DN-p85 significantly attenuated the stimulation effect of IL-3 on the transactivation activity of PU.1 (compare lanes 4 to 7 on the left side of Fig. 8B and lanes 8 to 11 on the right side). These results suggest that IL-3-enhanced transactivation activity of PU.1 is mediated through a p38α-dependent pathway.
FIG. 8.
IL-3 stimulates the transactivation activity of PU.1 on SIE-containing reporters via a p38α-dependent pathway. (A) Ba/F3 cells transfected with various mcl-1 reporter genes along with a control or PU.1 expression vector, as indicated, were stimulated with or without IL-3 for 12 h before the cell lysates were prepared and analyzed for luciferase activity. The data shown here are representative results of three or four independent experiments performed in duplicate. (B) Same conditions as in lanes 5 to 8 of panel A, except with the inclusion of vectors expressing DN-p38α, DN-p38β, and DN-p85 (lanes 5 to 7, respectively) during the transfection step. The data shown are means ± standard deviations of duplicates from an experiment that was repeated two to four times with similar results. Lanes 8 to 11 illustrate the average normalized results of four independent transfection experiments as described for lanes 4 to 7. For this normalization method, the percent increase in the transactivation activity of PU.1 caused by IL-3 treatment was assigned a value of 100 (i.e., the ratio of activity shown in lane 4 over that shown in lane 3). The activities shown in lanes 5 to 7 were each transformed by the same method, and the resultant quotient was normalized to that obtained from the results shown in lanes 3 and 4. The result shown in lane 9 is statistically significantly different from that shown in lane 8 (*, P = 0.002).
We next employed a type of assay similar to that shown in Fig. 8 to determine whether phosphorylation at serine 142 is responsible for the IL-3-enhanced transcriptional activity of PU.1 on SIE-containing reporters. For this experiment, we first examined whether mutation of ser-142, ser-148, or both residues would affect their transcriptional activities. Figure 9 shows that none of these mutations had a significant effect on their abilities to transactivate the SIE-containing reporter (compare panels A and B). However, the transactivation activity of the S142A or the SS142/148AA double mutant form on the SIE-containing reporters, i.e., p(−203/+10)mcl-luc (panel C) and −203/+10dlC (panel D), was enhanced by IL-3 significantly less than that of the wild-type or S148A mutant form. The nearly equal effects of all four proteins on the −203/+10mS reporter (panel E) further confirmed the specificity of this type of experiment. Taken together, these results suggest that the p38MAPK-dependent phosphorylation at serine 142 of PU.1 contributes, at least partially, to IL-3-stimulated mcl-1 promoter activity through the SIE motif.
FIG. 9.
Mutation of serine 142 to alanine attenuates IL-3-enhanced transactivation activity of PU.1. (A and B) The S142A, S148A, and SS142/148AA mutant forms all have transactivation activities similar to that of the wild-type (Wt or wt) protein on the SIE-containing reporter. Reporter gene assays were carried out with cells transfected with the p(−203/+10)mcl-luc (A) or −203/+10mS (B) reporter along with a pcDNA3 control or a PU.1 (wild-type or mutant)-expressing vector, as indicated. Both transfection assays were performed in the absence of IL-3. The luciferase activities obtained from cells transfected with the control vector were assigned a value of 100%. (C to E) Reporter gene assays carried out with cells transfected with the indicated reporter gene along with a wild-type or mutant PU.1 expression vector, as indicated. After transfection, cells were split into two groups; one remained untreated, and the other was stimulated with IL-3. Twelve hours after transfection, cells were lysed and analyzed for luciferase activity. The data shown here are normalized results to illustrate the average increase in the transactivation activity of wild-type or mutant PU.1 following IL-3 treatment. The normalization method used is described in the legend to Fig. 8B. The differences between the results shown in lanes 1 and 2 and those between the results shown in lanes 1 and 4 of panels C and D are statistically significant (*, P < 0.001; **, P < 0.01, n = 6).
