Abstract
Transcriptional repression is often correlated with the alteration of chromatin structure through modifications of the nucleosomes in the promoter region, such as by deacetylation of the N-terminal histone tails. This is presumed to make the promoter region inaccessible to other regulatory factors and the general transcription machinery. To accomplish this, histone deacetylases are recruited to specific promoters via DNA-binding proteins and tethering factors. We have previously reported the requirement for the NAD+-dependent histone deacetylase Hst1 and the DNA-binding protein Sum1 for vegetative repression of many middle sporulation genes in Saccharomyces cerevisiae. Here we report the identification of a novel tethering factor, Rfm1, that is required for Hst1-mediated repression. Rfm1 interacts with both Sum1 and Hst1 and is required for the Sum1-Hst1 interaction. DNA microarray and Northern blot analyses showed that Rfm1 is required for repression of the same subset of Sum1-repressed genes that require Hst1. These results suggest that Rfm1 is a specificity factor that targets the Hst1 deacetylase to a subset of Sum1-regulated genes.
The expression of a large number of genes in eukaryotes is controlled by repressor proteins that bind to specific DNA sites to prevent transcription. There are two major subclasses of transcriptional repression. One involves gene-specific repression, while the other involves transcriptional silencing of large regions of the chromosome in a gene-independent manner (6). Gene-specific repression is often mediated by DNA-binding proteins that bind to conserved DNA sites found in promoters of target genes. Once bound, these DNA-binding proteins usually recruit corepressors, such as deacetylases, that modify the histone tails in nucleosomes positioned adjacent to the repressor-binding site. The deacetylation state of the histone tails has been strongly correlated with the repressed state of the promoter (13). Transcriptional silencing, in contrast, is not limited to single genes but represses large chromosomal domains containing multiple genes. Silencing in yeast occurs at the HM mating type loci, telomeres, and the rRNA gene (rDNA) loci and requires the Sir2 NAD+-dependent histone deacetylase (10, 17). Although there is a general requirement for Sir2 at all of the silenced loci in yeast, the ability of Sir2 to associate at the different silenced loci requires distinct cofactors. For example, the ability of Sir2 to associate at the telomeres, HML, and HMR requires Sir3 and Sir4 while Net1 is required for Sir2 to localize to the rDNA (11, 21, 22). This indicates that Sir2 is unable to localize to the DNA on its own and that Net1 and Sir3/4 are specificity factors that direct Sir2 to the distinct silenced regions.
Homologs of the Sir2 histone deacetylase have been found in many organisms, and four orthologs, called Hst1-4, have been identified in yeast (2, 8). Although the exact function and targets of some of these Sir2 homologs are not known, it has been shown that Hst1, along with the DNA-binding protein Sum1, is required for repression of middle sporulation genes during vegetative growth and early meiosis in yeast (26). The Sum1 protein binds specifically to the middle sporulation element (MSE) found in the promoters of many middle sporulation genes and acts as a transcriptional repressor. Given the strong sequence similarity between Hst1 and Sir2 (63% identity and 76% similarity), it is possible that Hst1 behaves in a manner similar to that of the Sir2 protein to repress middle sporulation genes. Both proteins are NAD+-dependent deacetylases, and mutations in NPT1, which decreases cellular NAD+ levels, negatively affect the ability of both Sir2 and Hst1 to function (20, 23). In addition, Hst1, like Sir2, is known to be associated with two distinct repressor complexes, each requiring unique cofactors. Hst1 has been found to be part of the Set3c repressor complex, which contains the Hos2 histone deacetylase and the SET domain protein Set3 (18). Hst1 has also been shown to exist in a separate complex with the Sum1 DNA-binding protein (19). Since the ability of Sir2 to associate with the different silenced loci requires specific cofactors that tether it to those regions, we reasoned that there must be a cofactor that directs Hst1 to the promoters of middle sporulation genes by tethering it to Sum1. To identify cofactors that direct Hst1 to the Sum1 DNA-binding protein, we screened for mutants that were defective in Sum1-Hst1-mediated repression. We identified a previously uncharacterized open reading frame (ORF), YOR279C, which we have named RFM1. Deletion of RFM1 results in derepression of the same subset of middle sporulation genes that require Hst1. We also show that Rfm1 mediates the interaction between Hst1 and Sum1. In addition, we show that mutations in critical components of the Set3c complex do not affect Sum1-Rfm1-Hst1-mediated repression. These results suggest that, like that of Sir2, the ability of Hst1 to assemble into distinct repressor complexes depends upon unique tethering factors, such as Rfm1, that recruit it to specific regions of the chromosome.
MATERIALS AND METHODS
Plasmids.
The construction of pAV124 and pJX43 was described previously (9, 26). pMP192 was constructed by PCR amplifying the SUM1 ORF, including the promoter region, and cloning it into a vector containing 13 Myc epitope tags. This clone was then digested with BglII, which removes part of the SUM1 ORF and the 13 Myc tags, and this fragment was subcloned into the BglII sites of pJX62 (a genomic subclone of SUM1), creating a C-terminally tagged Sum1 protein in pRS415 (CEN LEU2). pMP194 was constructed as described above, except that the endogenous stop codon was maintained to prevent inclusion of the 13 Myc epitope tags. pMP206 and pMP208 were created by digesting pMP192 and pMP194 with SacII and XhoI and cloning this fragment into the SacII/XhoI sites of pRS416 (CEN URA3). pRAM15 was created by digesting a genomic clone of HST1 with SacII, which removed the HST1 ORF along with the endogenous promoter. This fragment was cloned into the SacII site of pRS425 (2μm LEU2). pRAM29 was created by digesting plasmid pRAM23, which contains HST1 with a C-terminal V5 epitope under the control of the endogenous promoter with XhoI and NotI and subcloning this fragment in pRS425. pRAM41 was created by subcloning the XhoI/NotI fragment from pRAM23 into pRS426 (2μm URA3). pSW106 was cloned by PCR amplifying the YOR279C ORF, including the promoter, and cloning the PCR product into the ApaI and SacII sites of pRS425 (2μm LEU). pSW155 was created by inserting an SpeI site adjacent to the stop codon in pSW106 and inserting an oligonucleotide containing a three-hemagglutinin epitope (HA) tag into this site.
