Abstract
The splicing of the c-src exon N1 is controlled by an intricate combination of positive and negative RNA elements. Most previous work on these sequences focused on intronic elements found upstream and downstream of exon N1. However, it was demonstrated that the 5′ half of the N1 exon itself acts as a splicing enhancer in vivo. Here we examine the function of this regulatory element in vitro. We show that a mutation in this sequence decreases splicing of the N1 exon in vitro. Proteins binding to this element were identified as hnRNP A1, hnRNP H, hnRNP F, and SF2/ASF by site-specific cross-linking and immunoprecipitation. The binding of these proteins to the RNA was eliminated by a mutation in the exonic element. The activities of hnRNP A1 and SF2/ASF on N1 splicing were examined by adding purified protein to in vitro splicing reactions. SF2/ASF and another SR protein, SC35, are both able to stimulate splicing of c-src pre-mRNA. However, splicing activation by SF2/ASF is dependent on the N1 exon enhancer element whereas activation by SC35 is not. In contrast to SF2/ASF and in agreement with other systems, hnRNP A1 repressed c-src splicing in vitro. The negative activity of hnRNP A1 on splicing was compared with that of PTB, a protein previously demonstrated to repress splicing in this system. Both proteins repress exon N1 splicing, and both counteract the enhancing activity of the SR proteins. Removal of the PTB binding sites upstream of N1 prevents PTB-mediated repression but does not affect A1-mediated repression. Thus, hnRNP A1 and PTB use different mechanisms to repress c-src splicing. Our results link the activity of these well-known exonic splicing regulators, SF2/ASF and hnRNP A1, to the splicing of an exon primarily controlled by intronic factors.
A common mechanism of the regulation of gene expression in metazoans is the alternative splicing of pre-mRNA (6, 11, 22, 23, 34). Through alternative splicing, multiple mRNAs are generated from the same primary RNA transcript. Changes in splice site choice are regulated by proteins that bind to the pre-mRNA and affect spliceosome assembly. One well-studied family of splicing regulatory factors is the SR proteins that function in both constitutive and alternative splicing and can act at various stages of spliceosome assembly (3, 10, 24, 48). These proteins are perhaps best known for binding exonic splicing enhancer (ESE) elements and thus stimulating spliceosome assembly at the adjacent splice sites. Some exons bind multiple SR proteins, each of which can activate splicing. Other exons are dependent on a single SR protein. Two of the best-characterized SR protein family members that bind ESEs and activate splicing are SF2/ASF and SC35.
Another large group of proteins that bind to nascent pre-mRNAs are the heterogeneous nuclear ribonucleoproteins (hnRNPs) (19, 30). At least in vitro, spliceosome assembly occurs after hnRNP binding and some hnRNPs are implicated in splicing regulation. Two of these, hnRNP A1 and polypyrimidine tract binding protein (PTB or hnRNP I) bind to exonic splicing silencer or intronic splicing silencer elements and thus repress the splicing of certain alternatively spliced exons (10, 46). The mechanisms that hnRNP A1 and PTB use to mediate splicing repression are not fully understood. Several models have been proposed (10, 50). One mechanism involves direct interactions between positive and negative factors at adjacent binding sites (9, 14, 18, 57). The concentration of the factors and their relative affinities for their binding sites are what determine exon inclusion or skipping. In other systems, the assembly of multimeric A1 or PTB complexes onto sites surrounding an alternatively spliced exon may cause the intervening region of RNA to loop out and thus prevent splice site recognition (2, 17, 22, 52). Another possible mechanism of repression by hnRNP A1 involves binding to a high-affinity site within an exon and then promoting the assembly of additional hnRNP A1 molecules along the pre-mRNA (57). This is thought to create a repressed zone of RNA refractory to the binding of the general splicing machinery at the splice sites. SF2/ASF can block this cooperative propagation of hnRNP A1 along the exon, thereby activating splicing. Both SR proteins and hnRNPs vary in concentration between different cell types, and it is thought that this differential expression of hnRNPs and SR proteins affects the alternative splicing of many pre-mRNAs (7, 25, 28, 37, 38, 56).
The N1 exon of c-src provides a model system for the study of neuron-specific splicing regulation. The 18-nucleotide exon, N1, is included in the mRNA in neurons but is skipped in nonneuronal cells (31, 36). Two intronic sequences are required for regulation of N1 splicing (1, 12, 42, 43). The 3′ splice site region upstream of the exon is required for repression of N1 in nonneuronal cells. It has been shown previously that PTB binds to CUCUCU elements (CU) within this sequence and is required for splicing repression in vitro (13, 17). The second intronic sequence, between 17 and 142 nucleotides downstream of N1, acts as a splicing enhancer and is also required for splicing repression (17). Within this element is a highly conserved region between nucleotides 37 and 70 called the downstream control sequence (DCS). The DCS contains a CU element that is required for splicing repression. Flanking this CU element are the elements GGGGGCUG and UGCAUG, which are needed for enhancer activity. Each of these elements binds to specific RNA binding proteins: hnRNP F and hnRNP H bind to the G-rich element (16, 35, 40), KH-type splicing-regulatory protein binds to the UGCAUG element (35, 41), and PTB or its neural homolog, nPTB, binds to the CU element (17, 35).
Studies of splicing regulation, including those on src N1, have generally focused on either intronic elements that bind PTB and other factors or on exonic elements that bind SR proteins, hnRNP A1, and other proteins. However, it is likely that in many systems the exonic and intronic elements are functioning together. Indeed, in addition to the intronic elements, an element within the N1 exon itself also acts as a splicing enhancer element. When this purine-rich sequence from exon N1, GAGGAAGGUG, is placed within a similarly sized test exon of a heterologous minigene, it activates splicing in vivo (42, 43). Splicing of the test exon was strongest when both intronic and exonic elements were placed together in the construct, suggesting that they normally work in combination (42). The enhancement of the test exon by these elements varied between the different neuronal and nonneuronal cell lines tested, but they do not appear to be highly neuron specific.
In this study, we further examine the role of the exonic enhancer in the regulation of exon N1 splicing. We identify the proteins SF2/ASF, hnRNP A1, hnRNP A1B, hnRNP H, and hnRNP F as binding to the element in splicing extracts. We show that SF2/ASF and SC35 can activate splicing of c-src pre-mRNA and that SF2/ASF enhances the splicing pathway leading to inclusion of N1. Both hnRNP A1 and PTB can counteract the activity of the SR proteins to repress splicing but apparently do so by different mechanisms.
MATERIALS AND METHODS
DNA constructs and in vitro transcription.
The BS3, BS7, BS9, and BS27 constructs have been previously described (12). BS7MUT, BS27MUT, BS9MUT, BS303, and BS304 were generated by PCR from the BS7, BS27, and BS3 constructs by standard methods. Plasmids were linearized with NotI (New England Biolabs) in src exon 4 for T7 RNA polymerase transcription.