DISCUSSION
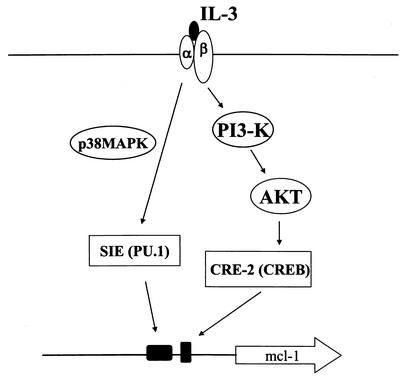
IL-3 activation of mcl-1 gene transcription is mediated through two promoter elements designated the CRE-2 and SIE motifs. In our previous reports, we stated that the CRE-2-binding complex contains the CREB protein and is activated by IL-3 through a PI3-K/Akt-dependent pathway (49). In the present study, we demonstrated that the Ets family of transcription factor PU.1 is one component of the SIE-binding complex. Unlike the case with the CREB protein, which binds to the CRE-2 motif in an IL-3-inducible manner, PU.1 binds to the SIE element constitutively but its transactivation activity is increased upon IL-3 stimulation of cells. Furthermore, we showed that IL-3 stimulation of mcl-1 gene transcription through the SIE motif involves phosphorylation of PU.1 at serine 142 by a p38MAPK-dependent pathway. These results, together with those in our previous report (49), indicate that the PI3-K/Akt/CREB and p38MAPK/PU.1 pathways cross talk at the mcl-1 gene locus (Fig. 10). Considering the fact that the core sequences of the CRE-2 and SIE motifs are only separated by 9 bp, there may be some protein-protein interactions between CREB- and PU.1-containing complexes that, via an unknown mechanism, stimulate transcription of the mcl-1 gene. More experiments are required to investigate this possibility.
FIG. 10.
Schematic representation of molecules involved in the IL-3 signaling pathways that lead to mcl-1 gene transcription through the SIE and CRE-2 elements (see text for details).
PU.1 is a hematopoietic, lineage-specific transcription factor with some characterized functional domains. These include multiple N-terminal acidic and glutamine-rich transactivation domains (11, 19, 20, 21, 43), a central proline-, glutamic-acid-, serine-, and threonine-rich domain that is important for protein-protein interactions (34, 35), and a C-terminal Ets DNA-binding domain (19, 20). The activity of PU.1 is primarily regulated posttranscriptionally by phosphorylation (reviewed in reference 26). While phosphorylation of two serine residues (positions 41 and 45) located at one of the acidic transactivation domains is necessary for macrophage colony-stimulating factor-dependent proliferation of bone marrow macrophages (4), a serine phosphorylation at position 148 in the proline-, glutamic-acid-, serine-, and threonine-rich domain mediates the interaction between PU.1 and the B-cell enhancer factor (NF-EM5). The latter interaction is required for optimal transactivation of the 3′ enhancer elements located in the immunoglobulin κ and λ light-chain genes (35). The serine 148 phosphorylation was also found to be required for the lipopolysaccharide-induced transactivation function of PU.1 (27). In contrast, in this study, we demonstrated that serine phosphorylation of PU.1 at position 142 instead of position 148 mediates, at least partially, the SIE-dependent IL-3 stimulation of mcl-1 reporter activity. While casein kinase II has been suggested to be responsible for phosphorylation of PU.1 at serine 148 (27, 35), our data indicate that the serine 142 phosphorylation is induced via activation of a p38MAPK-dependent pathway. The IL-3-induced in vivo phosphorylation of PU.1 is inhibited by SB203580, suggesting that p38α, p38β, or both mediate this phosphorylation event. However, the fact that IL-3-stimulated transactivation activity of PU.1 is inhibited by the dominant negative mutant form of p38α but not that of p38β (Fig. 8B) suggests that serine 142 phosphorylation is induced via a p38α-dependent pathway. On the other hand, while our results presented in Fig. 7 cannot distinguish whether PU.1 is directly phosphorylated by p38MAPK or by another associated kinase in the p38MAPK immunocomplex, the lack of phosphorylation of PU.1 at Ser-142 in an in vitro kinase assay using bacterium-produced p38α (data not shown) suggests that PU.1 is an indirect substrate of p38α. More experiments are required to reveal the identity of the putative p38α-associated kinase that catalyzes the phosphorylation of PU.1 at Ser-142 in response to IL-3 stimulation.