Strains.
The yeast strains used in this study are listed in Table 1. Strain YCM5, harboring the rfm1-1 mutation, was created during the EMS mutagenesis screen. To construct null mutations for each gene of interest in the desired strain backgrounds, we obtained the diploid KanMX null mutant strains from Research Genetics. Primers were designed to generate a fragment via a PCR that included the KanMX replacement of the gene of interest and 500 bp of homology upstream and downstream. These fragments were then transformed into the various strain backgrounds, and the integration of the KanMX null mutation was verified by using primers internal and external to the transformed fragment. The mutant strains were tested for MSE-mediated repression by using lacZ reporter plasmid pJX43 and by Northern blot analysis as described previously (26).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATaade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 | L. Neigeborn |
| W303-1B | MATα ade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 | L. Neigeborn |
| LNY433 | MATatrp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 | L. Neigeborn |
| LNY385 | MATα ade2-1 trp1-1 can1-100 ura3-1 leu2-3,112 | L. Neigeborn |
| YCM5 | MATα ade2-1 trp1-1 can1-100 ura3-1 leu2-3,112 rfm1-1 | This study |
| YCM6 | MATα ade2-1 trp1-1 can1-100 ura3-1 leu2-3,112 rfm1-2 | This study |
| YCM7 | MATα ade2-1 trp1-1 can1-100 ura3-1 leu2-3,112 rfm1-3 | This study |
| YCM11 | MATatrp1-1 can1-100 ura3-1 leu2-3,112 rfm1-1 | This study |
| JXY3 | Isogenic to W303-1A sum1 Δ::kanMX4 | J. Xie |
| JXY5 | Isogenic to W303-1A hst1 Δ::kanMX4 | J. Xie |
| JXY19 | Isogenic to W3031-B sir2Δ::kanMX4 | J. Xie |
| MPY11 | Isogenic to W303-1A rfm1Δ::kanMX4 | This study |
| MPY13 | Isogenic to W303-1B rfm1Δ::kanMX4 | This study |
| RMY8 | Isogenic to W303-1B rfm1Δ::kanMX4 hst1Δ::kanMX | This study |
| RMY6 | Isogenic to W303-1B npt1Δ::kanMX4 | This study |
| RMY40 | Isogenic to W303-1A set3Δ::kanMX4 | This study |
| RMY41 | Isogenic to W303-1A hos2Δ::kanMX4 | This study |
| RMY42 | Isogenic to W303-1A hos3Δ::kanMX4 | This study |
| RMY43 | Isogenic to W303-1A hos1Δ::kanMX4 | This study |
| RMY44 | Isogenic to W303-1A set2Δ::kanMX4 | This study |
| RMY45 | Isogenic to W303-1A set4Δ::kanMX4 | This study |
| RMY46 | Isogenic to W303-1A set5Δ::kanMX4 | This study |
| RMY47 | Isogenic to W303-1A set6Δ::kanMX4 | This study |
| JRY3178 | MATaade2-1 trp1-1 his3-11,15 ura3-1 leu2-3,11 hmr::TRPI | J. Rine |
| JRY3182 | MATaade2-1 trp1-1 his3-11,15 ura3-1 sir3::LEU2 hmr::TRPI | J. Rine |
| RMY34 | Isogenic to JRY3178 rfm1Δ::kanMX4 | This study |
| DN1004 | MATathr4 | D. Norris |
| MC57 | MATα ade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 GAL+sir2::HIS3 SUM1-1 | D. Shore |
| RMY23 | Isogenic to MC57 npt1Δ::kanMX4 | This study |
| RMY25 | Isogenic to MC57 rfm1Δ::kanMX4 | This study |
| RMY27 | Isogenic to MC57 hst1Δ::kanMX4 | This study |
Screen for mutants that fail to repress the SMK1 MSE.
Strain LNY385 (Table 1), containing pJX43, a hop1-lacZ transcription reporter vector under the control of the SMK1 MSE, was mutagenized, and colonies that turned blue on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates were rescreened by X-Gal filter assay (26). The level of derepression by each mutant was measured by liquid β-galactosidase assays. Each mutant was mated to wild-type strain LNY433, and the resulting diploids were tested for lacZ expression by the X-Gal filter assay to verify that the mutations were recessive. To test if the mutants contained mutations in SUM1 or HST1, they were transformed with pJX62, a SUM1 genomic subclone in pRS415; p42A-33A, a plasmid containing a genomic copy of HST1; and pRS415, a blank LEU2 cloning vector, and assayed for repression by using SMK1 MSE reporter plasmid pJX43. To determine complementation among the three mutants that were not SUM1 or HST1 mutants, we created a mutant of the opposite mating type (MATa) by sporulating the heterozygous diploid strain generated from YCM6 and screening the spores for the a mating type and mutant phenotype (YCM11). This strain was mated with YCM5, YCM6, and YCM7, each harboring pJX43, and the resulting diploids were assayed for repression of the reporter. Plasmids that complemented the MSE repression defect in YCM5 were cloned from a genomic plasmid library as previously described (26). The isolated plasmids were purified and retransformed into the original mutant to verify that the plasmid could complement the mutation. The end points of each insert were determined by sequencing, and the ORFs on each plasmid were identified by a BLASTN search of the Saccharomyces Genome Database (Stanford University).
Coimmunoprecipitation experiments.