In vitro splicing.
Nuclear extracts were prepared and in vitro splicing reactions were conducted as previously described (12), and the reaction mixtures contained either ∼100 μg of Weri-1 nuclear extract or ∼300 μg of Weri-1 S100 extract plus the proteins described in the text in a total volume of 25 μl.
Site-specific labeling and UV cross-linking.
Site-specifically labeled c-src pre-mRNA containing a single 32P-labeled guanosine was synthesized (44). The reaction conditions for UV cross-linking were the same as those for splicing. The site-specifically labeled RNAs were incubated between 1 and 120 min. After incubation, heparin sulfate was added to a concentration of 0.25 mg/ml and incubated for an additional 5 min on ice. The samples were then irradiated with 254-nm-wavelength UV light on ice for 15 min by using a handheld UV lamp (UVP). The samples were then treated with 1 μg of RNase A for 30 min at 30°C. The cross-linked proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% or 4 to 20% polyacrylamide) and visualized by PhosphorImager (Molecular Dynamics).
Immunoprecipitation.
The immunoprecipitation experiments used rabbit polyclonal antibodies raised against the hnRNP A1 C-terminal peptide, the AK-96 mouse monoclonal SF2/ASF antibody (Zymed), rabbit polyclonal antibodies against the C-terminal peptide of hnRNP H, rabbit polyclonal antibodies against the C-terminal peptide of hnRNP F, rabbit polyclonal antibodies against the C-terminal peptide of U170K, and the 4F4 mouse monoclonal hnRNP C1/C2 (a gift from G. Dreyfuss). In a typical experiment, 20 μl of GammaBind Plus Sepharose (Amersham Pharmacia), preequilibrated with immunoprecipitation buffer (IP buffer) (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100) and blocked with 2.5 mg of bovine serum albumin per ml in IP buffer, was preincubated with 5 to 20 μl of antibody for 1 h at 4°C. The beads were washed three times with 1 ml of IP buffer to remove unbound antibody. The UV-irradiated, RNase-digested splicing reaction mixture was incubated with the beads for 1 h at 4°C. The beads were then washed again three times with 1 ml of IP buffer. Proteins were eluted from the beads with SDS-PAGE loading buffer. Twenty microliters of rat anti-mouse beads (Dynal) were used in the mAb104 (a gift from M. Roth) immunoprecipitation experiment.
Protein purification.
hnRNP A1 cDNA (a gift from A. Mayeda) was fused to a six-His tag by using standard PCR methods. hnRNP A1-His was expressed in BL21-pLysS bacteria (Novagen) and was purified by using standard techniques (Qiagen). His-tagged La was expressed in BL21 bacteria (Novagen) and was similarly purified. PTB was purified from 10 ml of HeLa nuclear extract (∼200 mg of protein) as previously described (35). SC35 and SF2/ASF were isolated by using the baculovirus expression system. SC35 baculovirus (a gift from X.-D. Fu) was used to infect 50 ml of Sf9 cells and was purified as previously described (55). SF2/ASF was cloned into the pFastBac1 insect expression vector (Invitrogen) after PCR amplification from Weri-1 poly(A)+ RNA by using primers 5′-GATCGGATCCATATGTCGGGAGGTGGTGTG-3′ and 5′-CTAGGAATTCTTATGTACGAGAGCGAGATCTGCTATG-3′. Baculovirus containing the SF2/ASF cDNA was used to infect Sf9 insect cells. Purification of SF2/ASF from 50 ml of SF9 cell culture was performed as previously described (55). The activity of the SF2/ASF protein varied between preparations and was optimized for each experiment.
RESULTS
Identification of proteins binding to the purine-rich 5′ half of exon N1.
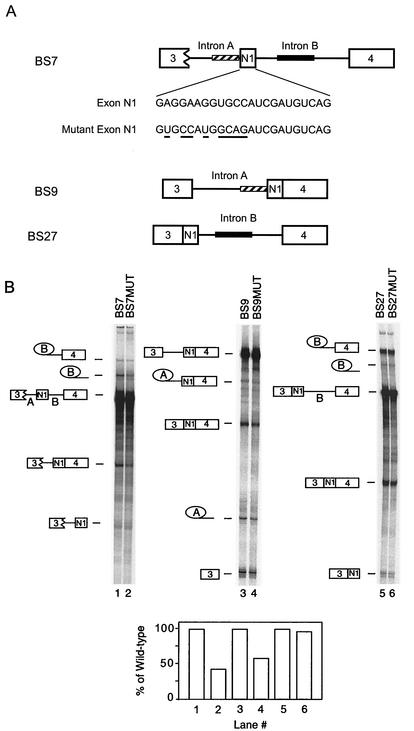
To determine whether the exonic element affected c-src splicing in vitro as it did in vivo, the element was mutated in several pre-mRNAs. The nucleotides chosen to be mutated were within the minimal element needed for splicing activation in vivo (42) and one adjacent nucleotide downstream of the element. The wild-type and mutant RNAs were incubated in neuronal (Weri-1 cell) splicing reaction mixtures, and the RNAs were extracted and separated by denaturing PAGE. Figure 1A shows the pre-mRNAs tested in vitro and the nucleotide changes made in N1. BS7 contains the N1 exon and downstream exon 4 as well as the regulatory sequences in the downstream intron and in the 3′ splice site upstream of N1. This substrate shows proper regulation of the exon when tested in Weri-1 and HeLa splicing extracts. BS9 and BS27 have the downstream and upstream exons, respectively, prespliced to exon N1. This deletes one of the two essential intronic regulatory sequences in each construct. Both of these RNAs show unregulated or constitutive splicing in both HeLa and Weri-1 nuclear extracts (1, 12). Mutation of the exonic enhancer element in BS7 reduced the splicing of intron B downstream of N1 by 57% (Fig. 1B, lanes 1 and 2). The same mutation in BS9 reduced the splicing of intron A upstream of N1 by 41%. Interestingly, the mutation had almost no effect on intron B in BS27, which is missing the repressor sequences upstream of N1 (Fig. 1B, compare lanes 3 and 4 and lanes 5 and 6). The enhancer appears to affect the 3′ splice site upstream of the N1 exon, either to prevent its repression activity in BS7 or to activate its splicing in BS9.
FIG. 1.