PU.1 binds to a cis element with a 5′-GGAA/T-3′ core sequence. Its binding to some gene promoters is constitutive (44, 46), whereas in some other cases, its binding activity is stimulated by extracellular stimuli or cytokines (18, 42). PU.1 binds to the SIE motif of the mcl-1 promoter irrespective of IL-3 stimulation, and IL-3 stimulates its phosphorylation-and-transactivation function. These results suggest that IL-3 stimulates the phosphorylation of PU.1 at serine 142, which either directly increases the transactivation activity of PU.1 or increases PU.1's ability to recruit another transcriptional activator and activates mcl-1 gene transcription in an indirect manner. PU.1 was shown to interact with many transcription factors or coactivators, including NF-EM5, Ets-1, NF-IL-6, HMG-I(Y), c-Jun, IRF-1, ICSBP, AML1, and CBP (2, 8, 25, 33, 35, 36, 51, 52). It remains to be determined what other factors are associated with PU.1 and mediate the SIE-dependent IL-3 stimulation of mcl-1 gene transcription. In the gel shift assay with the SIE probe, in addition to the B2 complex that contains PU.1, another specific B1 complex was consistently detected in the Ba/F3 cell extracts (Fig. 1). The formation of this B1 complex, like that of B2, is not influenced by the presence or absence of IL-3. It is not clear whether the B1 complex plays any role in the SIE-mediated IL-3 induction of mcl-1 gene transcription. If it does play a role in this process, the inability of the PU.1 antibody to supershift this complex suggests either that B1 does not contain PU.1 or that the PU.1 protein present in the B1 complex exists in a conformation that cannot be recognized by the antibody used in the assay. More experiments are required to address this issue.
An SIE-like element (termed SRE) is present in the human mcl-1 gene promoter (between nucleotides −105 and −92) (47). This DNA element was shown to be recognized by a protein complex containing serum response factor and Elk-1. Both serum response factor and Elk-1 act coordinately to affect both the basal activity and tetradecanoyl phorbol acetate inducibility of the human mcl-1 gene in human K-562 cells. Unlike the SIE motif reported in this study, the tetradecanoyl phorbol acetate inducibility of SRE in the human mcl-1 promoter was shown to be mediated through the extracellular signal-regulated kinase pathway (47). On the other hand, the STAT3 protein in the peripheral blood mononuclear cell extracts isolated from patients with large granular lymphocyte leukemia has been reported to bind to the murine SIE motif (9). In contrast, although STAT3 is activated by IL-3 in Ba/F3 cells (30; our unpublished data), no evidence suggests that it binds to SIE or plays a role in the IL-3 regulation of murine mcl-1 gene transcription (our unpublished results). Furthermore, Mcl-1 has been found to be expressed in many other cell types of nonhematopoietic origin (23, 39). Taken together, these results suggest that SIE-dependent mcl-1 gene transcription is likely to be regulated by different transcriptional factors in a cell type- and stimulation signal-dependent manner.
The function of PU.1 is pivotal to myeloid and lymphoid development, as many genes essential for these two processes are regulated by this transcription factor. Our identification of PU.1 as being involved in IL-3 regulation of mcl-1 gene expression suggests that Mcl-1 may also contribute to the development of myeloid and/or lymphoid cells. Mcl-1-deficient embryos did not survive beyond the peri-implantation stage of mouse development (39). This early embryo lethality precludes a direct assessment of Mcl-1 function during myeloid and lymphoid development. It would be interesting to investigate how severe mcl-1 gene expression is affected in PU.1-null embryos and whether overexpression of Mcl-1 would rescue any defects of these mutant phenotypes.