A 50-ml culture of each yeast strain was grown to mid-log phase (optical density at 600 nm of 0.5) in appropriate medium. Cells were harvested by centrifugation and resuspended in 100 μl of lysis buffer (250 mM NaCl, 50 mM Tris [pH 7.4], 0.1% IPEGAL (Sigma), 5 mM EDTA, 1.5 mM dithiothreitol, 0.1 mM tosylsulfonyl phenylalanyl chloromethyl ketone [TPCK], 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1× complete protease inhibitors [Boehringer Mannheim]). A 100-μl volume of glass beads (Sigma) was added, and each sample was vortexed at 4°C for 30 s and then placed on ice for 1 min (repeated eight times). After lysis, the total volume was increased to 500 μl by addition of 400 μl of fresh lysis buffer. Each sample was then clarified by centrifugation at 16,000 × g for 20 min at 4°C, and the supernatant was transferred to a new tube. For immunoprecipitation reactions, 200 μl of clarified lysate was diluted by addition of 200 μl of fresh lysis buffer and incubated for 3 h at 4°C with the appropriate antibody (anti-Myc [Covance], anti-HA [Roche], or anti-V5 [Invitrogen]). A 50-μl volume of protein G-agarose beads (Gibco BRL) was then added to each reaction mixture, and incubation was continued at 4°C for 1 h. The immunocomplexes were collected by centrifugation, washed twice in lysis buffer, resuspended in 25 μl of 2× sodium dodecyl sulfate (SDS) buffer, and placed at 95°C for 5 min. The samples were loaded onto either a 12% SDS (anti-Myc coimmunoprecipitations)- or an 8% SDS (anti-HA, anti-V5 coimmunoprecipitations)-polyacrylamide gel for Western blot analysis. Proteins were blotted to membranes and probed with either rat anti-HA, mouse anti-V5, or mouse anti-Myc. Horseradish peroxidase-conjugated secondary antibodies and ECL chemiluminescence reagents (Amersham) were used for detection.
Microarray analysis.
Microarray data were collected at Expression Analysis, Inc., Durham, N.C. (www.expressionanalysis.com), by using Yeast Genome S98 GeneChips (Affymetrix). The quality and quantity of each RNA sample were assessed by using a 2100 BioAnalyzer (Agilent). Total RNA (10 μg) was converted into cDNA and used as a template for labeling with biotinylated ribonucleotides by using an in vitro transcription kit (Enzo Diagnostics). After hybridization and a series of washes, the microarrays were stained with streptavidin-phycoerythrin and the fluorescent signal was amplified by using a biotinylated antibody solution. Fluorescent images were detected in an Agilent GeneArray Scanner. Expression data was extracted by using the MicroArray Suite 5.0 software (Affymetrix). All GeneChips were scaled to a median intensity setting of 500. The assays were repeated twice for each mutant strain, and genes that showed greater than threefold derepression in each experiment were chosen for further analysis.
RESULTS
Rfm1 is required for MSE-mediated repression.
Although the Sum1 and Hst1 proteins are required for repression of middle sporulation genes, it seemed likely that other proteins might be involved in MSE-mediated repression. To identify additional cofactors that are involved in the repression of middle sporulation genes, we performed an extensive screen to identify mutants that fail to repress an MSE-dependent promoter during vegetative growth (26). The mutants isolated from this screen fell into three complementation groups. One of the complementation groups contained mutations in SUM1, and another complementation group contained mutations in HST1. However, three independent mutants fell into a third group that were complemented for repression of the reporter when mated with either an sum1Δ or an hst1Δ mutant strain and failed to be complemented when transformed with either SUM1 or HST1 on a plasmid (data not shown). One of the mutants in this third complementation group was used to screen a yeast genomic library for clones that would complement the defect in repression. Several independent transformants were isolated that harbored library plasmids whose genomic DNAs overlap the same region of chromosome XV. Deletion analysis of one of the clones showed that the previously uncharacterized ORF YOR279C was required to complement the mutant phenotype. This was verified by cloning a PCR-generated fragment that only contained the promoter region and ORF of YOR279C and showing that this plasmid complemented the mutant phenotype.
To confirm that the genomic mutation identified in the screen was YOR279C, we constructed a null mutation in the ORF by using the KanMX replacement cassette (24). The null mutation caused roughly the same level of derepression of the MSE-regulated reporter as the EMS-generated mutation. The KanMX null mutant was crossed with the original EMS-generated genomic mutant. The resulting diploid strain was unable to repress the MSE-mediated reporter, indicating that the mutants fail to complement each other for the defect in repression. Tetrad analysis following sporulation yielded a 4:0 segregation of the mutant phenotype in the haploid progeny. These results indicated that the mutation was in YOR279C and that this gene is required for repression of the MSE reporter. We have named this gene RFM1 for repression factor of MSEs.
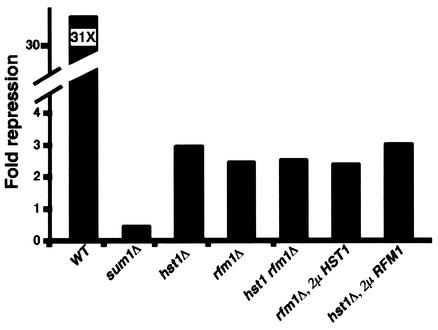
Given that Sum1 and Hst1 are also required for repression of the MSE-lacZ reporter, we wanted to determine if the level of derepression in the rfm1Δ mutant was similar to that in the hst1Δ mutant or the sum1 mutant. We found that the sum1Δ mutant had the highest level of derepression, while the levels of derepression in both the rfm1Δ and hst1Δ mutants were similar to each other and were not as severe as that in the sum1Δ mutant (Fig. 1). Since the hst1Δ and rfm1Δ mutants exhibited similar levels of derepression of the reporter, we were interested in determining whether these genes work together in the same pathway or whether they function in separate pathways. We found that the hst1Δ rfm1Δ double mutant was derepressed to roughly the same levels as either single mutant, suggesting that both genes are involved in the same aspect of MSE-mediated repression. We were therefore interested in determining whether overexpression of RFM1 or HST1 would be able to suppress the defect caused by the absence of the other. However, overexpression of Hst1 was unable to complement the repression defects of the rfm1 null mutation (Fig. 1). Likewise, overexpression of Rfm1 was unable to complement hst1 repression defects. These results suggest that these proteins have distinct activities and requirements in MSE-mediated repression.