The N1 exonic enhancer element functions in vitro. (A) The pre-mRNAs used in the experiment are diagrammed. The nucleotides in exon N1 and mutant N1 exon are also shown, and the nucleotide changes in the mutant exon are underlined. The wild-type exon contains an inserted ClaI site 3′ of the mutation. This insertion has been previously shown not to affect N1 splicing (1a). The striped bar represents the upstream acceptor sequence, and the black bar represents the downstream enhancer sequence. (B) Splicing of pre-mRNAs with either a wild-type exon or a mutated exon in Weri-1 nuclear extract. Lanes 1 and 2, BS7 and BS7MUT; lanes 3 and 4, BS9 and BS9MUT; lanes 5 and 6, BS27 and BS27MUT. The products and intermediates are shown to the side of the gels. The percentage of RNA converted to mRNA was determined and compared between pairs of substrates, with the value for the mutant shown as a percentage of that for the wild-type.
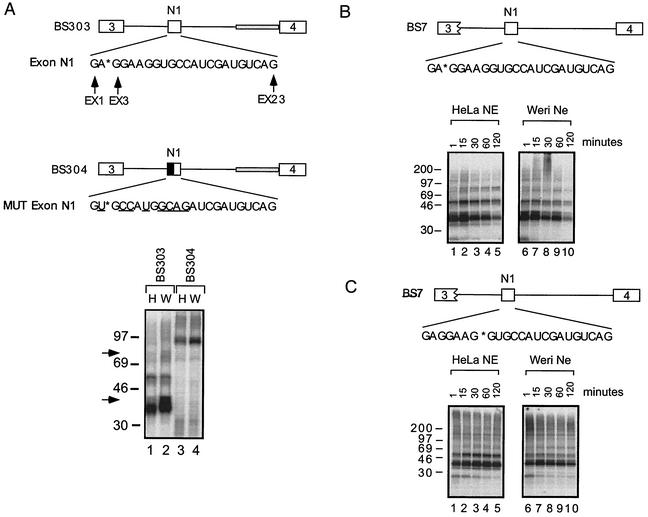
To identify proteins binding to the N1 enhancer sequence, UV cross-linking was performed on RNAs site-specifically labeled between nucleotides 2 and 3 of the exon (position EX3). BS303 contains three complete exons and shows regulated inclusion of the central exon N1 between the HeLa and Weri-1 extracts. BS303 pre-mRNA contains a wild-type exon N1, and BS304 contains the mutated exon N1. Two major groups of bands corresponding to proteins that are 33 to 40 kDa and 50 to 55 kDa in size are seen to cross-link to BS303. These proteins do not cross-link to BS304, where a 90- to 100-kDa protein is seen instead (Fig. 2A, compare lanes 1 and 2 with lanes 3 and 4). Interestingly in the BS303 reactions, a strong band at about 38 to 40 kDa and a weaker band at 75 kDa are enriched in the Weri-1 extract compared with the HeLa extract (Fig. 2A, compare lanes 1 and 2).
FIG. 2.
Proteins cross-link specifically to N1. (A) UV cross-linking to BS303 and BS304 site-specifically labeled at position EX3 in HeLa (H) and Weri-1 (W) nuclear extracts. The grey bar in intron B in BS303 and BS304 represents adenovirus acceptor sequence. This sequence has previously been shown to increase overall splicing levels but not to affect neural specificity. The nucleotides in the exon are designated EX1 through EX23. The mutated nucleotides are underlined. The asterisk denotes the site of the radioactive phosphate in the exon N1 sequence. Arrows highlight protein bands enriched in Weri-1 extract in lane 2. (B) UV cross-linking to BS7 labeled at position EX3 in HeLa and Weri-1 nuclear extracts. (C) UV cross-linking to BS7 site-specifically labeled at EX8 in HeLa and Weri-1 nuclear extracts.
We examined the binding of the proteins to different exon positions and over time by using a site-specifically labeled BS7 RNA. Cross-linking of BS7 RNA labeled at either position EX3 or EX8 again resulted in prominent bands at 35 and 55 kDa in both HeLa and Weri-1 nuclear extracts (Fig. 2B and C). These bands appeared within 1 min of incubation and peaked by 15 min. For the EX3 label, the bands decreased at longer incubation times. Although there were differences in some of the minor bands, the major cross-linked bands were the same for the two label positions.
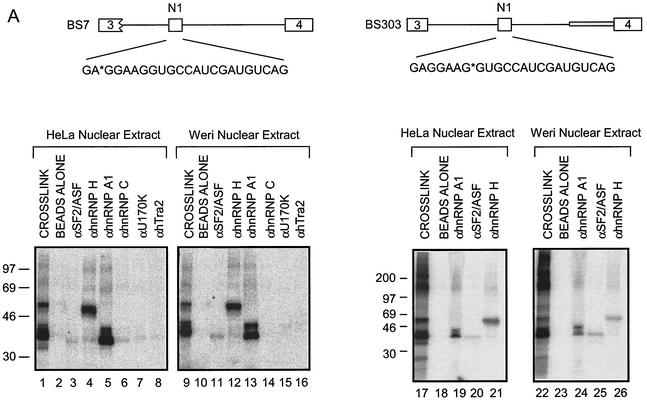
To identify the proteins cross-linking to exon N1, immunoprecipitation experiments were performed after cross-linking. The immunoprecipitated proteins were resolved by SDS-PAGE. The 35- and 38-kDa proteins that cross-link to the RNA labeled at EX3 were immunoprecipitated with antibodies to hnRNP A1 (Fig. 3A, lanes 5 and 13). Thus, the most prominent bands in this size range are likely hnRNP A1 and its longer isoform, hnRNP A1B (5). Antibody to hnRNP H immunoprecipitates the 55-kDa band (Fig. 3A, lanes 4 and 12), and antibody to hnRNP F immunoprecipitates a lighter 53-kDa band (data not shown). Given its activity as a splicing enhancer, it was likely that the exonic element bound to an SR protein. Indeed, immunoprecipitation of the cross-linked proteins identified SF2/ASF as another component of the 33- to 35-kDa band. This band was lighter than that of the comigrating hnRNP A1. This could be due to a lower stoichiometry of binding, poorer cross-linking, or poorer immunoprecipitation with this antibody. The immunoprecipitation experiments were repeated with RNA labeled at position EX8. The proteins cross-linking in the vicinity of this radiolabel were again immunoprecipitated with antibodies to hnRNP A1, SF2/ASF, and hnRNP H (Fig. 3A, lanes 19 to 21 and 24 to 26).
FIG. 3.