Acknowledgments
We thank Jiahuai Han for the p38 antibody used in the in vitro kinase assay and the plasmids expressing DN-p38α and DN-p38β; Koichi Nakajima and Toshio Hirano for the plasmid expressing DN-STAT3; Masato Kasuga for the Δp85 (DN-p85) expression vector; and Tzu-Hao Wang for the DN-JNK, DN-ERK1, and DN-ERK2 expression vectors.
This work was supported by intramural funds from Academia Sinica. J.-M. Wang was supported by a postdoctoral fellowship awarded by Academia Sinica.
REFERENCES
- 1.Bae, J., C. P. Leo, S. Y. Hsu, and A. J. Hsueh. 2000. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J. Biol. Chem. 275:25255-25261. [DOI] [PubMed] [Google Scholar]
- 2.Behre, G., A. J. Whitmarsh, M. P. Coghlan, T. Hoang, C. L. Carpenter, D. E. Zhang, R. J. Davis, and D. G. Tenen. 1999. c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J. Biol. Chem. 274:4939-4946. [DOI] [PubMed] [Google Scholar]
- 3.Bingle, C. D., R. W. Craig, B. M. Swales, V. Singleton, P. Zhou, and M. K. Whyte. 2000. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J. Biol. Chem. 275:22136-22146. [DOI] [PubMed] [Google Scholar]
- 4.Celada, A., F. E. Borras, C. Soler, J. Lloberas, M. Klemsz, C. van Beveren, S. McKercher, and R. A. Maki. 1996. The transcription factor PU.1 is involved in macrophage proliferation. J. Exp. Med. 184:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, J.-R., J.-M. Wang, S.-F. Lee, H.-W. Peng, Y.-H. Lin, C.-H. Chou, J.-C. Li, H.-M. Huang, C.-K. Chou, M.-L. Kuo, J. J.-Y. Yen, and H.-F. Yang-Yen. 1998. mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathway and is one component of the GM-CSF viability response. Mol. Cell. Biol. 18:4883-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H. M., P. Zhang, M. T. Voso, S. Hohaus, D. A. Gonzalez, C. K. Glass, D. E. Zhang, and D. G. Tenen. 1995. Neutrophils and monocytes express high levels of PU.1 (Spi-1) but not Spi-B. Blood 85:2918-2928. [PubMed] [Google Scholar]
- 7.DeKoter, R. P., H. J. Lee, and H. Singh. 2002. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity 16:297-309. [DOI] [PubMed] [Google Scholar]
- 8.Eklund, E. A., A. Jalava, and R. Kakar. 1998. PU.1, interferon regulatory factor 1, and interferon consensus sequence-binding protein cooperate to increase gp91(phox) expression. J. Biol. Chem. 273:13957-13965. [DOI] [PubMed] [Google Scholar]
- 9.Epling-Burnette, P. K., J. H. Liu, R. Catlett-Falcone, J. Turkson, M. Oshiro, R. Kothapalli, Y. Li, J.-M. Wang, H.-F. Yang-Yen, J. Karras, R. Jove, and T. P. Loughran, Jr. 2001. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J. Clin. Investig. 107:351-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galson, D. L., J. O. Hensold, T. R. Bishop, M. Schalling, A. D. D'Andrea, C. Jones, P. E. Auron, and D. E. Housman. 1993. Mouse β-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol. Cell. Biol. 13:2929-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagemeier, C., A. J. Bannister, A. Cook, and T. Kouzarides. 1993. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 90:1580-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hromas, R., A. Orazi, R. S. Neiman, R. Maki, C. Van Beveran, J. Moore, and M. Klemsz. 1993. Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1. Blood 82:2998-3004. [PubMed] [Google Scholar]
- 13.Huang, H.-M., C.-J. Huang, and J. J.-Y. Yen. 2000. Mcl-1 is a common target of stem cell factor and interleukin-5 for apoptosis prevention activity via MEK/MAPK and PI-3K/Akt pathways. Blood 96:1764-1771. [PubMed] [Google Scholar]