FIG. 1.
Rfm1 is required for MSE-mediated repression. The fold repression of the MSE-lacZ promoter reporter in each strain background is presented as a ratio of β-galactosidase activity of the reporter lacking an MSE, pAV124 (50 U in the wild-type [WT] strain), compared to activity of a reporter that contains an MSE, pJX43 (0.4 U in the wild-type strain). Repression was measured in the wild type (W303-1A), in the sum1Δ mutant (JXY3), in the hst1Δ mutant (JXY5), in the rfm1Δ mutant (MPY13), in the hst1Δ rfm1Δ mutant (RMY8), in the rfm1Δ mutant (MPY13) transformed with pRAM15 (2μm HST1), and in the hst1Δ mutant (JXY5) transformed with pSW106 (2μm RFM1).
Differential regulation of MSE-mediated genes.
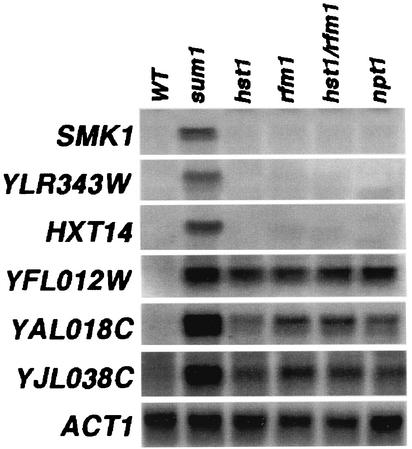
We have previously shown there are two subsets of MSE-repressed genes: one set that appears to require only Sum1 and one set that requires Sum1 and Hst1 (26). To further investigate whether Rfm1 works in conjunction with Hst1 to regulate this same subset of MSE-repressed genes, we prepared RNAs from sum1Δ, hst1Δ, rfm1Δ, hst1Δ rfm1Δ, and npt1Δ mutant strains and examined the expression levels of several middle sporulation-specific genes during vegetative growth by Northern blot analysis. All of the middle sporulation genes we examined were derepressed in the sum1Δ mutant (Fig. 2). In contrast, only a subset (YFL012W, YAL018C, and YJL038C) required Hst1 and Rfm1. The hst1Δ rfm1Δ double mutant had roughly the same level of derepression of these genes as either single mutant. In addition, the double mutant did not derepress any of the genes that only require Sum1. This correlates with our lacZ reporter expression data and provides further evidence that Hst1 and Rfm1 work in the same pathway to repress the transcription of a specific subset of Sum1-regulated genes.
FIG. 2.
Differential regulation of middle sporulation genes by Sum1, Hst1, Rfm1, and Npt1. RNAs were prepared from the wild-type (WT; W303-1A), sum1Δ (JXY3), hst1Δ (JXY5), rfm1Δ (MPY13), rfm1Δ hst1Δ (RMY8), and npt1Δ (RMY6) strains grown under vegetative conditions and used for Northern blot analysis. The same blot was hybridized with radiolabeled DNA fragments specific for the coding regions of the SMK1, YLR343W, HXT14, YFL012W, YAL018C, YJL038C, and ACT1 genes. An ACT1 probe was used as a loading control.
It has been shown that mutations in genes involved in the NAD+ salvage pathway, such as NPT1, cause a decrease in silencing by the NAD+-dependent histone deacetylase Sir2 (20). We found that genes requiring Rfm1 and Hst1 for repression also required Npt1, indicating that a decrease in cellular NAD+ levels negatively affects the repression of these genes (Fig. 2). However, Sum1-repressed genes that do not require Rfm1 and Hst1, such as SMK1, YLR343W, and HXT14, do not require Npt1. These results suggest that Sum1-repressed genes that are independent of Hst1 and Rfm1 are unlikely to be repressed by an NAD+-dependent histone deacetylase.
The SET2-6 and HOS genes are not required for MSE-mediated repression.
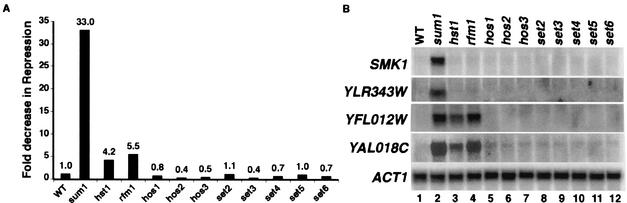
The Hst1 protein was recently found in a complex that is separate from Rfm1 and Sum1 (18). This complex included HOS2, a member of the HOS family of deacetylases, and SET3, a member of the SET family of methyltransferases. To determine if any of these genes are involved in MSE-mediated repression, we created null mutations in a number of the SET genes, as well as the deacetylase genes HOS1, HOS2, and HOS3, and assayed repression of the MSE-lacZ reporter during vegetative growth. Although the reporter was derepressed in the sum1Δ, hst1Δ, and rfm1Δ mutants, all of the setΔ and hosΔ mutants showed wild-type levels of repression, indicating that these genes are not required for repression of this reporter (Fig. 3A). To verify that these proteins are not required for MSE-mediated repression during vegetative growth, we prepared RNAs from the wild-type and mutant strains and performed a Northern blot analysis to monitor the derepression of middle sporulation genes. Deletion of the SET or HOS gene had no effect on the regulation of either subclass of MSE-regulated genes during vegetative growth (Fig. 3B). These results show that although Set3 and Hos2 were found in complex with Hst1, these proteins are not required for MSE-mediated repression, nor are other members of the SET and HOS gene families.