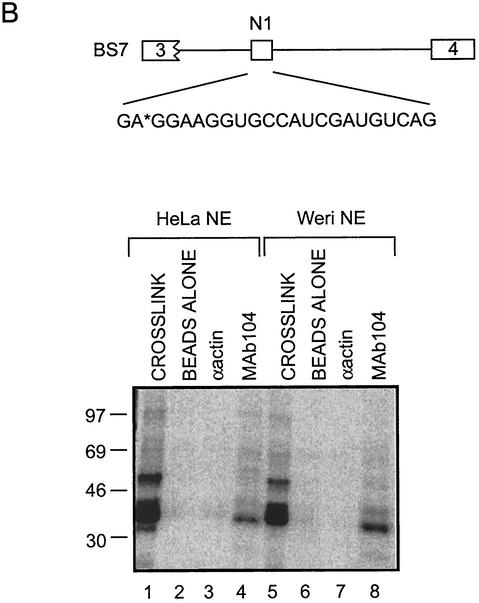
SR proteins and hnRNPs bind to exon N1. (A) Immunoprecipitation of proteins cross-linked to positions X3 and X8. Lanes 1, 9, 17, and 22 contain 1/10 of the total cross-linking reaction mixture. Other lanes contain the bound material from immunoprecipitates using the antibodies indicated. (B) A 35-kDa SR protein binds to exon N1. MAb104 was used to immunoprecipitate RS domain proteins that cross-link to position EX3 in BS7.
The immunoprecipitated SF2/ASF band was relatively weak with the SF2/ASF-specific antibody. To confirm that an SR protein binds to the N1 enhancer, the immunoprecipitation was repeated by using mAb104, an antibody that recognizes the common epitope of SR proteins (45). This antibody again immunoprecipitated the 35-kDa band (Fig. 3B, lanes 4 and 8). This confirms the binding of SF2/ASF at the enhancer, although other RS domain proteins could also be present. Antibodies to U2AF35 and U2AF65 were also tested in these experiments but failed to immunoprecipitate any protein (data not shown). Two-dimensional electrophoresis of the cross-linked proteins demonstrated that hnRNP A1 represents the majority of the band migrating at 35 kDa (data not shown).
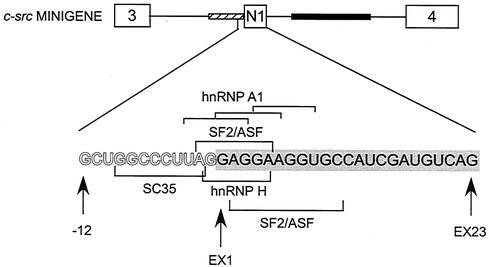
The binding of SF2/ASF and hnRNPs A1, H, and F to exon N1 was not unexpected. SF2/ASF and hnRNPs A1 and H have been implicated in splicing regulation through exonic elements (9, 15, 18, 27, 47, 49, 50, 57). Moreover, examination of the exon N1 sequence and the nucleotides just upstream reveals the sequence AG/GAGGA. This is identical to an SF2/ASF-dependent exonic enhancer identified in a functional SELEX screen by Liu et al.(33) (Fig. 4). In the c-src pre-mRNA, this SF2/ASF motif is not wholly exonic, overlapping two nucleotides of the 3′ splice site. A sequence starting slightly downstream is an 8-of-10-nucleotide match to AGGACRRAGC, another SF2/ASF high-affinity binding site identified by SELEX (49). Interestingly, in the intron upstream of these SF2/ASF binding motifs is an element with a 7-of-8-nucleotide match to the SC35 ESE consensus sequence, GRYYMCYR (R = A/G, Y = C/U, and M = A/C) (32). Overlapping all of these SR protein binding motifs is a series of sequences that each show a 4-of-6-nucleotide match to an hnRNP A1 binding sequence, UAGGGA/U (4). Also, within the exon, there are sequences showing 3-of-4-nucleotide match to the GGGA element identified as an hnRNP H/F binding site (8).
FIG. 4.
Diagram of the 3′ splice site and exon N1. Potential binding sites for hnRNP A1, SF2/ASF, SC35, and hnRNP H in the 3′ splice site of exon N1 and within the exon itself are bracketed. Intron nucleotides at the 3′ splice site are in outline. The nucleotides of exon N1 are filled and encompassed in a grey bar. In the c-src minigene diagram, the upstream acceptor sequence is represented by a striped bar and the downstream enhancer sequence is represented by a solid bar.
SR proteins affect exon N1 splicing in vitro.
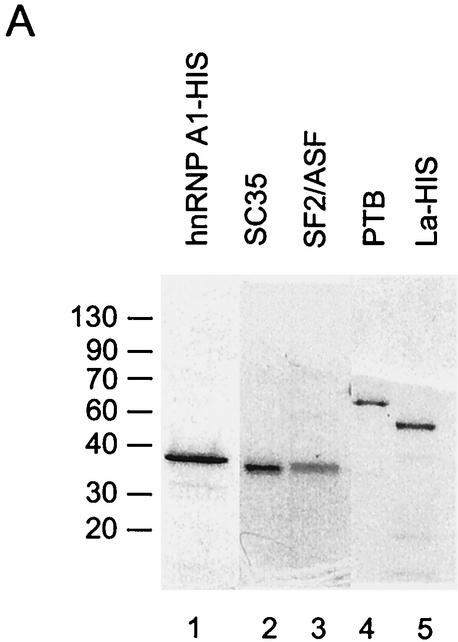
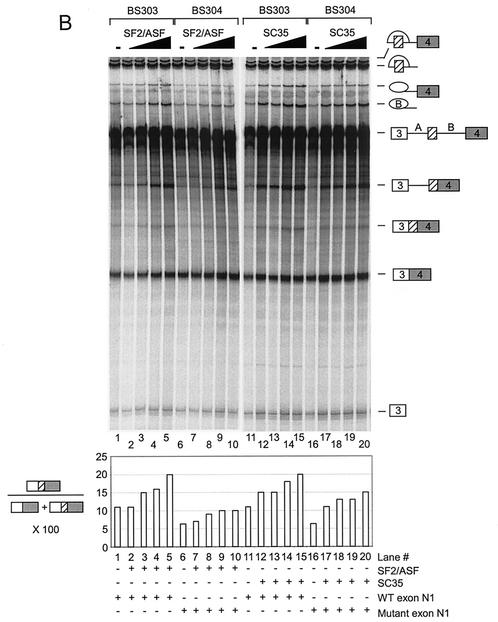
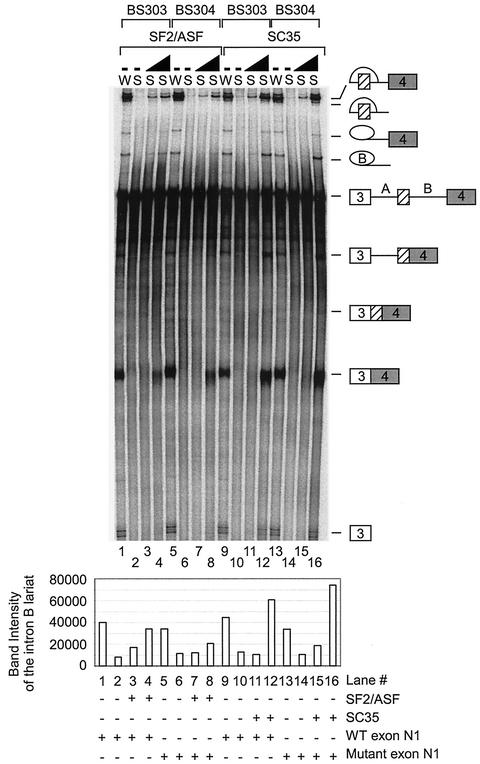
To analyze the roles of SF2/ASF and SC35, we tested the effect of the purified proteins on N1 exon splicing in vitro. The purified proteins are shown in Fig. 5A. Pre-mRNAs (BS303 and BS304) containing three exons and two introns were used to follow the splicing pathways leading to either the inclusion or skipping of exon N1. Increasing concentrations of purified SF2/ASF were added to Weri-1 in vitro splicing reaction mixtures containing either BS303 or BS304. The addition of SF2/ASF to BS303 splicing reaction mixtures enhanced the inclusion of exon N1 relative to the exon-skipping pathway. Mutation of the enhancer sequence in BS304 RNA significantly reduced the stimulation by SF2/ASF (Fig. 5B, compare lanes 2 to 5 with lanes 7 to 10). This suggests that SF2/ASF binding to the exon sequence can specifically enhance splicing of exon N1 over the exon-skipping pathway. The residual enhancement by SF2/ASF on BS304 likely implies that there are additional binding sites for SF2/ASF in the transcript. However, splicing of the mutant RNA at any SF2/ASF concentration is below the level for the wild-type RNA in the absence of added SF2/ASF. SF2/ASF also enhanced splicing in HeLa extracts and showed the same difference in activity between BS303 and BS304 (data not shown).