- 14.Huang, S., Y. Jiang, Z. Li, E. Nishida, P. Mathias, S. Lin, R. J. Ulevitch, G. R. Nemerow, and J. Han. 1997. Apoptosis signaling pathway in T cells is composed of ICE/Ced-3 family proteases and MAP kinase kinase 6b. Immunity 6:739-749. [DOI] [PubMed] [Google Scholar]
- 15.Jourdan, M., J. D. Vos, N. Mechti, and B. Klein. 2000. Regulation of Bcl-2-family proteins in myeloma cells by three myeloma survival factors: interleukin-6, interferon-alpha and insulin-like growth factor 1. Cell Death Differ. 7:1244-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karim, F. D., L. D. Urness, C. S. Thummel, M. J. Klemsz, S. R. McKercher, A. Celada, C. Van Beveren, R. A. Maki, C. V. Gunther, and J. A. Nye. 1990. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 4:1451-1453. [DOI] [PubMed] [Google Scholar]
- 17.Khaled, A. R., A. N. Moor, A. Li, K. Kim, D. K. Ferris, K. Muegge, R. J. Fisher, L. Fliegel, and S. K. Durum. 2001. Trophic factor withdrawal: p38 mitogen-activated protein kinase activates NHE1, which induces intracellular alkalinization. Mol. Cell. Biol. 21:7545-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, Y. M., H. S. Kang, S. G. Paik, K. H. Pyun, K. L. Anderson, B. E. Torbett, and I. Choi. 1999. Roles of IFN consensus sequence binding protein and PU.1 in regulating IL-18 gene expression. J. Immunol. 163:2000-2007. [PubMed] [Google Scholar]
- 19.Klemsz, M. J., S. R. McKercher, A. Celada, C. Van Beveren, and R. A. Maki. 1990. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell 61:113-124. [DOI] [PubMed] [Google Scholar]
- 20.Klemsz, M. J., and R. A. Maki. 1996. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol. Cell. Biol. 16:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kominato, Y., D. Galson, W. R. Waterman, A. C. Webb, and P. E. Auron. 1995. Monocyte expression of the human prointerleukin 1β gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1. Mol. Cell. Biol. 15:59-68. [PMC free article] [PubMed] [Google Scholar]
- 22.Kozopas, K. M., T. Yang, H. L. Buchan, P. Zhou, and R. W. Craig. 1993. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc. Natl. Acad. Sci. USA 90:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krajewski, S., S. Bodrug, M. Krajewska, A. Shabaik, R. Gascoyne, K. Berean, and J. C. Reed. 1995. Immunohistochemical analysis of Mcl-1 protein in human tissues: differential regulation of Mcl-1 and Bcl-2 protein production suggests a unique role for Mcl-1 in control of programmed cell death in vivo. Am. J. Pathol. 146:1309-1319. [PMC free article] [PubMed] [Google Scholar]
- 24.Leu, C. M., C. Chang, and C. Hu. 2000. Epidermal growth factor (EGF) suppresses staurosporine-induced apoptosis by inducing mcl-1 via the mitogen-activated protein kinase pathway. Oncogene 19:1665-1675. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, R. T., A. Andreucci, and B. S. Nikolajczyk. 2001. PU.1-mediated transcription is enhanced by HMG-I(Y)-dependent structural mechanisms. J. Biol. Chem. 276:9550-9557. [DOI] [PubMed] [Google Scholar]
- 26.Lloberas, J., C. Soler, and A. Celada. 1999. The key role of PU.1/SPI-1 in B cells, myeloid cells and macrophages. Immunol. Today 20:184-189. [DOI] [PubMed] [Google Scholar]