FIG. 3.
SET2-6 and the HOS genes are not required for MSE-mediated repression. (A) Fold repression of the MSE-lacZ promoter reporter in each strain background was calculated as described in the legend to Fig. 1. (B) RNAs were prepared from the wild-type (WT; W303-1A), sum1Δ (JXY3), hst1Δ (JXY5), rfm1Δ (MPY13), hos1Δ (RMY43), hos2Δ (RMY41), hos3Δ (RMY42), set2Δ (RMY44), set3Δ (RMY40), set4Δ (RMY45), set5Δ (RMY46), and set6Δ (RMY47) strains grown under vegetative conditions. The same blot was hybridized with radiolabeled DNA fragments specific for the coding regions of the SMK1, YLR343W, YFL012W, YAL018C, and ACT1 genes.
Microarray analysis of sum1Δ, rfm1Δ, and hst1Δ mutants.
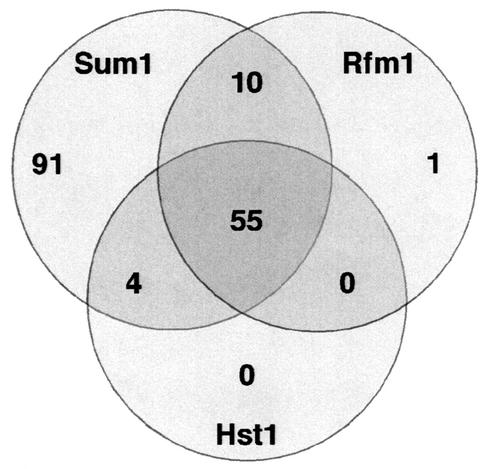
Our Northern blot analysis showed that there are two distinct classes of MSE-mediated repression: one set that requires only Sum1 and one set that requires Sum1, Rfm1, and Hst1. Among the genes tested by Northern blot analysis, Rfm1 and Hst1 are required for repression of the same subset of genes. These results raised the questions of whether there are genes that require only Hst1 or Rfm1 for repression and whether Hst1 or Rfm1 represses a subset of genes independently of Sum1. To address these questions, we performed a microarray analysis with RNAs prepared from isogenic sum1Δ, rfm1Δ, and hst1Δ mutant strains during vegetative growth. In both the rfm1Δ and hst1Δ mutants, there were roughly the same number of genes (66 and 59, respectively) that were derepressed at least threefold compared to the wild type (Fig. 4). Comparison of the genes derepressed in the rfm1Δ and hst1Δ mutants revealed that the data sets were nearly identical, with 55 genes that were derepressed in both mutants. Many of the 11 genes in the rfm1Δ mutant data set that were not present in the hst1Δ mutant data set showed moderate (1.5- to 2.9-fold) derepression in the hst1Δ mutant, suggesting that these genes are weakly coregulated by Rfm1 and Hst1. The microarray results correlate well with the Northern blot analysis results and indicate that Hst1 and Rfm1 are required to repress the same set of genes and do not act independently of each of other.
FIG. 4.
Microarray analysis of sum1Δ, rfm1Δ, and hst1Δ mutants. A Venn diagram comparing genes up-regulated more than threefold in sum1Δ, rfm1Δ, and hst1Δ mutant strains compared to the wild type is shown. Of the genes derepressed in the sum1Δ mutant, 91 are regulated independently of Rfm1 or Hst1. Genes regulated by both Rfm1 and Hst1 are nearly identical, with 55 coregulated genes. All 11 genes regulated by Rfm1 and not by Hst1 were weakly (less than threefold) derepressed in the hst1Δ mutant and therefore were not included in the set of Rfm1/Hst1-coregulated genes.
To determine if the genes derepressed in the rfm1Δ and hst1Δ mutants are a subset of genes derepressed in the sum1Δ mutant, we compared the rfm1Δ and hst1Δ mutant data sets to the list of genes derepressed in an isogenic sum1Δ mutant. Virtually all of the genes derepressed in the rfm1Δ and hst1Δ mutants were derepressed in the sum1Δ mutant (Fig. 4). However, there were 91 genes derepressed in the sum1Δ mutant strain that were not derepressed in either the rfm1Δ or the hst1Δ mutant. Many of these genes are strongly derepressed in the sum1Δ mutant, indicating that the differences between the number of genes repressed by Sum1 and those regulated by Hst1 and Rfm1 are significant. This finding correlates with our Northern blot analysis showing that there is a set of genes that require Sum1 for repression but are independent of Hst1 and Rfm1 (Fig. 2). The results of the genomewide analysis of the sum1Δ, rfm1Δ, and hst1Δ mutants demonstrate that the primary function of Rfm1 and Hst1 in vegetative cells is to work together to repress a subset of Sum1-repressed genes.
Rfm1 is required for Sum1-1 suppression of sir2 silencing defects at HMR.