FIG. 5.
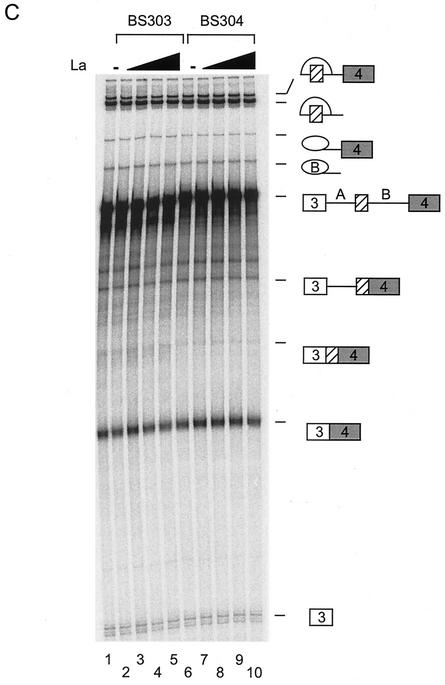
SF2/ASF and SC35 differ in their enhancement of N1. (A) Coomassie staining of purified proteins used in the experiment and separated in an SDS polyacrylamide gel. (B) In vitro splicing reactions in the presence of added SR proteins. Splicing of BS303 (lanes 1 to 5 and 11 to 15) and BS304 (lanes 6 to 10 and 16 to 20) was done in the presence of 100 μg of Weri-1 nuclear extract. Lanes 2 to 5 and 7 to 10 both contain 50, 100, 200, and 400 ng of SF2/ASF, respectively. Lanes 12 to 15 and 17 to 20 both contain 50, 100, 200, and 400 ng of SC35, respectively. Lanes 1, 6, 11, and 16 contain Weri-1 extract without any additional factors. Quantification of the products is shown below the gel. (C) La does not affect splicing of BS303 or BS304. Splicing of BS303 and BS304 was done in the presence of 100 μg of Weri-1 nuclear extract. Lanes 2 to 5 and 7 to 10 both contain 0.200, 0.400, 0.800, and 2 μg of La, respectively. Lanes 1 and 6 contain Weri-1 extract without any additional factors.
Increasing concentrations of SC35 were also added to the Weri-1 in vitro splicing reaction mixtures containing either BS303 or BS304. Interestingly, SC35 strongly enhanced exon N1 inclusion for both BS303 and BS304 (Fig. 5B, lanes 12 to 15 and 17 to 20). The BS304 mutation reduced N1 activation by SC35 somewhat but not as consistently as for SF2/ASF (Fig. 5B, compare lanes 15 and 20). In this case, the added SC35 could activate the mutant exon splicing to levels above that of the wild-type splicing in extract alone (Fig. 5B, lanes 12 to 15 and 17 to 20). SC35 also activated splicing by the exon-skipping pathway more strongly than did SF2/ASF. Some of the splicing enhancement by SC35 is likely due to protein binding to the predicted SC35 site upstream of N1, as mutation of this site reduces SC35 activation (data not shown).
To demonstrate that these changes in splicing pattern were specific to SR proteins, the RNA binding protein, La, was added to Weri-1 splicing reaction mixtures. The addition of La did not change the splicing of either BS303 or BS304 at any concentration tested (Fig. 5C). These experiments were repeated multiple times in different extracts and with different preparations of SR proteins. Although the overall level of splicing varied between experiments, the same relative stimulation activities of SF2/ASF and SC35 were always observed.
Weri-1 extract contains both SF2/ASF and SC35, as determined by Western blotting (data not shown). S100 extracts are depleted for SR proteins and can be supplemented with individual SR protein family members to activate splicing (29). This complementation assay was used to confirm whether either SF2/ASF or SC35 alone can activate c-src splicing. Increasing concentrations of SF2/ASF were added to Weri-1 S100 reaction mixtures containing either BS303 or BS304. In reaction mixtures containing BS303, SF2/ASF activated splicing pathways leading to both inclusion and skipping of exon N1 (Fig. 6, lanes 3 and 4). In the reactions containing BS304, SF2/ASF activated splicing by the exon-skipping pathway more strongly than N1 exon inclusion (Fig. 6, lanes 7 and 8). The addition of SC35 to Weri-1 S100 activated splicing through both pathways regardless of whether the substrate was BS303 or BS304. These results confirm that the exonic enhancer element affects splicing activation by SF2/ASF.
FIG. 6.
SF2/ASF and SC35 activate splicing of N1 in S100 extract. Splicing of BS303 (lanes 1 to 4 and 9 to 12) and BS304 (lanes 5 to 8 and 13 to 16) was done in the presence of 100 μg of either Weri-1 nuclear extract (lanes 1, 5, 9, and 13) or Weri-1 S100 extract (lanes 2 to 4, 6 to 8, 10 to 12, and 14 to 16). Lanes 3 and 7 contain 400 ng of SF2/ASF; lanes 4 and 8 contain 800 ng of SF2/ASF; lanes 11 and 15 contain 300 ng of SC35; lanes 12 and 16 contain 600 ng of SC35; and lanes 2, 6, 10, and 14 contain Weri S100 extract without any additional factors. Quantification of the intron B lariat is shown below the gel.
hnRNP A1 represses c-src splicing in vitro.