- 27.Lodie, T. A., R. Savedra, Jr., D. T. Golenbock, C. P. Van Beveren, R. A. Maki, and M. J. Fenton. 1997. Stimulation of macrophages by lipopolysaccharide alters the phosphorylation state, conformation, and function of PU.1 via activation of casein kinase II. J. Immunol. 158:1848-1856. [PubMed] [Google Scholar]
- 28.Macleod, K., D. Leprince, and D. Stehelin. 1992. The ets gene family. Trends Biochem. Sci. 17:251-256. [DOI] [PubMed] [Google Scholar]
- 29.McKercher, S. R., B. E. Torbett, K. L. Anderson, G. W. Henkel, D. J. Vestal, H. Baribault, M. Klemsz, A. J. Feeney, G. E. Wu, C. J. Paige, and R. A. Maki. 1996. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15:5647-5658. [PMC free article] [PubMed] [Google Scholar]
- 30.Nagata, Y., and K. Todokoro. 1996. Interleukin 3 activates not only JAK2 and STAT5, but also Tyk2, STAT1, and STAT3. Biochem. Biophys. Res. Commun. 221:785-789. [DOI] [PubMed] [Google Scholar]
- 31.Nye, J. A., J. M. Petersen, C. V. Gunther, M. D. Jonsen, and B. J. Graves. 1992. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 6:975-990. [DOI] [PubMed] [Google Scholar]
- 32.Petersson, M., C. Sundstrom, K. Nilsson, and L. G. Larsson. 1995. The hematopoietic transcription factor PU.1 is downregulated in human multiple myeloma cell lines. Blood 86:2747-2753. [PubMed] [Google Scholar]
- 33.Petrovick, M. S., S. W. Hiebert, A. D. Friedman, C. J. Hetherington, D. G. Tenen, and D. E. Zhang. 1998. Multiple functional domains of AML1: PU.1 and C/EBPα synergize with different regions of AML1. Mol. Cell. Biol. 18:3915-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pongubala, J. M., S. Nagulapalli, M. J. Klemsz, S. R. McKercher, R. A. Maki, and M. L. Atchison. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin κ 3′ enhancer activity. Mol. Cell. Biol. 12:368-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pongubala, J. M., C. Van Beveren, S. Nagulapalli, M. J. Klemsz, S. R. McKercher, R. A. Maki, and M. L. Atchison. 1993. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science 259:1622-1625. [DOI] [PubMed] [Google Scholar]
- 36.Rao, E., W. Dang, G. Tian, and R. Sen. 1997. A three-protein-DNA complex on a B cell-specific domain of the immunoglobulin mu heavy chain gene enhancer. J. Biol. Chem. 272:6722-6732. [DOI] [PubMed] [Google Scholar]
- 37.Ray, D., R. Bosselut, J. Ghysdael, M. G. Mattei, A. Tavitian, and F. Moreau-Gachelin. 1992. Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU.1. Mol. Cell. Biol. 12:4297-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds, J. E., T. Yang, L. Qian, J. D. Jenkinson, P. Zhou, A. Eastman, and R. W. Craig. 1994. Mcl-1, a member of the Bcl-2 family, delays apoptosis induced by c-Myc overexpression in Chinese hamster ovary cells. Cancer Res. 54:6348-6352. [PubMed] [Google Scholar]
- 39.Rinkenberger, J. L., S. Horning, B. Klocke, K. Roth, and S. J. Korsmeyer. 2000. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev. 14:23-27. [PMC free article] [PubMed] [Google Scholar]
- 40.Saccani, S., S. Pantano, and G. Natoli. 2001. Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med. 193:1351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott, E. W., M. C. Simon, J. Anastasi, and H. Singh. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265:1573-1577. [DOI] [PubMed] [Google Scholar]
- 42.Shackelford, R., D. O. Adams, and S. P. Johnson. 1995. IFN-γ and lipopolysaccharide induce DNA binding of transcription factor PU.1 in murine tissue macrophages. J. Immunol. 154:1374-1382. [PubMed] [Google Scholar]