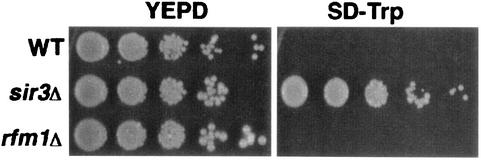
Sum1 was originally identified as a dominant allele, SUM1-1, that was able to suppress the silencing defects of sir2Δ mutants at HMR (4, 14, 15). On the basis of the finding that Sum1 and Hst1 are both involved in MSE-mediated repression, several groups have shown that the ability of SUM1-1 to suppress these silencing defects is dependent on Hst1 (19, 23). We were interested in whether SUM1-1 suppression of sir2 silencing defects at HMR is also dependent on Rfm1 or if Rfm1 is involved in Sir-mediated silencing at HMR. To determine if Rfm1 is required for silencing, an rfm1Δ mutant strain with a TRP1 marker integrated at the HMR locus was constructed and assayed for silencing defects. We compared this strain to a wild-type strain and a sir3Δ mutant strain with the same integrated reporter at HMR. Although all of the strains grew equally well on yeast complete medium (YEPD), HMR was silenced in a wild-type cell and therefore the TRP1 marker was not expressed and the strain was unable to grow on minimal medium lacking tryptophan (SD-Trp) (Fig. 5). However, the loss of silencing at HMR in a sir3 mutant allowed expression of TRP1, supporting growth on this medium. The rfm1Δ mutant strain did not grow on synthetic SD-Trp medium, indicating that the HMR locus was transcriptionally silenced. Moreover, rfm1Δ mutant strains were able to mate with cells of the opposite mating type, further indicating that silencing at the mating type loci is unaffected (data not shown). Taken together, these results suggest that RFM1 is not required for silencing at the mating type loci.
FIG. 5.
Rfm1 is not required for silencing at HMR. The wild-type (WT; W303-1A), sir3Δ (JRY3182), and rfm1Δ (RMY25) strains contain TRP1 integrated at HMR. The strains were grown to mid-log phase, normalized for concentration, and serially diluted fivefold, and then equivalent quantities of all of the dilutions were spotted onto YEPD and SD-Trp plates.
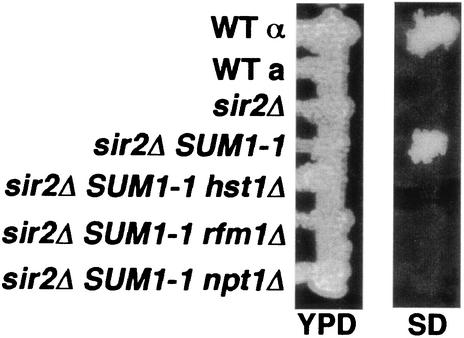
To investigate if RFM1 is required for SUM1-1 suppression of the silencing defects at HMR in a sir2Δ mutant, we constructed rfm1Δ, hst1Δ, and npt1Δ mutations in an SUM1-1 sir2 MATα strain background. We then tested the ability of each of these strains to mate by screening for the formation of a diploid cell that was able to grow on SD medium lacking amino acids. The sir2Δ mutant strain showed no growth on SD medium, indicating that it was unable to mate as a result of derepression of the silent mating type loci (Fig. 6). However, the wild-type α cell and the SUM1-1 sir2 strain were able to mate as a result of the ability to silence HMR. As was previously reported, SUM1-1 suppression of sir2 silencing defects is dependent on HST1 and NPT1 and these strains were unable to mate (19, 23). Our results show that RFM1 is also required for SUM1-1 suppression of the sir2Δ mutant. This result suggests that Sum1-1, Rfm1, and Hst1 form a complex at the silent loci.
FIG. 6.
Rfm1 is required for SUM1-1 suppression of sir silencing defects. The indicated strains (from the top, W303-1B, W303-1A, JXY19, MC57, RMY27, RMY25, and RMY23) were mated with a MATa mating type tester strain (DN1004) by patching on YPD and grown overnight at 30°C. To assay mating, the formation of the diploid stain was monitored by growth of cells from a replica plate of minimal SD medium. WT, wild type.
Protein interactions required for the Sum1-Rfm1-Hst1 repression complex.
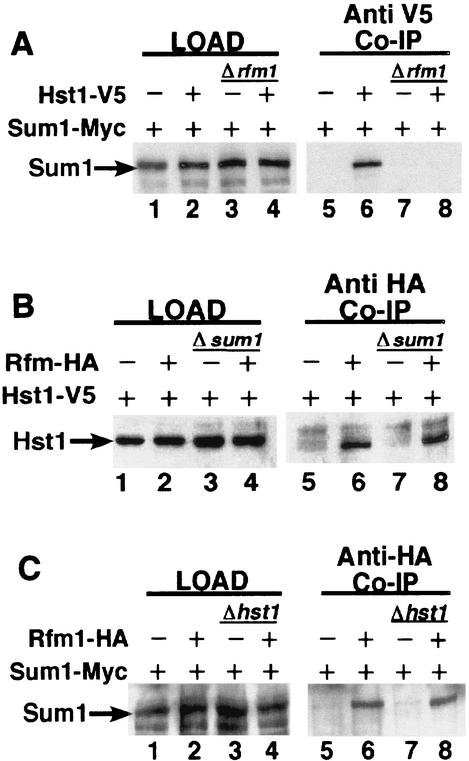
The Sum1, Hst1, and Rfm1 proteins associate in a complex that can be specifically immunoprecipitated with a TAP-tagged version of Hst1 (18). Given that this complex is required for MSE-mediated repression of many middle sporulation genes, we were interested in the protein-protein interactions that are required to form this complex. To investigate these interactions, we performed coimmunoprecipitation experiments using Myc-tagged Sum1, V5-tagged Hst1, and HA-tagged Rfm1. We found that Hst1-V5 was able to immunoprecipitate Sum1-Myc and that both Hst1-V5 and Sum1-Myc coimmunoprecipitated with HA-tagged Rfm1 (Fig. 7). We also found that both Hst1-V5 and Rfm1-HA immunoprecipitated with Sum1-Myc and that Hst1-V5 was able to immunoprecipitate Rfm1-HA. These results support the model in which these proteins exist in a trimeric complex (18).
FIG. 7.