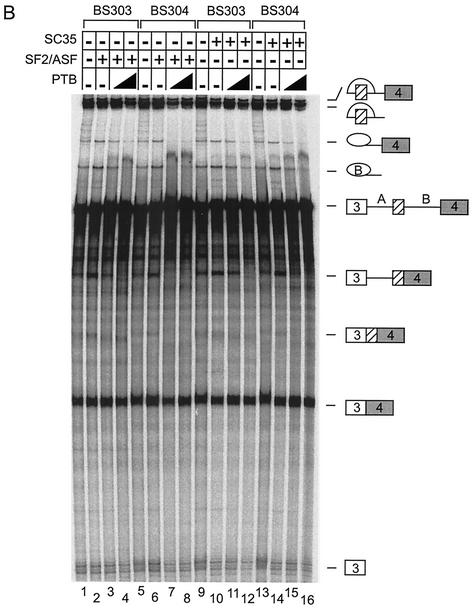
hnRNP A1 has been described as a splicing repressor that is antagonistic to splicing activation by SR proteins (37, 38, 57). To determine whether this also occurs for the N1 exon, hnRNP A1 was added to splicing reactions activated by SF2/ASF or SC35. The addition of increasing concentrations of hnRNP A1 to BS303 or BS304 splicing reactions, activated by SF2/ASF, repressed the inclusion of exon N1 (Fig. 7A, lanes 3, 4, 7, and 8). Note that under some conditions the mRNA product skipping N1 is less stable than the other spliced exon products. By following the lariats and the 5′ exon intermediate, it is clear that hnRNP A1 is more inhibitory for exon N1 splicing than for the exon-skipping pathway.
FIG. 7.
HnRNP A1 can repress splicing of exon N1. (A) Splicing of BS303 (lanes 1 to 4 and 9 to 12) and BS304 (lanes 5 to 8 and 13 to 16) in 100 μg of Weri-1 nuclear extract. Lanes 2 to 4 and 6 to 8 contain 400 ng of SF2/ASF. Lanes 10 to 12 and 14 to 16 contain 300 ng of SC35. Lanes 3 and 4, 7 and 8, 11 and 12, and 15 and 16 contain 100 and 200 ng, respectively, of hnRNP A1. Lanes 1, 5, 9, and 13 contain Weri-1 nuclear extract without additional factors. (B) Splicing of BS303 (lanes 1 to 4 and 9 to 12) and BS304 (lanes 5 to 8 and 13 to 16) in 100 μg of Weri-1 nuclear extract. Lanes 2 to 4 and 6 to 8 contain 400 ng of SF2/ASF. Lanes 10 to 12 and 14 to 16 contain 300 ng of SC35. Lanes 3 and 4, 7 and 8, 11 and 12, and 15 and 16 contain 200, and 1 μg, respectively, of PTB. Lanes 1, 5, 9, and 13 contain Weri-1 nuclear extract without additional factors.
The addition of hnRNP A1 also repressed N1 splicing in reactions activated by SC35 (Fig. 7A, lanes 11, 12, 15, and 16). Since BS304 has a mutation in the known hnRNP A1 binding site in the N1 exon, this site is evidently not essential to splicing repression by A1. Thus, although the inhibition by hnRNP A1 is directed to the exon inclusion pathway, its site of action is not clear. Additional cross-linking experiments to RNAs containing wild-type or mutant N1 exons indicated that although the predominant site of A1 binding is in the 5′ end sequence, there is additional binding elsewhere in the exon (data not shown). HnRNP A1 addition also repressed splicing of a three-exon pre-mRNA derived from β-globin. Examination of both the src and β-globin gene sequences identified multiple potential hnRNP A1 binding sites in the exons and introns. The multitude of potential hnRNP A1 binding sites makes it difficult to dissect the specific role played by the protein in the N1-splicing pathway. It is likely similar to the repression seen in other systems. However, the A1 may be distributed over multiple weak sites without the single strong site seen in systems such as human immunodeficiency virus (HIV) Tat exon 3 (57).
PTB is also known to repress N1 splicing (13, 17). Adding PTB instead of hnRNP A1 to the reactions gave similar results. In reactions activated by either SF2/ASF or SC35, PTB repressed splicing of exon N1 (Fig. 7B, lanes 3, 4, 7, and 8). N1 splicing was most easily repressed by PTB in reactions containing BS304 and SF2/ASF. Thus, hnRNP A1 and PTB can each repress N1 exon splicing that has been stimulated by SR proteins. In neither case is the 5′ element of the N1 exon required for the repression activity.
hnRNP A1 and PTB use different mechanisms to repress exon N1 splicing.
Repression of N1 splicing by PTB requires PTB binding sites upstream and downstream of the exon (13, 17). To demonstrate a difference in activity between PTB and hnRNP A1, we compared the effect of these proteins on BS7 RNA containing all the PTB binding sites with that on BS27 RNA which lacks the upstream sites and is not repressed by PTB. The addition of PTB to reactions containing BS7 repressed splicing as expected (Fig. 8, lanes 2 to 5). Also as expected, PTB did not repress the splicing of BS27 (Fig. 8, lanes 7 to 10). The addition of hnRNP A1 repressed splicing of BS7 (Fig. 8, lanes 12 to 15). In contrast to PTB, hnRNP A1 also repressed splicing of BS27, although at higher concentrations of protein (Fig. 8, lanes 17 to 20). Thus, hnRNP A1 and PTB have different sequence requirements in repressing N1 splicing.
FIG. 8.
hnRNP A1 and PTB use different mechanisms to repress exon N1 splicing. Splicing of BS7 (lanes 1 to 5 and 11 to 15) and BS27 (lanes 6 to 10 and 16 to 20) was done in the presence of 100 μg of Weri-1 nuclear extract. Lanes 2 to 5 and 7 to 10 both contain 50, 100, 200, and 500 ng of PTB, respectively. Lanes 1, 6, 11, and 16 contain Weri-1 nuclear extract without additional factors. Lanes 12 to 15 and 17 to 20 both contain 25, 50, 100 and 200 ng of hnRNP A1, respectively.
DISCUSSION
Systems of tissue-specific splicing are regulated in a combinatorial manner, with multiple positive and negative inputs. One long-term goal in analyzing the c-src N1 exon is to identify all of the positive and negative factors that control its splicing. In previous work, we focused on intronic repressor elements bound by PTB and an intronic enhancer sequence bound by a large complex of proteins. Here we examine a sequence within the N1 exon that acts as a moderate enhancer of splicing in vivo and in vitro. The characterization of the proteins that bind to the N1 exon will allow us to connect their activities to those of the intronic regulators.
hnRNP A1 and SR proteins affect N1 splicing in vitro.