- 43.Shin, M. K., and M. E. Koshland. 1993. Ets-related protein PU.1 regulates expression of the immunoglobulin J-chain gene through a novel Ets-binding element. Genes Dev. 7:2006-2015. [DOI] [PubMed] [Google Scholar]
- 44.Smith, M. F., Jr., V. S. Carl, T. Lodie, and M. J. Fenton. 1998. Secretory interleukin-1 receptor antagonist gene expression requires both a PU.1 and a novel composite NF-κB/PU.1/GA-binding protein binding site. J. Biol. Chem. 273:24272-24279. [DOI] [PubMed] [Google Scholar]
- 45.Stephanou, A., T. M. Scarabelli, B. K. Brar, Y. Nakanishi, M. Matsumura, R. A. Knight, and D. S. Latchman. 2001. Induction of apoptosis and Fas receptor/Fas ligand expression by ischemia/reperfusion in cardiac myocytes requires serine 727 of the STAT-1 transcription factor but not tyrosine 701. J. Biol. Chem. 276:28340-28347. [DOI] [PubMed] [Google Scholar]
- 46.Stutz, A. M., and M. Woisetschlager. 1999. Functional synergism of STAT6 with either NF-κB or PU.1 to mediate IL-4-induced activation of IgE germline gene transcription. J. Immunol. 163:4383-4391. [PubMed] [Google Scholar]
- 47.Townsend, K. J., P. Zhou, L. Qian, C. K. Bieszczad, C. H. Lowrey, A. Yen, and R. W. Craig. 1999. Regulation of MCL1 through a serum response factor/Elk-1-mediated mechanism links expression of a viability-promoting member of the BCL2 family to the induction of hematopoietic cell differentiation. J. Biol. Chem. 274:1801-1813. [DOI] [PubMed] [Google Scholar]
- 48.Visconti, R., M. Gadina, M. Chiariello, E. H. Chen, L. F. Stancato, J. S. Gutkind, and J. J. O'Shea. 2000. Importance of the MKK6/p38 pathway for interleukin-12-induced STAT4 serine phosphorylation and transcriptional activity. Blood 96:1844-1852. [PubMed] [Google Scholar]
- 49.Wang, J.-M., J.-R. Chao, W. Chen, M.-L. Kuo, J. J.-Y. Yen, and H.-F. Yang-Yen. 1999. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol. Cell. Biol. 19:6195-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Y., S. Huang, V. P. Sah, J. Ross, Jr., J. H. Brown, J. Han, and K. R. Chien. 1998. Cardiac muscle cell hypertrophy and apoptosis induced by distinct members of the p38 mitogen-activated protein kinase family. J. Biol. Chem. 273:2161-2168. [DOI] [PubMed] [Google Scholar]
- 50a.Xing, J., J. M. Kornhauser, Z. Xia, E. A. Thiele, and M. E. Greenberg. 1998. Nerve growth factor activates extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways to stimulate CREB serine 133 phosphorylation. Mol. Cell. Biol. 18:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto, H., F. Kihara-Negishi, T. Yamada, Y. Hashimoto, and T. Oikawa. 1999. Physical and functional interactions between the transcription factor PU.1 and the coactivator CBP. Oncogene 18:1495-1501. [DOI] [PubMed] [Google Scholar]
- 52.Yang, Z., N. Wara-Aswapati, C. Chen, J. Tsukada, and P. E. Auron. 2000. NF-IL6 (C/EBPβ) vigorously activates il1b gene expression via a Spi-1 (PU.1) protein-protein tether. J. Biol. Chem. 275:21272-21277. [DOI] [PubMed] [Google Scholar]
- 53.Yordy, J. S., and R. C. Muise-Helmericks. 2000. Signal transduction and the Ets family of transcription factors. Oncogene 19:6503-6513. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, M., L. New, V. V. Kravchenko, Y. Kato, H. Gram, F. di Padova, E. N. Olson, R. J. Ulevitch, and J. Han. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, P., L. Qian, K. M. Kozopas, and R. W. Craig. 1997. Mcl-1, a Bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood 89:630-643. [PubMed] [Google Scholar]