Sum1, Hst1, and Rfm1 associate in a protein complex. (A) Extracts from strains of the genotype RFM1 (lanes 1 and 2) or rfm1Δ (lanes 3 and 4) were cotransformed with plasmids expressing Hst1 (lanes 1 and 3) or Hst1-V5 (lanes 2 and 4) and Sum1-Myc (lanes 1 to 4). Extracts from these strains were coimmunoprecipitated (Co-IP) with antibodies specific to the V5 epitope. The resulting samples were probed with antibodies to Sum1-Myc (lanes 5 to 8). (B) Extracts from strains of the genotype SUM1 (lanes 1 and 2) or sum1Δ (lanes 3 and 4) were cotransformed with plasmids expressing Rfm1 (lanes 1 and 3), Rfm1-HA (lanes 2 and 4), and Hst1-V5 (lanes 1 to 4). Extracts from these strains were coimmunoprecipitated with antibodies specific to the HA epitope. The resulting samples were probed with antibodies to Hst1-V5 (lanes 5 to 8). (C) Extracts from strains of the genotype HST1 (lanes 1 and 2) or hst1Δ (lanes 3 and 4) were cotransformed with plasmids expressing Rfm1 (lanes 1 and 3) or Rfm1-HA (lanes 2 and 4). Extracts from these strains were coimmunoprecipitated with antibodies specific to the HA epitope. The resulting samples were probed with antibodies to the Myc epitope (lanes 5 to 8).
To further investigate the protein-protein interactions required for this complex formation, we repeated these experiments with hst1Δ, rfm1Δ, and sum1Δ mutant strains. In the first experiment, we were interested in determining if Rfm1 is required for interaction of the Sum1 and Hst1 proteins. To address this question, plasmids bearing Myc-tagged Sum1 and V5-tagged Hst1 were cotransformed into both the wild-type strain and an rfm1Δ mutant strain. Sum1 and Hst1 coimmunoprecipitated in the wild-type strain but did not coimmunoprecipitate in the rfm1Δ strain (Fig. 7A, lane 6 versus lane 8). This shows that Rfm1 is required for the Sum1-Hst1 interaction. We then tested whether Sum1 is required for the Hst1-Rfm1 interaction by cotransforming plasmids bearing V5-tagged Hst1 and HA-tagged Rfm1 into the wild-type and sum1Δ strains. We found that Hst1 and Rfm1 coimmunoprecipitated in both the wild-type and sum1Δ null strains, indicating that Sum1 is not required for Hst1-Rfm1 interaction (Fig. 7B). Finally, we examined whether Hst1 is required for the Sum1-Rfm1 interaction by cotransforming plasmids bearing Myc-tagged Sum1 and HA-tagged Rfm1 into wild-type and hst1Δ mutant strains. Rfm1-HA coimmunoprecipitated with Sum1-Myc in both the presence and the absence of Hst1, indicating that Hst1 is not required for the Sum1-Rfm1 interaction (Fig. 7C). Taken together, these results demonstrate that Rfm1 serves as an important link between Sum1 and Hst1 and is required for the formation of the Sum1-Rfm1-Hst1 repression complex.
DISCUSSION
Transcriptional regulation relies on the interplay of DNA-binding proteins and cofactors that act in concert to activate or repress genes in a timed and coordinated manner. We have shown that repression of many middle sporulation genes requires the coordinated action of Sum1, Rfm1, and Hst1. Rfm1 interacts with both Sum1 and Hst1 and appears to have an important role in forming a complex between Sum1, the site-specific DNA-binding protein, and Hst1, the NAD+-dependent histone deacetylase. Recruitment of Hst1 to the promoter presumably alters the local chromatin structure, thereby preventing transcriptional activation. These proteins share mechanistic aspects with both gene-specific repression and transcriptional silencing (19, 26).
A comparison of the DNA microarray expression data from the rfm1Δ and hst1Δ mutants with the sum1Δ data show that Rfm1 and Hst1 are only required for repression of a subset of genes that are repressed by Sum1. The Northern blot analysis verifies that a subset of middle sporulation genes are repressed by Sum1 independently of Rfm1 and Hst1, raising the possibility that Sum1 recruits different cofactors to these genes to repress transcription. In one appealing model, Sum1 recruits another member of the Sir2 family of NAD+-dependent deacetylases, such as Sir2, Hst2, Hst3, or Hst4. However, this is unlikely because mutations in these genes have no effect on MSE-mediated repression (26). This is further supported by the observation that this subset of Sum1-repressed genes is not derepressed in an npt1Δ mutant, suggesting that these NAD+-dependent deacetylases are not involved in repression of this set of genes. Examination of other histone deacetylases, such as Hos1, Hos2, Hos3, and Rpd3, indicates that these proteins are not required for repression of middle sporulation genes. In addition, microarray expression data from hda1Δ and rpd3Δ mutants do not show derepression of this set of Sum1-regulated genes (1, 25). Although we cannot rule out the possibility that some of these deacetylases may be redundant in function for Sum1-mediated repression, these results suggest that Sum1 either represses transcription on its own or recruits an as yet unidentified cofactor.
Acetylation is only one of many histone tail modifications that contribute to changes to chromatin structure and subsequently transcriptional regulation (13). Recently, it has been shown that methylation of lysine residues by SET domain proteins is required for both activation and repression of several genes (12). This has been shown at the rDNA loci, where the methyltransferase Set1 and the deacetylase Sir2 are required for silencing (3). It has recently been shown that Hst1 forms a complex with Set3 and Hos2, as well as with other proteins (18). It was therefore possible that this complex of proteins represses transcription in combination with Sum1. Although our data show that the Set proteins are not required for repression of Sum1-repressed middle sporulation genes, there is still a possibility that methylation of the histone tails occurs at these promoters.