Cross-linking and immunoprecipitation experiments identified the hnRNPs A1, A1B, F, and H and the SR protein SF2/ASF as binding to the exonic sequence. In spite of extensive previous characterization, SR proteins and hnRNP A1 have not previously been shown to be involved in the regulation of N1 splicing. The effect of SF2/ASF and hnRNP A1 on N1 splicing were examined by adding purified protein to splicing reactions. SF2/ASF and another SR protein, SC35, each stimulate N1 splicing in neuronal nuclear extract, and each is sufficient to activate splicing in S100 extract. SF2/ASF enhanced N1 splicing over the alternate pathway more specifically than did SC35. The SF2/ASF activity was also more dependent on the ESE than was SC35.
In contrast to SF2/ASF, hnRNP A1 repressed splicing of c-src transcripts when added to in vitro splicing reactions. As observed in other systems, SF2/ASF and hnRNP A1 play antagonistic regulatory roles. However, repression of N1 splicing by A1 did not require the exonic element. There are many potential A1 binding sites elsewhere in the src transcript and in most other transcripts that we have looked at. Thus, it may be a negative factor in many different splicing events.
hnRNP A1 and PTB have generally been studied in separate systems. Here, both hnRNP A1 and PTB were found to repress N1-splicing reactions activated by SR proteins, but this activity differed for the two proteins. A c-src pre-mRNA that has no PTB binding sites upstream of N1 is no longer repressed by PTB but is still repressed by hnRNP A1. Moreover, PTB was more specific than hnRNP A1 in inhibiting the N1 inclusion pathway over the exon-skipping pathway. The different sequence requirements and patterns of inhibition of these two proteins likely reflect different mechanisms of action. In other experiments, the two proteins in combination did not seem to synergize in silencing splicing (data not shown).
Models for N1 splicing regulation.
Previous results showed that PTB binding to the introns upstream and downstream of N1 prevents splicing in nonneuronal cells, most likely by looping out the RNA and blocking spliceosome assembly (Fig. 9). In neuronal cells, PTB dissociates from the RNA at the 3′ splice site, and this may allow proteins binding to the downstream enhancer sequence to activate splicing (17). We now need to fit hnRNP A1 and SF2/ASF into this model.
FIG. 9.
Model for regulation of exon N1. PTB binds to CU repeat sequences in the upstream and downstream introns to repress splicing of N1 in nonneuronal cells. In neuronal cells, PTB is removed to derepress splicing and the enhancer elements are able to activate splicing. Some hnRNP A1 molecules may be replaced with hnRNP A1B molecules in neuronal cells to assist in derepression of N1. SF2/ASF likely enhances splicing at both adjacent splice sites in neuronal cells in cooperation with the proteins binding to the downstream enhancer sequence.
The N1 ESE stimulated both the upstream and downstream introns in different constructs. Thus, as in other systems, SF2/ASF may stimulate binding of spliceosomal components such as U2AF and U1snRNP to the splice sites. It may also antagonize the negative effect of inhibitory proteins, such as hnRNP A1 and PTB. In BS9, which is not repressed by PTB, splicing activation of the upstream intron may be through U2AF interactions with the exon-bound SF2/ASF. In BS7, in which the downstream intron is repressed by PTB binding upstream, this same U2AF-SF2/ASF interaction may help block PTB binding to allow splicing of the downstream intron. It thus seems likely that the N1 enhancer uses a combination of activation mechanisms.
One interesting feature of this exonic element in N1 is how closely juxtaposed the elements are. In most other systems, ESEs bound by SR proteins and exonic splicing silencers bound by hnRNP A1 are adjacent but separable and not overlapping sequences. In the N1 exon, the binding sites for all of these proteins are within the same 11 nucleotides. So far, we have not been able to separate the binding sites for the different proteins through mutagenesis or through different site-specific labels, as was done with the DCS (35). For the DCS complex, the binding of the individual proteins to the DCS RNA is strongly affected by the other proteins in the complex (35). Similarly, the PTB binding sites in the DCS also stimulate PTB binding upstream (17). We have not observed these cooperative effects in the binding of the proteins to the ESE. Likewise, purified PTB does not seem to affect the binding of purified hnRNP A1 or SF2/ASF to small RNA probes containing all the binding sites. It is thus not clear yet whether the ESE proteins bind in a single complex to the exon or in separate complexes, although because of the closeness of the binding sites, we favor the idea of separate complexes.
SF2/ASF binds more prominently in Weri-1 extracts, where N1 is spliced, than in HeLa extracts, where it is repressed. Similarly, binding of the A1B isoform of hnRNP A1 is more pronounced in the Weri-1 extract than in the HeLa extract. In vivo and in vitro studies have shown differences in the RNA binding and alternative splicing activities of hnRNP A1 and hnRNP A1B (39, 53). Thus, hnRNP A1B may be a factor in the binding of SF2/ASF and the derepression of N1 splicing in Weri-1 nuclear extracts, but this possibility will require further investigation.
In other systems, hnRNP H acts as either a positive or negative splicing regulator. It binds to enhancer elements in HIV type 1 (HIV-1) tev exon 6D and in the thyroid hormone receptor transcript, whereas its binding in HIV-1 tat exon 2, to exon 7 from the rat β-tropomyosin pre-mRNA, and to the negative regulator of splicing element from Rous sarcoma virus represses splicing (9, 15, 20, 26, 27). hnRNP F has been shown to interact with the cap binding complex and with the carboxyl-terminal domain of RNA polymerase II, and it is thought that the expression level of hnRNP F plays a role in the regulated use of alternative polyadenylation sites in lymphocytes (21, 51, 54). In the c-src system, hnRNPs H and F are components of the regulatory complex which forms on the DCS. Antibody inhibition and depletion experiments suggest a positive role for these proteins in N1 splicing (16, 40). However, it could not be shown in these earlier studies that the effect of hnRNP H was dependent on the intronic binding site (16). This may be due to the presence of the exonic binding site described here. The addition of purified hnRNP H to splicing reactions slightly reduces splicing activation by added SF2/ASF (data not shown). In general, hnRNP H had a much smaller effect on splicing by either pathway than did the SR proteins or hnRNP A1. Elucidating the role of the exonic H/F proteins will require further study.
In vivo experiments showed that the N1 exonic enhancer and the downstream enhancer work in combination to activate splicing. In experiments with individual elements, the exonic enhancer was weaker than the full downstream enhancer but stronger than a single copy of the core DCS element. In these in vitro experiments, the N1 exonic enhancer is relatively weak compared with other well-studied exonic enhancers. It seems to be a feature of alternative splicing regulatory systems that multiple elements and factors are combined to produce strong effects. A challenge for the future is to understand how these different exonic and intronic components of the regulation work together to produce such highly specific control.