The ability of Hst1 to associate with two separate repression complexes parallels that of the histone deacetylase Sir2. Sir2 associates with Sir3/4 to repress the HM loci and telomeres or with Net1 to silence the rDNA (11, 21, 22). Therefore, a specific deacetylase can be recruited to different sites of repression via interactions with different tethering factors and DNA-binding proteins. We have shown that Hst1 is recruited to the Sum1-Hst1 repressor complex through interactions with Rfm1, a novel tethering factor. Rfm1 was found only in complex with Sum1 and Hst1 and not in the Set3c complex (18), which argues that the role of Rfm1 is to recruit Hst1 to Sum1-repressed genes. The microarray data show that Hst1 is required for repression of the same subset of genes that require Rfm1 and that these are both subsets of genes that are repressed by Sum1. This result indicates that even though Hst1 may be part of the Set3c complex, it does not play an essential role in repression by this complex during vegetative growth. Hst1 interacts directly with the YIL112W protein of the Set3c complex (18). This protein is not found associated with Sum1, which raises the possibility that, like Rfm1, this protein may provide the specificity to recruit Hst1 to the Set3c complex. Even though both proteins interact with Hst1, there is no apparent homology between RFM1 and YIL112W. The Rfm1 and YIL112W proteins may therefore interact with different regions of Hst1. This is an intriguing possibility given that mutational analysis of Sir2 has defined distinct functional domains that are required for either telomere/HM silencing or rDNA silencing (7), suggesting that Sir2 may interact differently with the cofactors at these distinct loci. These data suggest a functional parallel between Hst1, a gene specific repressor, and the silencing factor Sir2. Both deacetylases may be involved in regulating different classes of genes on the basis of interactions with a combination of specific cofactors and DNA-binding proteins.
It has been previously shown that overexpression of HST1 partially suppresses an sir2Δ mutant and overexpression of SIR2 partially suppresses an hst1Δ mutant (2, 26). Preliminary experiments indicate that suppression of the hst1Δ mutant by SIR2 requires Rfm1 (R. McCord, unpublished data). Therefore, although Sir2 is not normally required for MSE repression, it is possible that the Rfm1 and Sir2 proteins may weakly interact in the absence of Hst1. This interaction may explain why we observe slightly lower levels of expression of some genes (YAL018C and YJL038C; Fig. 2) in the hst1Δ mutant than in the rfm1Δ mutant. Although there is no apparent homology among the cofactors, the ability of the Sir2 and Hst1 proteins to interact with each other's cofactors suggests that the interactions of Hst1 with Rfm1 may be somewhat similar to the interactions of Sir2 with some of its cofactors. These interactions are likely to be mediated in part through the conserved core region in Hst1 and Sir2.
The roles of Rfm1 and Hst1 appear to be specific for the repression of middle sporulation genes. Most of the genes that are derepressed in the hst1Δ and rfm1Δ mutants are middle sporulation genes, and nearly all of them are regulated by Sum1. The Rfm1/Hst1-regulated genes that are not middle sporulation specific are relatively weak at the level of derepression, and the effects of the rfm1Δ and hst1Δ mutations may therefore be indirect. Our findings suggests that the sole role of the Rfm1/Hst1 complex is to be recruited to middle sporulation promoters during vegetative growth via interactions with Sum1. Although our data suggest that the Rfm1/Hst1 complex interacts with the Sum1 DNA-binding protein during vegetative growth, it is possible that this complex interacts with other DNA-binding proteins under different conditions. For example, during the middle stages of sporulation, the Sum1 protein is specifically degraded and the levels of the protein decrease (16). In contrast, the levels of HST1 and RFM1 expression significantly increase (5). It is possible that, during this stage of sporulation, Rfm1 and Hst1 interact with a different cofactor, perhaps to reestablish repression of genes that were specifically induced during the earlier stages of meiosis.
Acknowledgments
We thank Expression Analysis for the microarray analysis.
Michael Pierce was a recipient of a Busch Graduate fellowship. Carolyn Mickel and Sandeep Wonkatal were recipients of Waksman undergraduate research fellowships. This research was supported by a grant from the National Institutes of Health (GM 58762).
REFERENCES
- 1.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 3.Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis, and F. Winston. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12:165-170. [DOI] [PubMed] [Google Scholar]
- 4.Chi, M.-H., and D. Shore. 1996. SUM1-1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol. 16:4281-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 6.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15:2786-2796. [DOI] [PubMed] [Google Scholar]
- 7.Cuperus, G., R. Shafaatian, and D. Shore. 2000. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 19:2641-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derbyshire, M. K., K. G. Weinstock, and J. N. Strathern. 1996. HST1, a new member of the SIR2 family of genes. Yeast 12:631-640. [DOI] [PubMed] [Google Scholar]
- 9.Gailus-Durner, V., C. Chintamaneni, R. Wilson, S. J. Brill, and A. K. Vershon. 1997. Analysis of a meiosis-specific URS1 site: sequence requirements and involvement of replication protein A. Mol. Cell. Biol. 17:3536-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasser, S. M., and M. M. Cockell. 2001. The molecular biology of the SIR proteins. Gene 279:1-16. [DOI] [PubMed] [Google Scholar]
- 11.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 12.Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 13.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 14.Klar, A. J., S. N. Kakar, J. M. Ivy, J. B. Hicks, G. P. Livi, and L. M. Miglio. 1985. SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics 111:745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurenson, P., and J. Rine. 1991. SUM1-1: a suppressor of silencing defects in Saccharomyces cerevisiae. Genetics 129:685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon, and E. Winter. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moazed, D. 2001. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13:232-238. [DOI] [PubMed] [Google Scholar]
- 18.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rusche, L. N., and J. Rine. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 22.Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies, A. D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245-256. [DOI] [PubMed] [Google Scholar]
- 23.Sutton, A., R. C. Heller, J. Landry, J. S. Choy, A. Sirko, and R. Sternglanz. 2001. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 21:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 25.Wu, J., K. van Riper, N. Kacherovsky, M. Vogelauer, E. T. Young, M. Grunstein, E. Di Mauro, M. Caserta, and D. Robyr. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. EMBO J. 21:1101-1111.11867538 [Google Scholar]
- 26.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]