Acknowledgments
We thank Akila Mayeda for providing hnRNP A1 cDNA, Gideon Dreyfuss, Stefan Stamm, and Mark Roth for providing antibodies, and Xiang-Dong Fu for providing baculovirus.
This work was supported by NIH grant GM R01 49662 to D.L.B. D.L.B. is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Black, D. L. 1992. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell 69:795-807. [DOI] [PubMed] [Google Scholar]
- 1a.Black, D. L. 1991. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 5:389-402. [DOI] [PubMed] [Google Scholar]
- 2.Blanchette, M., and B. Chabot. 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 18:1939-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 4.Burd, C. G., and G. Dreyfuss. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13:1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buvoli, M., F. Cobianchi, M. G. Bestagno, A. Mangiarotti, M. T. Bassi, G. Biamonti, and S. Riva. 1990. Alternative splicing in the human gene for the core protein A1 generates another hnRNP protein. EMBO J. 9:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 7.Caceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. [DOI] [PubMed] [Google Scholar]
- 8.Caputi, M., and A. M. Zahler. 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 276:43850-43859. [DOI] [PubMed] [Google Scholar]
- 9.Caputi, M., and A. M. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 11.Chabot, B. 1996. Directing alternative splicing: cast and scenarios. Trends Genet. 12:472-478. [DOI] [PubMed] [Google Scholar]
- 12.Chan, R. C., and D. L. Black. 1995. Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol. Cell. Biol. 15:6377-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, R. C., and D. L. Black. 1997. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 17:4667-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlet-B, N., P. Logan, G. Singh, and T. A. Cooper. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9:649-658. [DOI] [PubMed] [Google Scholar]
- 15.Chen, C. D., R. Kobayashi, and D. M. Helfman. 1999. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 13:593-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou, M. Y., N. Rooke, C. W. Turck, and D. L. Black. 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 19:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou, M. Y., J. G. Underwood, J. Nikolic, M. H. Luu, and D. L. Black. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5:949-957. [DOI] [PubMed] [Google Scholar]
- 18.Del Gatto-Konczak, F., M. Olive, M. C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 20.Fogel, B. L., and M. T. McNally. 2000. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 275:32371-32378. [DOI] [PubMed] [Google Scholar]
- 21.Gamberi, C., E. Izaurralde, C. Beisel, and I. W. Mattaj. 1997. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol. Cell. Biol. 17:2587-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabowski, P. J., and D. L. Black. 2001. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 65:289-308. [DOI] [PubMed] [Google Scholar]
- 23.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 24.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanamura, A., J. F. Caceres, A. Mayeda, B. R. Franza, Jr., and A. R. Krainer. 1998. Regulated tissue specific expression of antagonistic pre-mRNA splicing factors. RNA 4:430-444. [PMC free article] [PubMed] [Google Scholar]
- 26.Hastings, M. L., C. M. Wilson, and S. H. Munroe. 2001. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA 7:859-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacquenet, S., A. Mereau, P. S. Bilodeau, L. Damier, C. M. Stoltzfus, and C. Branlant. 2001. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem. 276:40464-40475. [DOI] [PubMed] [Google Scholar]
- 28.Kamma, H., D. S. Portman, and G. Dreyfuss. 1995. Cell type-specific expression of hnRNP proteins. Exp. Cell Res. 221:187-196. [DOI] [PubMed] [Google Scholar]
- 29.Krainer, A. R., G. C. Conway, and D. Kozak. 1990. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4:1158-1171. [DOI] [PubMed] [Google Scholar]
- 30.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 31.Levy, J. B., T. Dorai, L. H. Wang, and J. S. Brugge. 1987. The structurally distinct form of pp60c-src detected in neuronal cells is encoded by a unique c-src mRNA. Mol. Cell. Biol. 7:4142-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, H. X., S. L. Chew, L. Cartegni, M. Q. Zhang, and A. R. Krainer. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, H. X., M. Zhang, and A. R. Krainer. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12:1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 35.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez, R., B. Mathey-Prevot, A. Bernards, and D. Baltimore. 1987. Neuronal pp60c-src contains a six-amino acid insertion relative to its non-neuronal counterpart. Science 237:411-415. [DOI] [PubMed] [Google Scholar]
- 37.Mayeda, A., D. M. Helfman, and A. R. Krainer. 1993. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell. Biol. 13:2993-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayeda, A., and A. R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68:365-375. [DOI] [PubMed] [Google Scholar]
- 39.Mayeda, A., S. H. Munroe, J. F. Caceres, and A. R. Krainer. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13:5483-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min, H., R. C. Chan, and D. L. Black. 1995. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 9:2659-2671. [DOI] [PubMed] [Google Scholar]
- 41.Min, H., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 42.Modafferi, E. F., and D. L. Black. 1999. Combinatorial control of a neuron-specific exon. RNA 5:687-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Modafferi, E. F., and D. L. Black. 1997. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol. 17:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore, M. J., and P. A. Sharp. 1992. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science 256:992-997. [DOI] [PubMed] [Google Scholar]
- 45.Roth, M. B., A. M. Zahler, and J. A. Stolk. 1991. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J. Cell Biol. 115:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 47.Sun, Q., A. Mayeda, R. K. Hampson, A. R. Krainer, and F. M. Rottman. 1993. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 7:2598-2608. [DOI] [PubMed] [Google Scholar]
- 48.Tacke, R., and J. L. Manley. 1999. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11:358-362. [DOI] [PubMed] [Google Scholar]
- 49.Tacke, R., and J. L. Manley. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14:3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tange, T. O., and J. Kjems. 2001. SF2/ASF binds to a splicing enhancer in the third HIV-1 tat exon and stimulates U2AF binding independently of the RS domain. J. Mol. Biol. 312:649-662. [DOI] [PubMed] [Google Scholar]
- 51.Veraldi, K. L., G. K. Arhin, K. Martincic, L.-H. Chung-Ganster, J. Wilusz, and C. Milcarek. 2001. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 21:1228-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, X., M. R. Bani, S. J. Lu, S. Rowan, Y. Ben-David, and B. Chabot. 1994. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl. Acad. Sci. USA 91:6924-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida, T., K. Kokura, Y. Makino, V. Ossipow, and T. Tamura. 1999. Heterogeneous nuclear RNA-ribonucleoprotein F binds to DNA via an oligo(dG)-motif and is associated with RNA polymerase II. Genes Cells 4:707-719. [DOI] [PubMed] [Google Scholar]
- 55.Zahler, A. M. 1999. Purification of SR protein splicing factors. Methods Mol. Biol. 118:419-432. [DOI] [PubMed] [Google Scholar]
- 56.Zahler, A. M., K. M. Neugebauer, W. S. Lane, and M. B. Roth. 1993. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science 260:219-222. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]